A Review of Artificial Intelligence-Based Down Syndrome Detection Techniques
Abstract
1. Introduction
2. Review Methodology
2.1. Research Questions
2.2. Search Strategy
2.3. Inclusion and Exclusion Criteria
2.4. Screening and Selection Process
2.5. Data Extraction and Critical Appraisal
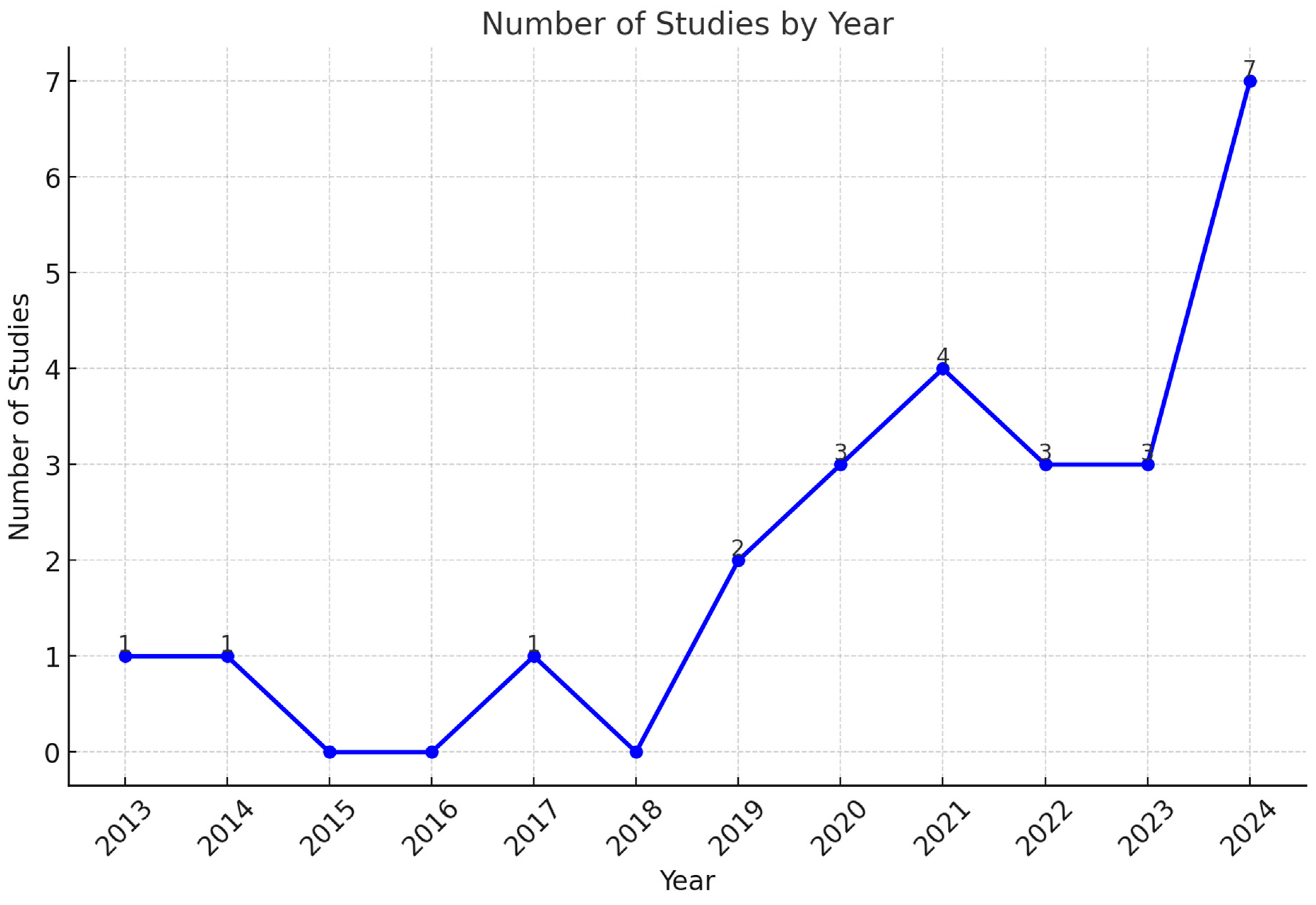
3. Results
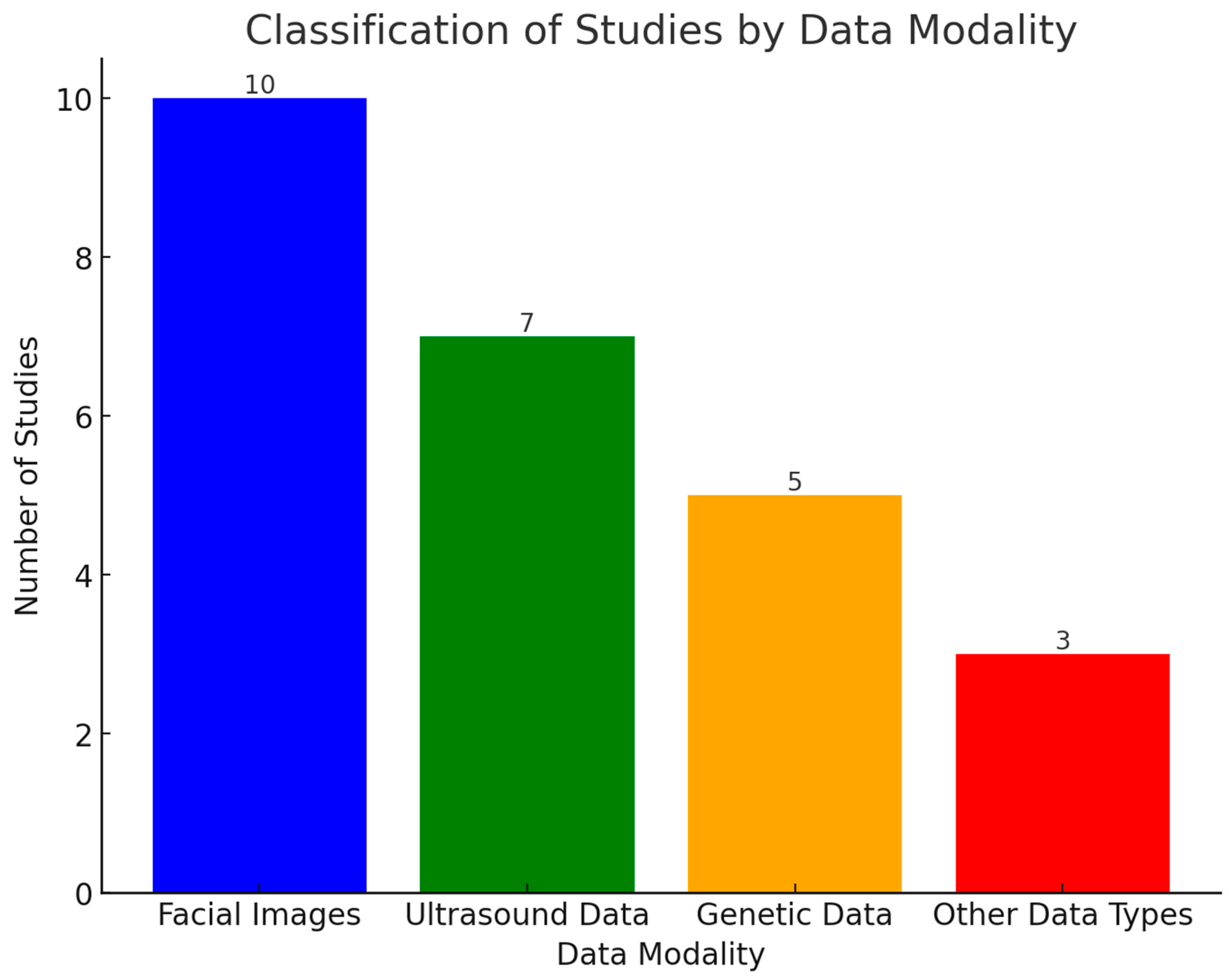
3.1. Data and Technologies with Their Performance
3.1.1. Facial Images
3.1.2. Ultrasound Scans
3.1.3. Genetics Data
3.1.4. Other Data Types
3.1.5. Classified Findings of Reviewed Studies
3.1.6. Challenges Faced by AI-Powered DS Diagnostic Models
3.2. Strategies for Reducing Biases and Improving Generalizability
4. Discussions
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Antonarakis, S.E.; Skotko, B.G.; Rafii, M.S.; Strydom, A.; Pape, S.E.; Bianchi, D.W.; Sherman, S.L.; Reeves, R.H. Down syndrome. Nat. Rev. Dis. Primers 2020, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Bull, M.J. Down syndrome. N. Engl. J. Med. 2020, 382, 2344–2352. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, J.A.; Amon, A.; Abbeduto, L.; Agiovlasitis, S.; Alsaied, T.; Anderson, H.A.; Bain, L.J.; Baumer, N.; Bhattacharyya, A.; Bogunovic, D.; et al. Opportunities, barriers, and recommendations in Down syndrome research. Transl. Sci. Rare Dis. 2021, 5, 99–129. [Google Scholar] [CrossRef] [PubMed]
- Bull, M.J.; Trotter, T.; Santoro, S.L.; Christensen, C.; Grout, R.W.; Council on Genetics. Health supervision for children and adolescents with Down syndrome. Pediatrics 2022, 149, e2022057010. [Google Scholar] [CrossRef]
- Startin, C.M.; D’Souza, H.; Ball, G.; Hamburg, S.; Hithersay, R.; Hughes, K.M.; Massand, E.; Karmiloff-Smith, A.; Thomas, M.S.; Strydom, A. Health comorbidities and cognitive abilities across the lifespan in Down syndrome. J. Neurodev. Disord. 2020, 12, 4. [Google Scholar] [CrossRef]
- Thurman, A.J.; del Hoyo Soriano, L. Down syndrome. In Handbook of Pragmatic Language Disorders: Complex and Underserved Populations; Springer: Cham, Switzerland, 2021; pp. 99–128. [Google Scholar]
- Morris-Rosendahl, D.J.; Crocq, M.A. Neurodevelopmental disorders—The history and future of a diagnostic concept. Dialogues Clin. Neurosci. 2020, 22, 65–72. [Google Scholar] [CrossRef]
- Ashton, N.J.; Janelidze, S.; Al Khleifat, A.; Leuzy, A.; van der Ende, E.L.; Karikari, T.K.; Benedet, A.L.; Pascoal, T.A.; Lleó, A.; Parnetti, L.; et al. A multicentre validation study of the diagnostic value of plasma neurofilament light. Nat. Commun. 2021, 12, 3400. [Google Scholar] [CrossRef]
- Jin, B.; Cruz, L.; Gonçalves, N. Deep facial diagnosis: Deep transfer learning from face recognition to facial diagnosis. IEEE Access 2020, 8, 123649–123661. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Sebire, E.; Rodrigo, C.H.; Bhattacharya, S.; Black, M.; Wood, R.; Vieira, R. The implementation and impact of non-invasive prenatal testing (NIPT) for Down’s syndrome into antenatal screening programmes: A systematic review and meta-analysis. PLoS ONE 2024, 19, e0298643. [Google Scholar] [CrossRef]
- Zerres, K.; Rudnik-Schöneborn, S.; Holzgreve, W. Do non-invasive prenatal tests promote discrimination against people with Down syndrome? What should be done? J. Perinat. Med. 2021, 49, 965–971. [Google Scholar] [CrossRef] [PubMed]
- Marchionni, E. Prenatal Exome Sequencing in Central Nervous System Anomalies: Diagnostic Yield and Challenges. Ph.D. Thesis, Università degli Studi di Roma “La Sapienza”, Rome, Italy, 2023. [Google Scholar]
