Abstract
Background: Cervical cancer (CC) is the fourth most common cancer diagnosis in women worldwide. Infection with high-risk human papillomavirus (HPV) is a critical but not determinative condition for CC development, as several co-factors modulate the progression of HPV-associated cervical lesions. Interleukin-8 (IL-8) and Interleukin-16 (IL-16) are chemokine-like interleukins involved in the pathogenesis of various cancers. Singular studies in Asian populations have suggested a potential role of IL-8 rs4073 (−251 A>T) and IL-16 rs1131445 (3′UTR T>C) in cervical carcinogenesis. Methods: A case-control study was conducted in a European cohort of 339 women, including 126 CC patients and 213 controls. Four common IL-8 SNPs, rs4073 (−251 A>T), rs2227306 (+781 C>T), rs1126647 (+2767 A>T), and rs2227543 (+1633 C>T), and four IL-16 polymorphism, rs4778889 (−295 T>C), rs11556218 (3441 T>G), rs4072111 (1300 C>T), and rs1131445 (3′UTR T>C), were assessed using RFLP-PCR and analyzed under seven inheritance models. Subgroup analyses were stratified by menopausal status (age threshold 51 years), disease stage, and histological subtype. Results: IL-16 rs4072111 was significantly associated with an increased CC risk in premenopausal women in the co-dominant (p = 0.038), dominant (p = 0.022), and heterozygote (p = 0.045) models, identifying the T allele as the risk allele (OR 2.31, CI95% 1.17–4.56; p = 0.017). In women aged over 51, IL-16 rs4778889 was associated with CC in the heterozygote (p = 0.048) and overdominant (p = 0.042) models but not in the co-dominant model (p = 0.092). None of the analyzed SNPs significantly increased CC risk in the entire cohort. Specifically, neither IL-16 rs1131445 nor IL-8 rs4073, previously reported as risk factors in Asian populations, were associated with CC risk in this European cohort. Conclusions: These findings highlight the role of age stage in immunity and cancer susceptibility, suggest that IL-8 and IL-16 SNPs may function differently in cervical carcinogenesis compared with other cancers, and emphasize the importance of ethnic background in cancer risk, warranting further research.
1. Introduction
Cervical cancer (CC) is the fourth most diagnosed cancer and the fourth leading cause of cancer-related death among women worldwide, following breast, colorectal, and lung cancers [1]. The lifetime risk of developing CC is approximately 1.5%, with an estimated 660,000 new cases and 350,000 deaths estimated globally in 2022. CC accounts for 6.8% of all female cancer cases and 8.1% of female cancer deaths [1]. Despite advances in early detection and prevention, the mortality-to-incidence ratio remains high at around 53% [1]. Histologically, 75–85% of CC cases are squamous carcinomas, 12–13% are adenocarcinomas, and 3–5% are adenosquamous carcinomas [2,3]. The remaining 5% include rare subtypes, such as neuroendocrine, clear cell, serous carcinomas, lymphomas, and sarcomas [3,4]. Surgery with stage-adapted radicality is the first-line option for early-stage CC, while systemic treatments (radiochemotherapy, chemotherapy) are the first-line options for advanced stages [5,6]. The overall 5-year survival rate decreases significantly with advanced FIGO stages: 88% for stage I, 82% for stage II, 74% for stage III, and 12% for stage IV. Survival also declines with patient age: 84% for those under 40, 80% for ages 40–65, and 37% for those over 65 [7].
CC is an infection-related cancer, as the human papillomavirus (HPV) infection is a necessary, but not sufficient, cause of the disease [1,5,8]. HPV is detected in 96% of CC cases, yet most HPV infections resolve spontaneously within 8–16 months [8,9,10]. During the ages of greatest sexual activity, the prevalence of subclinical HPV infections in women can reach 40%, with an annual infection rate of 10–15%. In women over 30, the prevalence drops to 5–10% [8]. While the incidence of new infections decreases with age, the persistence of existing infections increases [8]. Among 448 known HPV types, 25 are classified as high-risk HPV (hrHPV), including HPV16, HPV18, HPV31, HPV33, and HPV45 [9]. HPV16 and HPV18 alone are responsible for approximately 70% of cervical cancers [9,10]. The prevalence of hrHPV infection peaks twice: first between the ages of 15 and 26 and again around 45 years [10]. These patterns correspond to two peaks in CC incidence: the first between ages 30 and 45 and the second between 60 and 80 years [11,12].
Although most hrHPV infections are transient, around 10% persist and can progress to low-grade squamous intraepithelial lesions (LSILs), high-grade squamous intraepithelial lesions (HSILs), and eventually invasive CC. Infections with HPV16 and HPV33 carry a markedly increased risk (~25-fold) of progressing to preinvasive or invasive cancer, whereas HPV16 and HPV31 have the lowest likelihood of spontaneous clearance [13]. LSILs regress spontaneously in about 60% of cases, while HSILs have a lower regression rate of around 25%; up to 18% of HSILs progress to invasive CC if untreated [14].
The variability in HPV infection outcomes and CC progression arises from a complex interplay of viral and host factors. On the viral side, HPV genotype, viral genetic and epigenetic modifications, and viral load are critical determinants of disease progression [15]. The host immune system, both innate and adaptive, is crucial in determining whether HPV infections are cleared, persist, or progress to cancer [16]. Host-related co-factors primarily are related to sexual and reproductive behavior, e.g., multiple sexual partners, early sexual debut, long-term use of oral contraceptives, high parity, cervicovaginal microbiome alterations, sexually transmitted co-infections (e.g., HIV and Chlamydia trachomatis), and further including lifestyle factors such as smoking [9,10,15]. Immunological changes have been documented in premalignant and malignant cervical lesions, including alterations in cytokine expression by cervical epithelial cells and by infiltrating leukocytes. These immune factors are detectable in biopsies from patients with HPV infections and different stages of CIN or CC. Additionally, leukocytes in the tumor microenvironment release signaling molecules that modulate processes like angiogenesis, chemotaxis, and apoptosis [16,17].
Family aggregation studies and heritability estimates indicate a substantial genetic component to CC susceptibility [15]. A genome-wide association study (GWAS) estimated that 24% of the variation in CC risk is due to common autosomal single nucleotide polymorphisms (SNPs), slightly lower than the 27% heritability estimate from family studies. Genetic variants associated with CC development identified in GWAS include lymphotoxin alpha (LTA), tumor necrosis factor (TNF), Paired Box 8 (PAX8), Cleft Lip and Palate Transmembrane Protein 1-like (CLPTM1L), and Human Leukocyte Antigen (HLA) genes, indicating a potential disruption in apoptotic and immune function pathways [15,18]. However, variants identified by GWAS explained only 2.1% of phenotypic variance, implying that a significant proportion of heritability is tagged by common SNPs with small individual effects [18].
Interleukins, a class of cytokines originally named for their role in leukocyte communication, are now recognized to be produced by a wide range of cells and play various roles in carcinogenesis. Numerous candidate gene studies have shown that SNPs in interleukin genes, such as IL-1, IL-6, IL-10, and IL-12, are associated with altered risks of HSIL and CC [19,20,21,22]. Among 41 known interleukins, IL-8 and IL-16 are often classified together as “chemokine-like interleukins” for their role in attracting immune cells to inflammation sites [23]. IL-8, also known as CXCL8 (C-X-C motif chemokine ligand 8), is a pro-inflammatory and pro-angiogenic chemokine within the CXC chemokine superfamily [24]. It is produced by neutrophils, monocytes, fibroblasts, endothelial cells, smooth muscle cells, and epithelial cells [24,25]. High IL-8 expression and its receptor activation are observed in multiple tumor microenvironment components, including cancer cells, endothelial cells, and tumor-associated macrophages, promoting angiogenesis, cell proliferation, survival, and migration [24].