- Corsten-Janssen, N.; Bouman, K.; Diphoorn, J.C.; Scheper, A.J.; Kinds, R.; El Mecky, J.; Breet, H.; Verheij, J.B.; Suijkerbuijk, R.; Duin, L.K.; et al. A prospective study on rapid exome sequencing as a diagnostic test for multiple congenital anomalies on fetal ultrasound. Prenat. Diagn. 2020, 40, 1300–1309. [Google Scholar] [CrossRef] [PubMed]
- Deden, C.; Neveling, K.; Zafeiropopoulou, D.; Gilissen, C.; Pfundt, R.; Rinne, T.; de Leeuw, N.; Faas, B.; Gardeitchik, T.; Sallevelt, S.C.; et al. Rapid whole exome sequencing in pregnancies to identify the underlying genetic cause in fetuses with congenital anomalies detected by ultrasound imaging. Prenat. Diagn. 2020, 40, 972–983. [Google Scholar] [CrossRef] [PubMed]
- Leghari, I.M.; Ali, S.A. Artificial Intelligence Techniques to improve cognitive traits of Down Syndrome Individuals: An Analysis. Int. J. Softw. Sci. Comput. Intell. 2023, 15, 1–11. [Google Scholar] [CrossRef]
- Kaur, K.; Singh, C.; Kumar, Y. Diagnosis and detection of congenital diseases in new-borns or fetuses using artificial intelligence techniques: A systematic review. Arch. Comput. Methods Eng. 2023, 30, 3031–3058. [Google Scholar] [CrossRef]
- Taidi, L.; El Khamlichi, S. A comprehensive review of artificial intelligence techniques for timely and accurate prediction of Down syndrome. In Agile Security in the Digital Era; CRC Press: Boca Raton, FL, USA, 2024; pp. 179–194. [Google Scholar]
- Anitha Elavarasi, S.; Jayanthi, J. Role of Machine Learning and Deep Learning in Assisting the Special Children’s Learning Process. J. Algebr. Stat. 2022, 13, 2327–2334. [Google Scholar]
- Shukla, P.; Gupta, T.; Saini, A.; Singh, P.; Balasubramanian, R. A deep learning frame-work for recognizing developmental disorders. In Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 705–714. [Google Scholar]
- Agbolade, O.; Nazri, A.; Yaakob, R.; Ghani, A.A.; Cheah, Y.K. Down syndrome face recognition: A review. Symmetry 2020, 12, 1182. [Google Scholar] [CrossRef]
- de Barros, F.R.D.; da Silva, C.N.F.; de Castro Michelassi, G.; Brentani, H.; Nunes, F.L.; Machado-Lima, A. Computer aided diagnosis of neurodevelopmental disorders and genetic syndromes based on facial images—A systematic literature review. Heliyon 2023, 9, e20517. [Google Scholar] [CrossRef]
- Koul, A.; Ahmad, F.; Bhat, A.; Aein, Q.; Ahmad, A.; Reshi, A.; Kaul, R. Unraveling Down Syndrome: From Genetic Anomaly to Artificial Intelligence-Enhanced Diagnosis. Biomedicines 2023, 11, 3284. [Google Scholar] [CrossRef]
- Vidhyasagar, B.; Madamanchi, S.C.; Kalimuthu, S. Early Detection of Down Syndrome Through Ultrasound Imaging Using Deep Learning Strategies—A Review. In Proceedings of the 2024 Second International Conference on Emerging Trends in Information Technology and Engineering (ICETITE), Vellore, India, 22–23 February 2024; IEEE: Piscataway, NJ, USA, 2024; pp. 1–6. [Google Scholar]
- Shafi, S.; Reshi, A.; Wani, M.A.; Ahmad, F.; Shafi, S.; Bashir, S. Artificial Intelligence Driven Bibliometric Insights: Pioneering Down Syndrome Research. Biomedicines 2024, 12, 34. [Google Scholar] [CrossRef]
- Zhao, Q.; Rosenbaum, K.; Okada, K.; Zand, D.J.; Sze, R.; Summar, M.; Linguraru, M.G. Automated down syndrome detection using facial photographs. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 3670–3673. [Google Scholar]
- Zhao, Q.; Werghi, N.; Okada, K.; Rosenbaum, K.; Summar, M.; Linguraru, M.G. Ensemble learning for the detection of facial dysmorphology. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; IEEE: Piscataway, NJ, USA, 2014; pp. 754–757. [Google Scholar]