The IL8 gene, located on chromosome 4q12-q135, comprises four exons and three introns, encoding a 99-amino acid precursor protein processed into active isoforms. Its transcription is primarily regulated by NF-κB through TNF and TRAF6 pathways [24,26]. Polymorphisms in IL8 can affect gene expression or protein structure, altering binding affinities and downstream signaling [26]. The most studied IL-8 SNPs are rs4073 (−251 A>T) in the promoter, rs2227306 (+781 C>T) in intron 1, rs2227543 (+1633 C>T) in intron 3, and rs1126647 (+2767 A>T) in the 3′UTR [26,27]. The rs4073 T allele is linked to higher IL-8 expression, with two to five times greater transcriptional activity than the A allele [28]. This SNP correlates with elevated IL-8 plasma levels, showing the highest expression in AA, intermediate in AT, and lowest in TT genotypes [29]. Rs2227306 in intron 1 also affects transcription and regulation [30]. Beyond oncology, IL8 SNPs, especially rs4073, are linked to, e.g., asthma [30], acute coronary syndrome [29], and age-related macular degeneration [31]. In oncology, IL-8 SNPs are associated with glioma [32], osteosarcoma [33], and gastric [28], lung [34], hepatocellular [35], and nasopharyngeal cancers [36]. For the female genital tract, one study linked the TT genotypes of rs2227306 and rs1126647 to increased ovarian cancer (OC) risk [37]. In our previous study, three of the four SNPs (rs4073, rs2227306, and rs2227543) were associated with OC risk in postmenopausal but not premenopausal women. We also found that rs1126647 genotypes containing the T-allele were significantly linked to endometriosis-related OC subtypes, with TT homozygotes more frequent in these subtypes than in other OC subtypes (39% vs. 19%) [38].
IL-16, first identified in 1982 as a “lymphocyte chemoattractant factor” for CD4+ cells, is a multifunctional protein involved in immune regulation, cell migration, and cell cycle control [39,40]. It is produced by T lymphocytes, macrophages, dendritic cells, fibroblasts, mast cells, B cells, and bronchial epithelial cells [40,41,42]. While CD4 is its primary receptor, IL-16 can also bind to CD9 or function via CD4/CD9-independent mechanisms [43,44]. The IL16 gene on chromosome 15q26.3 has seven exons and six introns and encodes two precursor isoforms produced by alternative splicing: a 636-amino acid Pro-IL-16 in immune cells and a 1244-amino acid neuronal variant (nPro-IL-16) [39,40,42]. Caspase-3 cleaves both into mature IL-16, a 121-amino acid protein with chemotactic and growth factor activities [41,42,43]. Both precursor forms migrate to the nucleus, where they act as nuclear transcriptional repressors [41,44,45]. IL-16 is linked to inflammatory and autoimmune diseases, such as asthma, multiple sclerosis, systemic lupus erythematosus (SLE), and rheumatoid arthritis (RA) [42,46,47,48], as well as inflammation-driven conditions like susceptibility to viral infections, depression, and cardiovascular diseases [42,49,50]. In cancer, IL-16 attracts CD4+ cells, showing both tumor-promoting and suppressive effects. Its role varies: in cutaneous T-cell lymphoma (CTCL), a pro-IL-16 mutation reduces p27KIP1, enhancing cell growth; in multiple myeloma (MM), IL-16 overexpression drives plasma cell proliferation; and in breast cancer, it recruits pro-tumor macrophages [43,50].
The most studied IL16 variants are rs4778889, rs11556218, rs4072111, and rs1131445 [50]. The rs11556218 (T>G) missense mutation in exon 6 is linked to higher IL-16 levels in TG/GG genotypes [51,52], with the G allele increasing the risk of lung [53], oral [54], nasopharyngeal [52], gastric [50,55,56], and colorectal cancer [55], as well as osteosarcoma [51], endometriosis [57,58], cardiovascular disease [50], and SLE [59]. Another missense mutation, rs4072111 (C>T) in exon 6, is associated with an increased risk of Parkinson’s [60], Alzheimer’s [61], and SLE [59], though its link to endometriosis [57,58] and gastric cancer [55,62] remains inconsistent. Rs4778889 (T>C), located in the promoter region, reduces promoter activity, with the T allele increasing asthma risk [46], while the C allele (TC/CC genotypes) is linked to a higher risk of renal cell carcinoma [63], gastric cancer [50], nasopharyngeal cancer [52], endometriosis [64,65], and SLE [59]. Rs1131445 (T>C) in the 3′-untranslated region (3′UTR) affects the miR-135b binding site, disrupting miRNA suppression and upregulating IL-16 expression [48,66]. The C allele is associated with an increased risk of colorectal cancer [67], RA [48], and SLE [48]. In gynecology, IL16 genetic variants play a role in ovarian carcinogenesis. Our recent study found a strong association between OC risk and rs11556218 (G vs. T allele: OR 2.76, p < 0.0001) across all age groups, as well as with rs4778889 (C vs. T allele: OR 1.94, p = 0.016) in premenopausal women [68].
For CC, only one IL-8 SNP (rs4073) [69] and one IL-16 SNP (rs1131445) [66] have been studied, each in a single study, both limited to Chinese populations. For IL-8 rs4073, T-allele-containing genotypes (AT and TT) were significantly associated with CC, with TT homozygotes showing an increased risk of lymph node metastasis [69]. In the seminal study by Mi et al., rs1131445 in the miR-135b binding site of IL-16 3′-UTR was found to affect IL-16 protein expression by interfering with miR135b suppressive function and was significantly associated with the risk of CC. Patients carrying the rs1131445 C allele had higher serum IL-16 levels compared with non-carriers [66].
The remaining IL-8 SNPs (rs2227306, rs2227543, rs1126647) and IL-16 SNPs (rs4778889, rs11556218, rs4072111) have not been studied in CC. To fill this gap, we evaluated the association of four common IL-8 and four IL-16 polymorphisms with CC risk in a geographically and ethnically well-defined European cohort, including subgroup analyses by estimated menopausal status, histological subtype, and FIGO stage.
2. Materials and Methods
2.1. Study Design and Participant Characteristics
This case-control study evaluated four IL-8 SNPs, rs4073 (−251 A>T), rs2227306 (+781 C>T), rs1126647 (+2767 A>T), and rs2227543 (+1633 C>T), along with four IL-16 SNPs: rs4778889 (−295 T>C), rs11556218 (3441 T>G), rs4072111 (1300 C>T), and rs1131445 (3′UTR T>C). The analysis included blood samples from 339 Central European women, comprising 126 CC cases and 213 healthy controls. SNP genotyping was performed using the restriction fragment length polymorphism (PCR-RFLP) method.
The blood samples were sourced from the Molecular Oncology Group’s blood bank at the Medical University of Vienna, the coordinating center of a European biobanking project approved by the Ethics Committee of the Medical University of Vienna (EK-366/2003 and EK 1966/2020) and registered on ClinicalTrials.gov (NCT01763125). Blood samples for this study were selected from a collection obtained between 1996 and 2021, involving patients and controls recruited at the Medical University of Vienna and partner institutions across Europe. The samples used in the current analysis originated specifically from Austria, Poland, Germany, and Belgium. Women with metastatic cervical lesions and those with induced menopause (e.g., surgical or pharmacological) were excluded from the study. Menopausal status was estimated using an age cutoff of 51 years, reflecting the median age of menopause in Central Europe [70], and staging was adjusted to the revised 2018 FIGO classification [71]. Ethical compliance with the 1964 Helsinki Declaration and its amendments was ensured. Written informed consent was obtained from all participants, and data were anonymized and processed according to good scientific practice.