- Mittal, A.; Gaur, H.; Mishra, M. Detection of down syndrome using deep facial recognition. In Proceedings of the 3rd International Conference on Computer Vision and Image Processing: CVIP 2018, Jabalpur, India, 29 September–1 October 2018; Springer: Singapore, 2020; Volume 1, pp. 119–130. [Google Scholar]
- Pooch, E.H.P.; Alva, T.A.P.; Becker, C.D.L. A Computational Tool for Automated Detection of Genetic Syndrome Using Facial Images. In Intelligent Systems, Proceedings of the 9th Brazilian Conference, BRACIS 2020, Rio Grande, Brazil, 20–23 October 2020, Proceedings, Part I 9; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 361–370. [Google Scholar]
- Qin, B.; Liang, L.; Wu, J.; Quan, Q.; Wang, Z.; Li, D. Automatic identification of down syndrome using facial images with deep convolutional neural network. Diagnostics 2020, 10, 487. [Google Scholar] [CrossRef] [PubMed]
- Geremek, M.; Szklanny, K. Deep learning-based analysis of face images as a screening tool for genetic syndromes. Sensors 2021, 21, 6595. [Google Scholar] [CrossRef]
- Porras, A.R.; Rosenbaum, K.; Tor-Diez, C.; Summar, M.; Linguraru, M.G. Development and evaluation of a machine learning-based point-of-care screening tool for genetic syndromes in children: A multinational retrospective study. Lancet Digit. Health 2021, 3, e635–e643. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liang, B.; Zhao, L.; Chen, Y.; Fu, W.; Yu, P.; Chen, H.; Wang, H.; Xie, G.; Wu, T.; et al. Multiple genetic syndromes recognition based on a deep learning framework and cross-loss training. IEEE Access 2022, 10, 117084–117092. [Google Scholar] [CrossRef]
- Islam, T.U.; Shaikh, T.A. A lightweight attention-based deep learning facial recognition system for multiple genetic syndromes. Int. J. Data Sci. Anal. 2024, 29, 1–19. [Google Scholar] [CrossRef]
- Raza, A.; Munir, K.; Almutairi, M.S.; Sehar, R. Novel transfer learning based deep features for diagnosis of down syndrome in children using facial images. IEEE Access 2024, 12, 16386–16396. [Google Scholar] [CrossRef]
- Yekdast, R. An intelligent method for down syndrome detection in fetuses using ultrasound images and deep learning neural networks. Comput. Res. Prog. Appl. Sci. Eng. 2019, 5, 92–97. [Google Scholar]
- Zhang, H.; Jiang, Y.; Dai, S.; Li, L.; Hu, X.; Liu, R. Application of Intelligent Algorithms in Down Syndrome Screening During Second Trimester Pregnancy. World J. Clin. Cases 2021, 9, 4573–4584. [Google Scholar] [CrossRef]
- Thomas, M.C.; Arjunan, S.P. Deep learning measurement model to segment the nuchal translucency region for the early identification of down syndrome. Meas. Sci. Rev. 2022, 22, 187–192. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, D.; Sun, Y.; Hu, C.; Sun, C.; Wu, Q.; Tian, J. Development and validation of a deep learning model to screen for trisomy 21 during the first trimester from nuchal ultrasonographic images. JAMA Netw. Open 2022, 5, e2217854. [Google Scholar] [CrossRef]
- Tang, J.; Han, J.; Xue, J.; Zhen, L.; Yang, X.; Pan, M.; Hu, L.; Li, R.; Jiang, Y.; Zhang, Y.; et al. A deep-learning-based method can detect both common and rare genetic disorders in fetal ultrasound. Biomedicines 2023, 11, 1756. [Google Scholar] [CrossRef] [PubMed]