2.2. DNA Extraction and Genotyping
Peripheral blood was collected from all participants in EDTA tubes. Genomic DNA was isolated from white blood cells using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany) and then quantified using the QuantiFluor® dsDNA System (Promega, Alcobendas, Madrid, Spain) and the QuBit Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). The single nucleotide variants of IL-8 and IL-16 were determined by analyzing fragment length polymorphisms of the respective PCR products (PCR-RFLP). Details on primers, annealing temperatures, and restriction enzymes are provided in Table 1. The amplicons were generated from 25 ng genomic DNA as a template in a 25 µL reaction mix, containing 5 pmol of the respective forward and reverse primers and MangoMix™ (Bioline, London, UK) providing MangoTaq™ DNA polymerase, MgCl2, and dNTPs. Amplification began with a hot start at 95 °C for 5 min, followed by 45 cycles of 30 s denaturation at 95 °C, 30 s annealing at specific temperatures (Table 1), and 60 s extension at 72 °C. A final extension was performed at 72 °C for 7 min. PCR products were digested with the respective restriction endonuclease (New England Biolabs, Ipswich, MA, USA) under the conditions given in Table 1. The restriction fragments were separated with capillary electrophoresis using the Fragment Analyzer™ Automated CE System (Advanced Analytical, Ankeny, IA, USA) and the DNF-905 dsDNA Kit (Agilent, Santa Clara, CA, USA). The sizes of the fragments were assessed using the software PROSize® 3.0 version 3.0.1.6 (Advanced Analytical Technologies, Orangeburg, NY, USA).

Table 1.
PCR-RFLP of IL-8 and IL-16 SNPs. Primers for amplification, annealing temperature, and restriction enzyme for digestion.
2.3. Statistical Analysis
Genotype and allele frequencies were compared between cases and controls using the Chi-squared test, with Yates correction applied when cell counts were below five. Odds ratios (ORs) with 95% confidence intervals (CIs) and Fisher’s exact test were used to assess the effect of each SNP on CC risk. In the control group, Hardy–Weinberg equilibrium (HWE) was verified for all SNPs using the goodness-of-fit χ2 test. Age differences were analyzed with the Mann–Whitney test, considering a two-sided p-value ≤ 0.05 as statistically significant. Associations between SNPs and CC risk were evaluated using seven inheritance models (Table 2) [72,73]. All statistical calculations were conducted using the JASP statistical software v.19.0.0 for Windows [74] and the VassarStats Website for Statistical Computation [75].

Table 2.
Genetic models studied.
3. Results
Within the cohort of 339 women, 126 were diagnosed with CC, and 213 were healthy controls. The median age of the cases was 45.5 years (range: 25–79), while the median age of the controls was 51 years (range: 18–87). The proportion of premenopausal patients, defined as those younger than 51 years, was higher among cases (61%) than controls (46%; p = 0.006), as shown in Table 3. As shown in Figure A1 (Appendix A), the disease peaked between the ages of 35 and 40, followed by the 40–45 age group. After menopause, most patients were diagnosed between 50 and 55 years, followed by the 55–60 age group.

Table 3.
Study population characteristics.
As presented in Table 3, 50 out of 126 CC cases (40%) were diagnosed at an early FIGO stage (I-IIa), while 73/126 (58%) were diagnosed at advanced stages (IIb-IV). The most common histological type was squamous carcinoma, observed in 80% of patients (101/126), while 18% had non-squamous subtypes (adenocarcinoma or adenosquamous carcinoma). A detailed listing of histological subtypes and grades, as well as stratification by FIGO stage, is provided in Table A1.
The minor allele frequencies (MAFs) in the control group are presented in Table 4. As expected from other sources, the MAFs for IL-8 were typically high, with 46.7% for rs4073 (A allele), 43.7% for rs2227306 (T allele), 42.7% for rs2227543 (T allele), and 41.1% for rs1126647 (T allele). For IL-16 SNPs, the MAFs were 9.2% for rs11556218 (G allele), 13.6% for rs4778889 (C allele), 10.3% for rs4072111 (T allele), and 32.4% for rs1131445 (C allele). All MAFs in the control group were representative of the European population, as demonstrated by comparison with those reported in the gnomAD (Genome Aggregation Database) [76] and dbGaP Allele Frequency Aggregator (ALFA) databases [77]. The genotype distribution did not deviate from Hardy–Weinberg equilibrium in any case (see Table 4).

Table 4.
MAF values for the study cohort, p-values for Hardy–Weinberg equilibrium (HWE), and reference MAFs for European populations from gnomAD and/or the dbGaP ALFA project.
As shown in Table 5 and Table 6, in this ethnically homogeneous European cohort, no general association was observed between the IL-8 and IL-16 SNPs and the risk of CC. Specifically, no association was found for the G allele of rs11556218, which has been linked to many oncological and non-oncological conditions, nor for rs1131445 (T>C), the only IL-16 SNP previously investigated in relation to CC [66]. Additionally, in contrast to a solitary prior study investigating the IL-8 SNP rs4073 in CC [69], neither the genotype nor the allelic frequencies differed significantly between the CC group and the healthy controls.

Table 5.
Genotype and allele frequencies of IL-8 SNPs among CC cases and healthy controls.

Table 6.
Genotype and allele distribution of IL-16 SNPs rs11556218, rs4778889, rs4072111, and rs1131445.
A significant association, however, was observed between IL-16 SNP rs4072111 (C>T) and CC risk in premenopausal women. The association was especially strong (χ2 = 6.1) and significant (p = 0.014) in the allelic comparison, revealing the common C allele as protective and the minor T allele as a risk allele, with a corresponding OR of 2.31 (95% CI 1.17–4.56). Similarly, in the genotype analysis, being homozygous for the common CC genotype was associated with a significantly reduced CC risk compared with heterozygotes (CT).
Furthermore, regarding menopausal status, a weak association between IL-16 rs4778889 (T>C) and CC risk was noted for postmenopausal women in both the heterozygote (p = 0.048) and overdominant (p = 0.042) models, with the minor C allele being the risk allele. However, this association diminished below the significance level in the co-dominant model (p = 0.091) and allele frequency comparison (p = 0.164). These results are presented in Table 7.

Table 7.
Genotype and allele distribution of IL-16 SNPs rs 4072111 and rs4778889 broken down by menopausal status.
No further associations were found when broken down by the estimated menopausal status for IL-16 rs11556218 (T>G) and IL-16 rs1131445.
Notably, none of the studied SNPs showed any association with histological type (squamous vs. non-squamous) or stage of disease—early vs. advanced—at first diagnosis.
4. Discussion
Persistent hrHPV infections initiate carcinogenesis primarily through the expression of the viral oncogenes E6 and E7, which inactivate the tumor suppressor proteins p53 and retinoblastoma protein (pRB), disrupt cell cycle control and activate telomerase, leading to cellular immortalization. E6 and E7 are implicated in every stage of cervical carcinogenesis and contribute to the evasion of host immune responses, interference with key signaling pathways like MAPK and mTOR, and reprogramming of the host cellular environment [9,78]. The tumor microenvironment is critical in regulating tumor progression, angiogenesis, and metastasis. This environment includes tumor cells, endothelial cells, cancer-associated fibroblasts, and infiltrating inflammatory cells controlling local cytokine networks. Among these, IL-8 (CXCL8) is a chemokine that activates intracellular signaling, mediating pro-tumorigenic effects such as epithelial-mesenchymal transition, survival, proliferation, migration, invasion, angiogenesis, and resistance to apoptosis [79]. IL-8 mRNA and protein are upregulated in CC tissues and cell lines compared with normal cervical tissues and are associated with the proliferation and migration of cervical epithelial cell lines [80,81]. A recent study confirmed that CXCL1, CXCL2, CXCL3, and CXCL8 (IL-8) are regulated by HPV16 and HPV18 E6/E7 and are overexpressed in CC biopsies, with higher expression associated with worse survival [82]. IL-8 levels in liquid-based cervical samples increase with the progression of intraepithelial lesions from low grade to high grade [83]. Similarly, IL-6 and IL-8 levels measured in cervicovaginal washings are higher in patients with invasive CC than in those with cervical intraepithelial neoplasia [84]. In addition, IL-6 and IL-8 show greater expression in the LSIL and HSIL groups compared with normal controls, with melatonin modulating both cytokines’ effect on the progression of neoplastic lesions in HPV infection [85]. In addition to its immune functions, IL-8 is a pro-angiogenic factor that regulates vascular endothelial growth factor (VEGF), matrix metalloproteinases, and other mediators through autocrine and paracrine pathways [86]. IL-8 also enhances the angiogenic capability of CC cells via G protein-coupled lysophosphatidic acid (LPA) receptors 2 and 3, Gi-mediated PI3K-Akt, PKC pathways, and NF-kB activation [87]. In a xenograft model, IL-8 promoted tumor growth and metastasis in vivo, while IL-8 antibody treatment reduced tumor volume, decreased lymph node metastasis, and improved animal survival. Thus, IL-8 blockade shows promise as an alternative approach for CC treatment [88]. Several studies confirmed that IL-8 protein expression correlates with clinical stage, distant metastasis, histological type, and histological grade of CC and that high IL-8 expression is linked to shorter survival in CC patients [79,80,89,90].
Given the diverse role of IL-8 in cervical carcinogenesis and the documented impact of genetic variants in other pro-inflammatory and pro-angiogenic cytokine genes [19,20,21,91], we expected that common SNPs in the IL-8 gene might alter CC risk. In a Chinese study, the T-allele-containing genotypes (AT and TT) of IL-8 rs4073 (−251 A>T) were significantly more frequent in CC patients compared with controls, and the TT genotype was associated with an increased risk of lymph node metastasis (OR = 2.917, p = 0.035) [69]. However, in our European cohort, no association was found between any of the IL-8 SNPs and CC risk. This lack of association appears robust despite the relatively small sample size, given the high MAFs of the IL-8 SNPs (ranging from 41% to 47%). Common variants with high MAFs are less prone to false positives than rare variants, especially in studies with modest sample sizes, as they provide more statistical power for detecting associations [92,93]. Thus, the absence of significant associations suggests that these variants likely do not play a major role in cervical carcinogenesis.
Furthermore, differences in the prevalence and impact of genetic variants with small effects between geographically/ethnically distinct populations, particularly between Asian, African, and European populations, are well documented [94,95] and have also been reported in relation to CC [96]. Variations in allele frequencies can be attributed to population-specific genetic drift, natural selection, and historical demographic events [97], whereas the functional significance of certain immune-related genetic variants may arise from transcriptome and epigenetic alterations influenced regionally by environmental and lifestyle factors, as well as by exposure to pathogens and toxins. These influences are reflected in changes to immune cell function and disease susceptibility [98,99].
The role of IL-16 in relation to CC is less well researched as compared to IL-8. IL-16 SNPs have been implicated in several cancers [50], with the G allele of rs11556218 associated with an increased risk of gastric [50], lung [53], oral [54], and ovarian [68] cancers. However, little is known about the role of IL-16 in cervical HPV infection and CC, particularly regarding the influence of IL-16 genetic variants. A Chinese study demonstrated that rs1131445, located in the miR-135b binding site of the IL-16 3′-UTR, affects IL-16 protein expression by interfering with the suppressive function of miR-135b [66]. MicroRNAs are small (18–30 nucleotides) non-coding RNAs regulating the function of other genes primarily by “inhibiting the production of protein from mRNAs to which the microRNAs can bind by base pairing” [100]. As negative regulators of gene expression, they play a fundamental role in cell development and carcinogenesis (the discovery of micro RNAs has been awarded the 2024 Nobel Prize) [100]. Patients carrying the rs1131445 C allele have higher serum IL-16 levels than non-carriers. This interference is significantly associated with an increased risk of cervical cancer in Asian patients [66]. In contrast, in our European cohort, no association between rs1131445 and CC risk could be observed.
We are the first to report associations between two IL-16 SNPs, rs4072111 (C>T) and rs4778889 (T>C), when stratified by menopausal status. In premenopausal women, the common C allele is imposed as protective and the minor T allele as a risk allele regarding rs4072111 (C>T) and CC. In contrast, in postmenopausal women, we observed an association between IL-16 rs4778889 (T>C) and CC for individuals carrying the minor C allele. However, the latter observation was limited to heterozygote and overdominant models.
This result emphasizes menopause as a critical threshold in local and systemic immunological processes, including those relevant to HPV-associated carcinogenesis. Our previous studies clearly showed the different impacts of IL-8 and IL-16 SNPs depending on menopausal status [38,68]. Epidemiological data demonstrate a bimodal distribution of CC, with peaks occurring between 30–39 years of age and 60–69 years of age [12]. In the present study, the majority of CC cases were diagnosed as premenopausal, but 40% occurred in postmenopausal women. This bimodal distribution may reflect different peaks in HPV acquisition [11], as well as variations in epigenetic and environmental factors influencing CC development. Elderly-onset CC patients exhibit a significantly higher frequency of NOTCH1 and TP53 driver mutations compared with younger patients, along with a notably higher tumor mutational burden [101]. Additionally, patients aged 65 and older with squamous CC show a higher frequency of PIK3CA mutations, which are associated with increased mutation rates in other genes involved in key cancer-associated pathways, such as tyrosine kinase receptors, K-Ras/BRAF/MAPK and the Wnt/β-catenin pathway [102]. However, as the functional consequences of the rs4778889 genetic variants on IL-16 protein expression in CC are not known, and the association in our study was limited to two inheritance models in postmenopausal women, further investigation is warranted.
A stronger and more definitive association was observed, in contrast, in premenopausal women, where the common C allele was imposed as protective and the minor T allele as a risk allele of rs4072111 (C>T) regarding CC risk. This observation is particularly intriguing, as nPro-IL-16 (the larger isoform of the IL-16 precursor) was originally believed to be exclusively expressed in hippocampal and cerebellar neurons [45]. Similar to Pro-IL-16 in immune cells, nPro-IL-16 is cleaved by caspase-3 to release mature IL-16 [45]. In the nervous system, nPro-IL-16 plays a role in upregulating the transcription factor c-fos, promoting neurite outgrowth, and interacting with neurotransmitter receptors and neuronal ion channels [45]. Rs4072111 (C>T) is a missense mutation in exon 6, leading to a proline-to-serine substitution (Pro434Ser) in nPro-IL-16 [57,58]. The T allele has been linked to an increased risk of Parkinson’s disease [60] and Alzheimer’s disease [61]. More recently, nPro-IL-16 has also been detected in extraneuronal tissues, such as arthritic cartilage [103], indicating that its role beyond the nervous system is still not fully understood. Additionally, rs4072111 has been associated with gastric cancer [54,62] and endometriosis [57,58], with the T allele linked to severe endometriosis stages [57].
Our results suggesting a potential association between rs4072111 and CC are therefore noteworthy. The cervix and parametrium contain nerve fibers [6,104], and perineural invasion (PNI) is a critical factor in CC progression, establishing pathways for CC colonization and metastasis along nerves [105,106]. Molecules initially identified in nerve tissues are now recognized for their roles in tumor-nerve interactions, influencing PNI and directly affecting CC cell growth. For instance, neurotrophins like nerve growth factor (NGF) and brain-derived neurotrophic factor (BDNF) are overexpressed in CC cells and promote PNI through interactions with p75 and Trk receptors [105,107]. NGF induces CC cell proliferation and migration [108], and high levels of NGF and TrkA are associated with PNI in early-stage cervical cancer [109]. The neuropeptide neuromedin B (NMB), also produced by CC cells, induces PNI by reprogramming Schwann cells, driving morphological and transcriptional changes, promoting proliferation and migration, and initiating PNI via CCL2 secretion and stimulation of axon regeneration [110]. It can be hypothesized that nPro-IL-16, through its role in nerve growth and differentiation, may contribute to CC expansion similarly to neurotrophins. Additionally, the secreted form of IL-16, produced from nPro-IL-16 cleavage, could further influence cervical carcinogenesis. Therefore, the potential role of rs4072111 (C>T), including its impact on the extracellular functions of IL-16, warrants further investigation.
To conclude, our study has several strengths. First, it is the first to investigate IL-8 and IL-16 genetic variants and their impact on CC risk in a European population. Second, the differentiated approach, which considered menopausal status, revealed that some associations were specific to premenopausal or postmenopausal women. Third, the observed association of rs4072111 with CC may encourage further research into the role of IL-16 and its neuronal precursor molecule in carcinogenesis.