- Mavaluru, D.; Ravula, S.R.; Auguskani, J.P.L.; Dharmarajlu, S.M.; Chellathurai, A.; Ramakrishnan, J.; Mugaiahgari, B.K.M.; Ravishankar, N. Advancing Fetal Ultrasound Diagnostics: Innovative Methodologies for Improved Accuracy in Detecting Down Syndrome. Med. Eng. Phys. 2024, 126, 104132. [Google Scholar] [CrossRef] [PubMed]
- Reshi, A.A.; Shafi, S.; Qayoom, I.; Wani, M.; Parveen, S.; Ahmad, A. Deep Learning-Based Architecture for Down Syndrome Assessment During Early Pregnancy Using Fetal Ultrasound Images. Int. J. Exp. Res. Rev. 2024, 38, 182–193. [Google Scholar] [CrossRef]
- Feng, B.; Samuels, D.C.; Hoskins, W.; Guo, Y.; Zhang, Y.; Tang, J.; Meng, Z. Down syndrome prediction/screening model based on deep learning and illumina genotyping array. In Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas, MO, USA, 13–16 November 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 347–352. [Google Scholar]
- He, F.; Lin, B.; Mou, K.; Jin, L.; Liu, J. A machine learning model for the prediction of down syndrome in second trimester antenatal screening. Clin. Chim. Acta 2021, 521, 206–211. [Google Scholar] [CrossRef]
- Goh, C.J.; Kwon, H.J.; Kim, Y.; Jung, S.; Park, J.; Lee, I.K.; Park, B.R.; Kim, M.J.; Kim, M.J.; Lee, M.S. Improving CNV Detection Performance in Microarray Data Using a Machine Learning-Based Approach. Diagnostics 2023, 14, 84. [Google Scholar] [CrossRef]
- Baldo, F.; Piovesan, A.; Rakvin, M.; Ramacieri, G.; Locatelli, C.; Lanfranchi, S.; Onnivello, S.; Pulina, F.; Caracausi, M.; Antonaros, F.; et al. Machine learning based analysis for intellectual disability in Down syndrome. Heliyon 2023, 9, e19444. [Google Scholar] [CrossRef]
- Do, H.D.; Allison, J.J.; Nguyen, H.L.; Phung, H.N.; Tran, C.D.; Le, G.M.; Nguyen, T.T. Applying machine learning in screening for Down Syndrome in both trimesters for diverse healthcare scenarios. Heliyon 2024, 10, e34476. [Google Scholar] [CrossRef]
- Li, L.; Liu, W.; Zhang, H.; Jiang, Y.; Hu, X.; Liu, R. Down syndrome prediction using a cascaded machine learning framework designed for imbalanced and feature-correlated data. IEEE Access 2019, 7, 97582–97593. [Google Scholar] [CrossRef]
- Mal’e, J.; Fortea, J.; Padilla, N. Towards the Discovery of Down Syndrome Brain Biomarkers Using Generative Models. Biomedicines 2024, 12, 452. [Google Scholar]
- Galindo-Lopez, C.R.; Beltran, J.; Perez, C.B.; Caro, K.; Macias, A.; Castro, L.A. Deep Learning-Based Action Classification for Parents and Children with Down Syndrome in Educational Settings. Int. J. Hum. Comput. Interact. 2024, 24, 1–15. [Google Scholar] [CrossRef]
- Khalil, M.; Naeem, A.; Naqvi, R.A.; Zahra, K.; Muqarib, S.A.; Lee, S.W. Deep learning-based classification of abrasion and ischemic diabetic foot sores using camera-captured images. Mathematics 2023, 11, 3793. [Google Scholar] [CrossRef]
- Abidin, Z.U.; Naqvi, R.A.; Haider, A.; Kim, H.S.; Jeong, D.; Lee, S.W. Recent deep learning-based brain tumor segmentation models using multi-modality magnetic resonance imaging: A prospective survey. Front. Bioeng. Biotechnol. 2024, 12, 1392807. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, R.A.; Haider, A.; Kim, H.S.; Jeong, D.; Lee, S.W. Transformative Noise Reduction: Leveraging a Transformer-Based Deep Network for Medical Image Denoising. Mathematics 2024, 12, 2313. [Google Scholar] [CrossRef]
- Iqbal, M.S.; Naqvi, R.A.; Alizadehsani, R.; Hussain, S.; Moqurrab, S.A.; Lee, S.W. An adaptive ensemble deep learning framework for reliable detection of pandemic patients. Comput. Biol. Med. 2024, 168, 107836. [Google Scholar] [CrossRef] [PubMed]