However, our study also has limitations. The moderate sample size may have reduced the statistical power for SNPs with low MAF, particularly for IL-16. Additionally, as we did not have data on the expression levels of IL-8 and IL-16 in tissues or peripheral blood, our results, while assessing risk, do not allow us to determine the possible mechanisms underlying the observed or potentially missed associations. Third, we used an age threshold of <51/≥51 years as a proxy for menopausal status, a recommended approach when direct information on menstrual history is unavailable [111,112]. However, our data did not allow for distinguishing the effects of ovarian function cessation (menopause) from age-related processes, such as the progressive accumulation of DNA damage, reduced DNA repair capacity, increased cellular senescence, and accumulating epigenetic alterations [99,113]. Additionally, lifestyle factors (e.g., diet, physical activity, circadian disruption) and chronic low-grade systemic inflammation (“inflammaging”) further exacerbate age-related vulnerabilities to malignancies [114,115]. Thus, our findings that SNPs with low penetrance can impact CC risk differently in younger and older women may reflect influences of menopausal status, age, or their combined effects [99].
5. Conclusions
This study is the first to report a significant association between IL-16 rs4072111 and increased CC risk in women aged under 51 and between IL-16 rs4778889 and CC risk in women aged over 51. None of the eight analyzed SNPs significantly affected CC risk in the entire cohort. Specifically, neither IL-16 rs1131445 nor IL-8 rs4073, previously linked to CC risk in Asian populations, were linked to CC risk in this ethnically homogenous European cohort. Our results highlight the role of age-stratified analysis in cancer susceptibility and underscore the importance of ethnic background in assessing genetic risk factors. Additionally, they suggest that IL-8 and IL-16 SNPs may influence CC risk differently than other cancers, likely due to the specific relationship of CC carcinogenesis with HPV.
Author Contributions
Conceptualization, R.W. and R.Z.; methodology, R.W., E.O. and R.Z.; software, R.W. and E.O.; validation, G.H.; formal analysis, R.W.; investigation, R.W., E.O. and E.S.; resources, E.O., R.W., R.Z., M.B.F., S.P., T.V.G. and S.M.; data curation, E.O., E.S. and R.W.; writing—original draft preparation, R.W.; writing—review and editing, R.W., E.O. and R.Z.; visualization, R.W.; supervision, R.Z.; project administration, E.O.; funding acquisition, E.O., R.W. and R.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Ethics Committee of the Medical University of Vienna (EK 366/2003 and EK 1966/2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on reasonable request from the first (R.W.) or the corresponding (E.O.) author.
Acknowledgments
We are deeply grateful to the patients who donated blood samples to the Molecular Oncology Group’s blood bank, making studies like the present one possible.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A
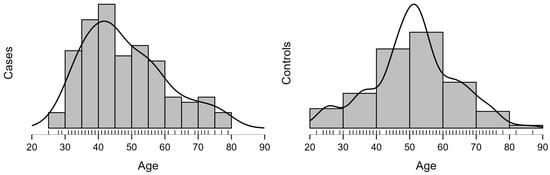
Figure A1.
Age distribution and density between CC cases (at first diagnosis) and controls.

Table A1.
Detailed clinicopathological characteristics of CC cases.
Table A1.
Detailed clinicopathological characteristics of CC cases.
| n | Percent | |
|---|---|---|
| Histology | ||
| Squamous | 101 | 80.2% |
| Adenocarcinoma | 18 | 14.3% |
| Adenosquamous | 5 | 4% |
| Missing | 2 | 1.6% |
| Grading | ||
| G1 | 10 | 7.9% |
| G2 | 53 | 42.1% |
| G3 | 40 | 31.8% |
| G-X | 23 | 18.3% |
| Pelvic lymph nodes | ||
| Positive | 36 | 28.6% |
| Negative | 72 | 57.1% |
| No PLND | 18 | 14.3% |
| Paraaortic lymph nodes | ||
| Positive | 3 | 2.4% |
| Negative | 85 | 67.5% |
| No PALND | 38 | 30.2% |
| FIGO (2018) | ||
| I A | 12 | 9.5% |
| I B | 36 | 28.6% |
| II A | 2 | 1.6% |
| II B | 25 | 19.8% |
| III A | 5 | 4% |
| III B | 6 | 4.8% |
| III C | 32 | 25.4% |
| IV A | 3 | 2.4% |
| IV B | 2 | 1.6% |
| Missing | 3 | 2.4% |
| Treatment | ||
| Surgery alone | 38 | 30.2% |
| Surgery followed by R(CH)T | 22 | 17.5% |
| LN-staging followed by R(CH)T | 48 | 38.1% |
| R(CH)T alone | 3 | 2.4% |
| Neoadjuvant CHT followed by surgery | 4 | 3.2% |
| RT followed by surgery | 1 | 0.8% |
| CHT alone | 1 | 0.8% |
| Missing | 9 | 7.1% |
PLND—pelvic lymphadenectomy, PALND—paraaortic lymphadenectomy, RT—radiotherapy, CHT—chemotherapy, R(CH)T—radiochemotherapy.
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Wang, M.; Huang, K.; Wong, M.C.S.; Huang, J.; Jin, Y.; Zheng, Z.-J. Global Cervical Cancer Incidence by Histological Subtype and Implications for Screening Methods. J. Epidemiol. Glob. Health 2024, 14, 94–101. [Google Scholar] [CrossRef]
- Cui, P.; Cong, X.; Chen, C.; Yang, L.; Liu, Z. Adenosquamous Carcinoma of the Cervix: A Population-Based Analysis. Front. Oncol. 2021, 11, 652850. [Google Scholar] [CrossRef]
- Watrowski, R.; Striepecke, E.; Jäger, C.; Bauknecht, T.; Horst, C. Papillary-Serous Adenocarcinoma of the Uterine Cervix during Tamoxifen Therapy after Bilateral Breast Cancer. Anticancer Res. 2012, 32, 5075–5078. [Google Scholar] [PubMed]
- Jung, L.; Klamminger, G.G.; Bier, B.; Eltze, E. From Satirical Poems and Invisible Poisons to Radical Surgery and Organized Cervical Cancer Screening-A Historical Outline of Cervical Carcinoma and Its Relation to HPV Infection. Life 2024, 14, 307. [Google Scholar] [CrossRef]
- Kostov, S.; Kornovski, Y.; Watrowski, R.; Yordanov, A.; Slavchev, S.; Ivanova, Y.; Yalcin, H.; Ivanov, I.; Selcuk, I. Revisiting Querleu-Morrow Radical Hysterectomy: How to Apply the Anatomy of Parametrium and Pelvic Autonomic Nerves to Cervical Cancer Surgery? Cancers 2024, 16, 2729. [Google Scholar] [CrossRef] [PubMed]
- Mangone, L.; Marinelli, F.; Bisceglia, I.; Roncaglia, F.; Mastrofilippo, V.; Morabito, F.; Neri, A.; Aguzzoli, L.; Mandato, V.D. Trends in Cervical Cancer: A Decade-long Analysis of Incidence, Survival and Demographic Disparities in a Northern Italian Province. Mol. Clin. Oncol. 2024, 21, 71. [Google Scholar] [CrossRef] [PubMed]
- Castellsagué, X. Natural History and Epidemiology of HPV Infection and Cervical Cancer. Gynecol. Oncol. 2008, 110, S4–S7. [Google Scholar] [CrossRef] [PubMed]
- Crosbie, E.J.; Einstein, M.H.; Franceschi, S.; Kitchener, H.C. Human Papillomavirus and Cervical Cancer. Lancet 2013, 382, 889–899. [Google Scholar] [CrossRef]
- Gardella, B.; Pasquali, M.F.; La Verde, M.; Cianci, S.; Torella, M.; Dominoni, M. The Complex Interplay between Vaginal Microbiota, HPV Infection, and Immunological Microenvironment in Cervical Intraepithelial Neoplasia: A Literature Review. Int. J. Mol. Sci. 2022, 23, 7174. [Google Scholar] [CrossRef] [PubMed]
- Vänskä, S.; Luostarinen, T.; Lagheden, C.; Eklund, C.; Kleppe, S.N.; Andrae, B.; Sparén, P.; Sundström, K.; Lehtinen, M.; Dillner, J. Differing Age-Specific Cervical Cancer Incidence Between Different Types of Human Papillomavirus: Implications for Predicting the Impact of Elimination Programs. Am. J. Epidemiol. 2021, 190, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Deutsch, I.; Horowitz, D.P.; Hershman, D.L.; Lewin, S.N.; Lu, Y.-S.; Neugut, A.I.; Herzog, T.J.; Chao, C.K.; Wright, J.D. Patterns of Care and Treatment Outcomes for Elderly Women with Cervical Cancer. Cancer 2012, 118, 3618–3626. [Google Scholar] [CrossRef] [PubMed]
- Jaisamrarn, U.; Castellsagué, X.; Garland, S.M.; Naud, P.; Palmroth, J.; Del Rosario-Raymundo, M.R.; Wheeler, C.M.; Salmerón, J.; Chow, S.-N.; Apter, D.; et al. Natural History of Progression of HPV Infection to Cervical Lesion or Clearance: Analysis of the Control Arm of the Large, Randomised PATRICIA Study. PLoS ONE 2013, 8, e79260. [Google Scholar] [CrossRef]
- Muntinga, C.L.P.; de Vos van Steenwijk, P.J.; Bekkers, R.L.M.; van Esch, E.M.G. Importance of the Immune Microenvironment in the Spontaneous Regression of Cervical Squamous Intraepithelial Lesions (cSIL) and Implications for Immunotherapy. J. Clin. Med. 2022, 11, 1432. [Google Scholar] [CrossRef] [PubMed]
- Bowden, S.J.; Bodinier, B.; Kalliala, I.; Zuber, V.; Vuckovic, D.; Doulgeraki, T.; Whitaker, M.D.; Wielscher, M.; Cartwright, R.; Tsilidis, K.K.; et al. Genetic Variation in Cervical Preinvasive and Invasive Disease: A Genome-Wide Association Study. Lancet Oncol. 2021, 22, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jiang, J.; Shen, H.; Hu, Z. Genetic Susceptibility of Cervical Cancer. J. Biomed. Res. 2011, 25, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Carrero, Y.N.; Callejas, D.E.; Mosquera, J.A. In Situ Immunopathological Events in Human Cervical Intraepithelial Neoplasia and Cervical Cancer: Review. Transl. Oncol. 2021, 14, 101058. [Google Scholar] [CrossRef]
- Chen, D.; Cui, T.; Ek, W.E.; Liu, H.; Wang, H.; Gyllensten, U. Analysis of the Genetic Architecture of Susceptibility to Cervical Cancer Indicates That Common SNPs Explain a Large Proportion of the Heritability. Carcinogenesis 2015, 36, 992–998. [Google Scholar] [CrossRef]
- Grimm, C.; Watrowski, R.; Baumühlner, K.; Natter, C.; Tong, D.; Wolf, A.; Zeillinger, R.; Leodolter, S.; Reinthaller, A.; Hefler, L. Genetic Variations of Interleukin-1 and -6 Genes and Risk of Cervical Intraepithelial Neoplasia. Gynecol. Oncol. 2011, 121, 537–541. [Google Scholar] [CrossRef]
- Karimi-Zarchi, M.; Abbasi, H.; Javaheri, A.; Hadadan, A.; Meibodi, B.; Tabatabaei, R.S.; Ghelmani, Y.; Neamatzadeh, H. Association of IL-12B Rs3212227 and IL-6 Rs1800795 Polymorphisms with Susceptibility to Cervical Cancer: A Systematic Review and Meta-Analysis. Asian Pac. J. Cancer Prev. 2020, 21, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Hu, G.; Chen, J.; Xie, G. Interleukin 1β and Interleukin 1 Receptor Antagonist Gene Polymorphisms and Cervical Cancer: A Meta-Analysis. Int. J. Gynecol. Cancer 2014, 24, 984–990. [Google Scholar] [CrossRef]
- Das, A.P.; Saini, S.; Agarwal, S.M. A Comprehensive Meta-Analysis of Non-Coding Polymorphisms Associated with Precancerous Lesions and Cervical Cancer. Genomics 2022, 114, 110323. [Google Scholar] [CrossRef] [PubMed]
- Akdis, M.; Aab, A.; Altunbulakli, C.; Azkur, K.; Costa, R.A.; Crameri, R.; Duan, S.; Eiwegger, T.; Eljaszewicz, A.; Ferstl, R.; et al. Interleukins (from IL-1 to IL-38), Interferons, Transforming Growth Factor β, and TNF-α: Receptors, Functions, and Roles in Diseases. J. Allergy Clin. Immunol. 2016, 138, 984–1010. [Google Scholar] [CrossRef] [PubMed]
- Waugh, D.J.J.; Wilson, C. The Interleukin-8 Pathway in Cancer. Clin. Cancer Res. 2008, 14, 6735–6741. [Google Scholar] [CrossRef] [PubMed]
- Qazi, B.S.; Tang, K.; Qazi, A. Recent Advances in Underlying Pathologies Provide Insight into Interleukin-8 Expression-Mediated Inflammation and Angiogenesis. Int. J. Inflam. 2011, 2011, 908468. [Google Scholar] [CrossRef]
- Dakal, T.C.; Kala, D.; Dhiman, G.; Yadav, V.; Krokhotin, A.; Dokholyan, N.V. Predicting the Functional Consequences of Non-Synonymous Single Nucleotide Polymorphisms in IL8 Gene. Sci. Rep. 2017, 7, 6525. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Yang, L.; Yin, S.; Zang, R.; Yang, G. The Polymorphism Interleukin-8 -251A/T Is Associated with a Significantly Increased Risk of Cancers from a Meta-Analysis. Tumour Biol. 2014, 35, 7115–7123. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-P.; Tai, D.-I.; Lan, K.-H.; Li, A.F.-Y.; Hsu, H.-C.; Lin, E.-J.; Lin, Y.-P.; Sheu, M.-L.; Li, C.-P.; Chang, F.-Y.; et al. The -251T Allele of the Interleukin-8 Promoter Is Associated with Increased Risk of Gastric Carcinoma Featuring Diffuse-Type Histopathology in Chinese Population. Clin. Cancer Res. 2005, 11, 6431–6441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, B.; Zhang, M.; Han, Y.; Zhao, Y.; Meng, Z.; Li, X.; Kang, J.; Yan, C. Interleukin-8 Gene Polymorphism Is Associated with Acute Coronary Syndrome in the Chinese Han Population. Cytokine 2011, 56, 188–191. [Google Scholar] [CrossRef] [PubMed]
- Charrad, R.; Kaabachi, W.; Rafrafi, A.; Berraies, A.; Hamzaoui, K.; Hamzaoui, A. IL-8 Gene Variants and Expression in Childhood Asthma. Lung 2017, 195, 749–757. [Google Scholar] [CrossRef]