- Uppal, M.; Gupta, D.; Juneja, S.; Gadekallu, T.R.; El Bayoumy, I.; Hussain, J.; Lee, S.W. Enhancing accuracy in brain stroke detection: Multi-layer perceptron with Adadelta, RMSProp and AdaMax optimizers. Front. Bioeng. Biotechnol. 2023, 11, 1257591. [Google Scholar] [CrossRef]
- Abdulrazzaq, M.M.; Ramaha, N.T.; Hameed, A.A.; Salman, M.; Yon, D.K.; Fitriyani, N.L.; Syafrudin, M.; Lee, S.W. Consequential Advancements of Self-Supervised Learning (SSL) in Deep Learning Contexts. Mathematics 2024, 12, 758. [Google Scholar] [CrossRef]
- Harms, J.; Lei, Y.; Wang, T.; Zhang, R.; Zhou, J.; Tang, X.; Curran, W.J.; Liu, T.; Yang, X. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography. Med. Phys. 2019, 46, 3998–4009. [Google Scholar] [CrossRef]
- Qu, Y.; Chen, Y.; Huang, J.; Xie, Y. Enhanced pix2pix dehazing network. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 8160–8168. [Google Scholar]


| Age Group | Phenotype Features | Common Health Comorbidities |
|---|---|---|
| Childhood (0–12 years) | Intellectual disability, delayed motor development, and hypotonia | Congenital heart disease, hearing loss, and gastrointestinal disorders |
| Young adulthood (13–35 years) | Short stature, mild to moderate intellectual impairment, and speech difficulties | Obesity, obstructive sleep apnea, and early-onset Alzheimer’s |
| Late adulthood (36+ years) | Premature aging, cognitive decline, and osteoporosis | High risk of Alzheimer’s disease and increased cardiac issues |
| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Peer-reviewed studies evaluating AI-based DS diagnostic approaches | Studies focused solely on non-AI diagnostic approaches |
| Studies encompass medical imaging and genetic data analysis using ML tools and techniques | Articles lacking quantitative metrics |
| Research addressing AI implementation into clinical workflows | Non-peer-reviewed studies |
| Studies presenting quantitative outcomes | Non-English studies and research with incomplete data |
| Article published in English |
| Authors | Dataset | Method | Performance | Strengths | Limitations |
|---|---|---|---|---|---|
| Zhao et al. (2013) [26] | 100 frontal facial images; ethnicity: not specified; and location: not specified | SVM, Linear SVM, K-NN, and RF | Accuracy = 94.6%, sensitivity = 95.5%, and specificity = 92.5% | Non-invasive methodology and innovative feature extraction | Limited dataset, non-standardized conditions, and limited validation |
| Zhao et al. (2014) [27] | 130 frontal facial images; ethnicity: Caucasian, African, and Asian; and location: National Medical Center, Abu Dhabi, United Arab Emirates | SVM, Linear SVM, RF, and linear discriminant analysis | Accuracy = 96.7%, sensitivity = 93.3%, specificity = 92.8%, and area under the receiver operating characteristic curve (AUROC) = 0.996 | Comprehensive feature set, diverse dataset, and validation across classifiers | Computational complexities, limited focus on real-world testing, and image variability |
| Mittal et al. (2020) [28] | 1706 images; ethnicity: multinational; and location: not specified | AlexNet with SVM | Accuracy = 98.5%, sensitivity = 96.2%, and specificity= 95.7% | Advanced feature extraction, scenario analysis, and focus on regional features | Gender-based variance, dependency on pre-trained models, and limited dataset |
| Pooch et al. (2020) [29] | 170 images; ethnicity: Brazilian; and location: Rio Grande, Brazil | Inception-ResNet-V2 and Linear SVM | Accuracy = 94.0%, sensitivity = 95.0%, and specificity = 92.0% | Scalability potential and framework comparisons | Lack of a public dataset, non-standardized image acquisition, and relatively small dataset |