- Ulhaq, Z.S.; Soraya, G.V. Roles of IL-8 -251A/T and +781C/T Polymorphisms, IL-8 Level, and the Risk of Age-Related Macular Degeneration. Arch. Soc. Esp. Oftalmol. (Engl. Ed.) 2021, 96, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Mao, P.; Xie, C.; Xie, W.; Wang, M.; Jiang, H. Association between Interleukin 8-251 T/A and +781 C/T Polymorphisms and Glioma Risk. Diagn. Pathol. 2015, 10, 138. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Y.; Liu, S.; Zhu, S.; Jiang, H.; Ding, J. Association between Interleukin 8 -251 A/T and +781 C/T Polymorphisms and Osteosarcoma Risk in Chinese Population: A Case-Control Study. Tumour Biol. 2016, 37, 6191–6196. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-H.; Yang, Y.-C.; Hsia, T.-C.; Shen, T.-C.; Shen, Y.-C.; Chang, W.-S.; Wang, Y.-C.; Tsai, C.-W.; Bau, D.-T. Association of Interleukin-8 Promoter Genotypes with Taiwan Lung Cancer Risk. Anticancer. Res. 2022, 42, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Chien, M.-H.; Yeh, C.-B.; Li, Y.-C.; Wei, L.-H.; Chang, J.-H.; Peng, Y.-T.; Yang, S.-F.; Kuo, W.-H. Relationship of Interleukin-8 Gene Polymorphisms with Hepatocellular Carcinoma Susceptibility and Pathological Development. J. Surg. Oncol. 2011, 104, 798–803. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Chang, W.-S.; Tsai, C.-W.; Hsia, T.-C.; Shen, T.-C.; Bau, D.-T.; Shui, H.-A. The Contribution of Interleukin-8 Genotypes and Expression to Nasopharyngeal Cancer Susceptibility in Taiwan. Medicine 2018, 97, e12135. [Google Scholar] [CrossRef]
- Koensgen, D.; Bruennert, D.; Ungureanu, S.; Sofroni, D.; Braicu, E.I.; Sehouli, J.; Sümnig, A.; Delogu, S.; Zygmunt, M.; Goyal, P.; et al. Polymorphism of the IL-8 Gene and the Risk of Ovarian Cancer. Cytokine 2015, 71, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Watrowski, R.; Schuster, E.; Hofstetter, G.; Fischer, M.B.; Mahner, S.; Van Gorp, T.; Polterauer, S.; Zeillinger, R.; Obermayr, E. Association of Four Interleukin-8 Polymorphisms (−251 A>T, +781 C>T, +1633 C>T, +2767 A>T) with Ovarian Cancer Risk: Focus on Menopausal Status and Endometriosis-Related Subtypes. Biomedicines 2024, 12, 321. [Google Scholar] [CrossRef]
- Cruikshank, W.W.; Center, D.M.; Nisar, N.; Wu, M.; Natke, B.; Theodore, A.C.; Kornfeld, H. Molecular and Functional Analysis of a Lymphocyte Chemoattractant Factor: Association of Biologic Function with CD4 Expression. Proc. Natl. Acad. Sci. USA 1994, 91, 5109–5113. [Google Scholar] [CrossRef]
- Cruikshank, W.W.; Kornfeld, H.; Center, D.M. Interleukin-16. J. Leukoc. Biol. 2000, 67, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.C.; Center, D.M.; Cruikshank, W.W. The Effect of Interleukin-16 and Its Precursor on T Lymphocyte Activation and Growth. Growth Factors 2004, 22, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Glass, W.G.; Sarisky, R.T.; Vecchio, A.M.D. Not-so-Sweet Sixteen: The Role of IL-16 in Infectious and Immune-Mediated Inflammatory Diseases. J. Interferon Cytokine Res. 2006, 26, 511–520. [Google Scholar] [CrossRef]
- Richmond, J.; Tuzova, M.; Cruikshank, W.; Center, D. Regulation of Cellular Processes by Interleukin-16 in Homeostasis and Cancer. J. Cell Physiol. 2014, 229, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Hridi, S.U.; Franssen, A.J.P.M.; Jiang, H.-R.; Bushell, T.J. Interleukin-16 Inhibits Sodium Channel Function and GluA1 Phosphorylation via CD4- and CD9-Independent Mechanisms to Reduce Hippocampal Neuronal Excitability and Synaptic Activity. Mol. Cell Neurosci. 2019, 95, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Kurschner, C.; Yuzaki, M. Neuronal Interleukin-16 (NIL-16): A Dual Function PDZ Domain Protein. J. Neurosci. 1999, 19, 7770–7780. [Google Scholar] [CrossRef] [PubMed]
- Burkart, K.M.; Barton, S.J.; Holloway, J.W.; Yang, I.A.; Cakebread, J.A.; Cruikshank, W.; Little, F.; Jin, X.; Farrer, L.A.; Clough, J.B.; et al. Association of Asthma with a Functional Promoter Polymorphism in the IL16 Gene. J. Allergy Clin. Immunol. 2006, 117, 86–91. [Google Scholar] [CrossRef]
- Skundric, D.S.; Cruikshank, W.W.; Montgomery, P.C.; Lisak, R.P.; Tse, H.Y. Emerging Role of IL-16 in Cytokine-Mediated Regulation of Multiple Sclerosis. Cytokine 2015, 75, 234–248. [Google Scholar] [CrossRef]
- Zeinalzadeh, S.; Kheradmand, N.; Rasouli, G.; Esmaeilzadeh, E.; Pakzad, B.; Behroozi, J.; Chamanara, M.; Zoshk, M.Y.; Ehtesham, N.; Sabet, M.N. Association of a miRNA-Binding Site Polymorphism in IL-16 Gene with Disease Risk and Clinical Characteristics of Rheumatoid Arthritis and Systemic Lupus Erythematosus. Clin. Rheumatol. 2022, 41, 2189–2196. [Google Scholar] [CrossRef] [PubMed]
- Almulla, A.F.; Abbas Abo Algon, A.; Tunvirachaisakul, C.; Al-Hakeim, H.K.; Maes, M. T Helper-1 Activation via Interleukin-16 Is a Key Phenomenon in the Acute Phase of Severe, First-Episode Major Depressive Disorder and Suicidal Behaviors. J. Adv. Res. 2024, 64, 171–181. [Google Scholar] [CrossRef]
- de Souza, V.H.; de Alencar, J.B.; Tiyo, B.T.; Alves, H.V.; Vendramini, E.C.L.; Sell, A.M.; Visentainer, J.E.L. Association of Functional IL16 Polymorphisms with Cancer and Cardiovascular Disease: A Meta-Analysis. Oncotarget 2020, 11, 3405–3417. [Google Scholar] [CrossRef]
- Tang, Y.-J.; Wang, J.-L.; Xie, K.-G.; Lan, C.-G. Association of Interleukin 16 Gene Polymorphisms and Plasma IL16 Level with Osteosarcoma Risk. Sci. Rep. 2016, 6, 34607. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Peng, Q.; Lao, X.; Chen, Z.; Lu, Y.; Lao, X.; Mo, C.; Sui, J.; Wu, J.; Zhai, L.; et al. The Association of Interleukin-16 Gene Polymorphisms with IL-16 Serum Levels and Risk of Nasopharyngeal Carcinoma in a Chinese Population. Tumour Biol. 2014, 35, 1917–1924. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-F.; Wang, Y.-C.; Shen, T.-C.; Chang, W.-S.; Li, H.-T.; Liao, C.-H.; Gong, C.-L.; Wang, Z.-H.; Tsai, C.-W.; Hsia, T.-C.; et al. Significant Association of Interleukin-16 Genetic Variations to Taiwanese Lung Cancer. In Vivo 2020, 34, 1117–1123. [Google Scholar] [CrossRef]
- Shih, L.-C.; Chang, W.-S.; Lee, H.-T.; Wang, Y.-C.; Wang, Z.-H.; Chao, C.-Y.; Yu, C.-C.; Lin, H.-Y.; Shen, T.-C.; Kuo, C.-C.; et al. Interaction of Interleukin-16 Genotypes with Betel Quid Chewing Behavior on Oral Cancer in Taiwan. In Vivo 2020, 34, 1759–1764. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.-B.; Rao, L.; Wang, Y.-Y.; Liang, W.-B.; Li, C.; Xue, H.; Zhou, B.; Sun, H.; Li, Y.; Lv, M.-L.; et al. The Association of Interleukin-16 Polymorphisms with IL-16 Serum Levels and Risk of Colorectal and Gastric Cancer. Carcinogenesis 2009, 30, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.-K.; Mong, M.-C.; Tzeng, H.-E.; Yang, M.-D.; Chen, J.-C.; Hsia, T.-C.; Hsia, N.-Y.; Tsai, C.-W.; Chang, W.-S.; Chen, C.-P.; et al. The Significant Contribution of Interleukin-16 Genotypes, Smoking, Alcohol Drinking, and Helicobacter Pylori Infection to Gastric Cancer. In Vivo 2024, 38, 90–97. [Google Scholar] [CrossRef]