| Qin et al. (2020) [30] | 405 images; ethnicity: Chinese; and location: China | Deep CNNs, SVM, and K-NN | Accuracy = 95.87%, sensitivity = 93.18, and specificity = 97.40% | Advanced deep CNN, effective use of transfer learning, and robust preprocessing | Dataset bias, limited clinical validation, and lack of model’s interpretability |
| Geremek and Szklanny (2021) [31] | 2101 images; ethnicity: European; and location: Poland | Arcface, Deepface, FaceNet, other pre-trained CNNs, and SVM | Accuracy = 96%, sensitivity = 94.9%, and specificity = 92.1% | Generative capability and compatibility with mobile platforms | The limited scope of geometric analysis and demographic bias |
| Porras et al. (2021) [32] | 1400 images; ethnicity: multinational; and location: not specified | Three neural networks models for DS risk identification | Accuracy = 88%, sensitivity = 90%, and specificity = 86% | Robust model, supports generalizability, and use of three distinct models | Limited validation and dependence on phenotypic features |
| Wang et al. (2022) [33] | 490,000 images of 10,000 different individuals; ethnicity: Chinese; and location: China | ResNet 64 with squeeze-and-excitation block | Accuracy = 93.5%, sensitivity = 87.0%, and specificity = 83.2% | Cross-loss training and robust preprocessing approach | High resource requirements and demographic biases |
| Islam and Shaikh (2024) [34] | 408 images; ethnicity: multinational; and location: multiple locations | CNNs with an improved attention mechanism | Accuracy = 95.4%, sensitivity = 88.1%, and specificity = 86.2% | Integrated attention mechanism and robust data preprocessing | Limited generalizability and high computational demands |
| Raza et al. (2024) [35] | 3009 images; ethnicity: Middle Eastern; and location: Saudi Arabia | VGG16 and light gradient boosting machine | Accuracy = 99.0%, sensitivity = 92.3%, and specificity = 91.5% | Highly effective ensemble approach and effective feature extraction | Dependence on pre-trained model may reduce the model’s performance |
| Authors | Dataset | Method | Performance | Strengths | Limitations |
|---|---|---|---|---|---|
| Yekdast (2019) [36] | 300 images; mother’s age range: 28–35 years; ethnicity: Iranian; and location: Iran | CNN, PSO, and multi-layer perceptron-based classification | Accuracy = 99.38%, sensitivity = 92.1%, and specificity = 94.3% | PSO-based hyperparameter tuning and adaptation to diverse medical imaging data | Single dataset source and requirement of substantial computational resources |
| Zhang et al. (2021) [37] | 4953 images; mother’s age range: 19–35 years; ethnicity: Chinese; and location: Jilin University, China | SVM and AdaBoost classification | Accuracy = 100%, sensitivity = 91.5%, and specificity = 94.3% | Integration of CART and AdaBoost and exploration of T21 risk cutoff thresholds | Reliance on serum markers and large computational resources requirements |
| Thomas and Arjunan (2022) [38] | 100 images; mother’s age range: 19–31 years; ethnicity: Indian; and location: India | VGG-16 and AlexNet-based feature classification | Accuracy = 91.7%, sensitivity = 85.7%, and specificity = 100% | Speckle noise reduction approach and fine-tuned AlexNet-based feature classification | Small dataset and requirement of extensive preprocessing |
| Zhang et al. (2022) [39] | 822 images; mother’s age range: 19–35 years; ethnicity: Chinese; and location: China | A shallow CNN with dropout layers | Accuracy = 90.5, sensitivity = 92.5%, and specificity = 90.6% | Robust data augmentation and visualization techniques | Reliance of image quality and limited dataset size |
| Tang et al. (2023) [40] | 1120 images; mother’s age range: 19–35 years; ethnicity: multinational; and location: multiple locations | ResNet-18 with Grad-CAM heatmap technology | Accuracy = 93.0%, sensitivity = 89.0%, and specificity = 96.0% | Identification of key biomarkers using Grad-CAM heatmap visualization, a technique in TorchCAM 0.3. | Lack of external validation and quality of images may influence the model’s performance |
| Mavaluru et al. (2024) [41] | 1075 images; mother’s age range: 28–35 years; ethnicity: Indian; and location: India | Feature extraction and segmentation approaches with SVM | Accuracy = 98.2%, sensitivity = 90.7%, and specificity = 92.8% | Integration of fuzzy gradient vector flow and wavelet-based segmentation | Limited generalization and segmentation challenges |
| Reshi et al. (2024) [42] | 1120 images; mother’s age range: 28–35 years; ethnicity: Middle Eastern; and location: Saudi Arabia | CNN with dropout layers | Accuracy = 94.5%, sensitivity = 91.8%, and specificity = 94.2% | Focus on early pregnancy and comprehensive framework | High computational requirements |
| Authors | Dataset | Method | Performance | Strengths | Limitations |
|---|---|---|---|---|---|
| Feng et al. (2017) [43] | 378 samples (only 63 DS samples); ethnicity: Chinese; and location: China | Customized CNN model based on genotyping array | Accuracy = 99.3%, sensitivity = 91.5%, and specificity = 88.6% | Dual-branch CNN design for visualizing feature maps | Relatively small sample size, limiting the model’s generalizability |
| He et al. (2021) [44] | 86,142 individuals’ data; ethnicity: Chinese; and location: China | RF and SVM model | Internal validation: accuracy = 66.7%, sensitivity = 59.8%, and specificity = 61.3%; external validation: accuracy = 85.2%, sensitivity = 72.6%, and specificity = 73.8% | High predictive performance and robust generalization | Population specificity and data imbalance |
| Goh et al. (2023) [45] | 742,750 SNP markers and 138 copy number variation (CNV)-related chromosomal disorders; ethnicity: Korean; and location: Seoul, South Korea | K-means clustering and K-neural networks | Accuracy = 100%, sensitivity = 95.8%, and specificity = 97.6% | Robust validation and enhanced CNN detection | Limited detection scope and dependence on genomic wave mitigation |
| Baldo et al. (2023) [46] | 106 individuals; ethnicity: Italian; and location: Italy | RF and gradient boosting model (GBM)-based classification | Accuracy = 67%, sensitivity = 61.4%, and specificity = 60.7% | Innovative preprocessing, feature importance analysis, and robust classification model | Complexity of data inter-relationships and bias in age-related variables |
| Do et al. (2024) [47] | 1035 individuals; ethnicity: Vietnamese; and location: Vietnam | RF and GBM-based classification | Accuracy = 90%, sensitivity = 83.2%, and specificity = 81.5% | Integration of multi-modality data and adaptations to diverse populations | High computational resources and dependence on data quality |
| Authors | Dataset | Method | Performance | Strengths | Limitations |
|---|---|---|---|---|---|
| Li et al. (2019) [48] | 100,252 features; ethnicity: Chinese; and location: China | Isolation forest model for detecting anomalies and refining predictions | Accuracy = 95.1%, specificity = 93.2%, and sensitivity = 95.0% | Efficient data imbalance handling and comprehensive feature selection approaches | Heavily relies on biochemical and ultrasound markers |
| Mal’e et al. (2024) [49] | Brain MRI scans, a total of 2113 images from multiple repositories, were used to train the model; ethnicity: Spanish; and location: Spain | Variational autoencoders and diffusion models were used to determine the subtle variations in MRI scans | Accuracy = 96.1%, specificity = 97.1%, and sensitivity = 93.0% | Biomarker discovery and quantitative volumetric analyses of subcortical structures | Relying on mid-sagittal 2D MRI scans may overlook critical spatial information |
| Galindo-Lopez et al. (2024) [50] | 12 parent–child interaction video recordings; ethnicity: Mexican; and location: Mexico | CNN–LSTM model | Physical approach: accuracy = 92.1%, specificity = 90.1%, and sensitivity = 91.1%; verbal expression: accuracy = 90.96%, specificity = 88.1%, and sensitivity = 86.2% | Innovative hybrid architecture and high accuracy in capturing physical activities | Imbalanced datasets, camera perspective dependence, and verbal expression challenges |
| Data Type | Sensitivity | Specificity | Accuracy | Risk Level |
|---|---|---|---|---|
| First-trimester ultrasound images | 85–92% | 94–100% | 90–100% | Low |
| Second-trimester ultrasound images | Low | |||
| Genetics data | 61–96% | 60–97% | 66–99% | Moderate |
| Facial images | 87–96% | 83–97% | 88–99% | Low |
| Other | 86–95% | 86–97% | 90–96% | Moderate |
| Research Gaps | Future Research Directions |
|---|---|
| Limited demographic diversity, leading to biases in model predictions | The multinational dataset contains increased dataset representation across different ethnicities |
| Reduced model’s sensitivity and specificity in real-world clinical applications | Developing domain adaptation techniques and conducting multi-center external validations |
| Focused on single modality [facial images, ultrasound scans, or genetics data] | Building multimodality-based AI models, improving diagnostic accuracy and robustness |
| Requirement of significant computational resources, limiting deployment in resource-constrained healthcare settings | Exploring lightweight architectures for implementing cloud-based AI solutions |
| Lack of decision-making explanations | Integration of explainable AI techniques to enhance transparency |
| Different studies used varied evaluation metrics | Establishing standardized benchmarks and reporting guidelines |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaikh, M.A.; Al-Rawashdeh, H.S.; Sait, A.R.W. A Review of Artificial Intelligence-Based Down Syndrome Detection Techniques. Life 2025, 15, 390. https://doi.org/10.3390/life15030390
Shaikh MA, Al-Rawashdeh HS, Sait ARW. A Review of Artificial Intelligence-Based Down Syndrome Detection Techniques. Life. 2025; 15(3):390. https://doi.org/10.3390/life15030390
Chicago/Turabian StyleShaikh, Mujeeb Ahmed, Hazim Saleh Al-Rawashdeh, and Abdul Rahaman Wahab Sait. 2025. "A Review of Artificial Intelligence-Based Down Syndrome Detection Techniques" Life 15, no. 3: 390. https://doi.org/10.3390/life15030390
APA StyleShaikh, M. A., Al-Rawashdeh, H. S., & Sait, A. R. W. (2025). A Review of Artificial Intelligence-Based Down Syndrome Detection Techniques. Life, 15(3), 390. https://doi.org/10.3390/life15030390