- Azimzadeh, P.; Khorram Khorshid, H.R.; Akhondi, M.M.; Shirazi, A. Association of Interleukin-16 Polymorphisms with Disease Progression and Susceptibility in Endometriosis. Int. J. Immunogenet. 2016, 43, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Matalliotakis, M.; Zervou, M.I.; Eliopoulos, E.; Matalliotaki, C.; Rahmioglu, N.; Kalogiannidis, I.; Zondervan, K.; Spandidos, D.A.; Matalliotakis, I.; Goulielmos, G.N. The Role of IL-16 Gene Polymorphisms in Endometriosis. Int. J. Mol. Med. 2018, 41, 1469–1476. [Google Scholar] [CrossRef]
- Xue, H.; Gao, L.; Wu, Y.; Fang, W.; Wang, L.; Li, C.; Li, Y.; Liang, W.; Zhang, L. The IL-16 Gene Polymorphisms and the Risk of the Systemic Lupus Erythematosus. Clin. Chim. Acta 2009, 403, 223–225. [Google Scholar] [CrossRef]
- Zhang, X.; Xue, L.; Sun, X.; Jin, J.; Bai, X.; Xie, A. Association of IL-16 Gene Polymorphisms with Sporadic Parkinson’s Disease in a Han Chinese Population. Neurosci. Lett. 2020, 724, 134877. [Google Scholar] [CrossRef]
- Anvar, N.E.; Saliminejad, K.; Ohadi, M.; Kamali, K.; Daneshmand, P.; Khorshid, H.R.K. Association between Polymorphisms in Interleukin-16 Gene and Risk of Late-Onset Alzheimer’s Disease. J. Neurol. Sci. 2015, 358, 324–327. [Google Scholar] [CrossRef] [PubMed]
- Kashfi, S.M.H.; Behboudi Farahbakhsh, F.; Nazemalhosseini Mojarad, E.; Mashayekhi, K.; Azimzadeh, P.; Romani, S.; Derakhshani, S.; Malekpour, H.; Asadzadeh Aghdaei, H.; Zali, M.R. Interleukin-16 Polymorphisms as New Promising Biomarkers for Risk of Gastric Cancer. Tumour Biol. 2016, 37, 2119–2126. [Google Scholar] [CrossRef]
- Yang, S.X.; Chen, F.; Zhang, J.W.; Sun, Z.Q.; Chen, B.P. IL-16 Rs4778889 Polymorphism Contribution to the Development of Renal Cell Cancer in a Chinese Population. Genet. Mol. Res. 2016, 15, 1–7. [Google Scholar] [CrossRef]
- Gan, X.-L.; Lin, Y.-H.; Zhang, Y.; Yu, T.-H.; Hu, L.-N. Association of an Interleukin-16 Gene Polymorphism with the Risk and Pain Phenotype of Endometriosis. DNA Cell Biol. 2010, 29, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Babah, O.A.; Ojewunmi, O.O.; Onwuamah, C.K.; Udenze, I.C.; Osuntoki, A.A.; Afolabi, B.B. Serum Concentrations of IL-16 and Its Genetic Polymorphism Rs4778889 Affect the Susceptibility and Severity of Endometriosis in Nigerian Women. BMC Womens Health 2023, 23, 253. [Google Scholar] [CrossRef]
- Mi, Y.; Wang, L.; Zong, L.; Pei, M.; Lu, Q.; Huang, P. Genetic Variants in microRNA Target Sites of 37 Selected Cancer-Related Genes and the Risk of Cervical Cancer. PLoS ONE 2014, 9, e86061. [Google Scholar] [CrossRef]
- Azimzadeh, P.; Romani, S.; Mohebbi, S.R.; Mahmoudi, T.; Vahedi, M.; Fatemi, S.R.; Zali, N.; Zali, M.R. Association of Polymorphisms in microRNA-Binding Sites and Colorectal Cancer in an Iranian Population. Cancer Genet. 2012, 205, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Watrowski, R.; Schuster, E.; Van Gorp, T.; Hofstetter, G.; Fischer, M.B.; Mahner, S.; Polterauer, S.; Zeillinger, R.; Obermayr, E. Association of the Single Nucleotide Polymorphisms Rs11556218, Rs4778889, Rs4072111, and Rs1131445 of the Interleukin-16 Gene with Ovarian Cancer. Int. J. Mol. Sci. 2024, 25, 10272. [Google Scholar] [CrossRef]
- Wu, S.; Lu, S.; Tao, H.; Zhang, L.; Lin, W.; Shang, H.; Xie, J. Correlation of Polymorphism of IL-8 and MMP-7 with Occurrence and Lymph Node Metastasis of Early Stage Cervical Cancer. J. Huazhong Univ. Sci. Technol. Med. Sci. 2011, 31, 114–119. [Google Scholar] [CrossRef]
- Kaczmarek, M. The Timing of Natural Menopause in Poland and Associated Factors. Maturitas 2007, 57, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Bhatla, N.; Berek, J.S.; Cuello Fredes, M.; Denny, L.A.; Grenman, S.; Karunaratne, K.; Kehoe, S.T.; Konishi, I.; Olawaiye, A.B.; Prat, J.; et al. Revised FIGO Staging for Carcinoma of the Cervix Uteri. Int. J. Gynaecol. Obstet. 2019, 145, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.M. Genetic Association Studies: Design, Analysis and Interpretation. Brief. Bioinform. 2002, 3, 146–153. [Google Scholar] [CrossRef]
- Horita, N.; Kaneko, T. Genetic Model Selection for a Case-Control Study and a Meta-Analysis. Meta Gene 2015, 5, 1–8. [Google Scholar] [CrossRef]
- JASP, Version 19.0.0; JASP Team: Amsterdam, The Netherlands, 2024. Available online: https://jasp-stats.org/ (accessed on 28 August 2024).
- VassarStats: Website for Statistical Computation. Copyright: Richard Lowry 1998–2023. Available online: http://www.vassarstats.net (accessed on 28 August 2024).
- The Genome Aggregation Database (gnomAD). Available online: https://gnomad.broadinstitute.org (accessed on 4 September 2024).
- dbGaP Allele Frequency Aggregator (ALFA) Database. Available online: https://www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa/ (accessed on 4 September 2024).
- Yugawa, T.; Kiyono, T. Molecular Mechanisms of Cervical Carcinogenesis by High-Risk Human Papillomaviruses: Novel Functions of E6 and E7 Oncoproteins. Rev. Med. Virol. 2009, 19, 97–113. [Google Scholar] [CrossRef]
- Kim, W.; Pyo, J.; Noh, B.-J.; Jeong, J.-W.; Lee, J.; Kim, J.-E. CCAR2 Negatively Regulates IL-8 Production in Cervical Cancer Cells. Oncotarget 2018, 9, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-J.; Xue, J.-M.; Li, J.; Wan, L.-H.; Zhu, Y.-X. Identification of Key Genes and Pathways of Diagnosis and Prognosis in Cervical Cancer by Bioinformatics Analysis. Mol. Genet. Genom. Med. 2020, 8, e1200. [Google Scholar] [CrossRef]
- Jia, L.; Li, F.; Shao, M.; Zhang, W.; Zhang, C.; Zhao, X.; Luan, H.; Qi, Y.; Zhang, P.; Liang, L.; et al. IL-8 Is Upregulated in Cervical Cancer Tissues and Is Associated with the Proliferation and Migration of HeLa Cervical Cancer Cells. Oncol. Lett. 2018, 15, 1350–1356. [Google Scholar] [CrossRef]
- Fernandez-Avila, L.; Castro-Amaya, A.M.; Molina-Pineda, A.; Hernández-Gutiérrez, R.; Jave-Suarez, L.F.; Aguilar-Lemarroy, A. The Value of CXCL1, CXCL2, CXCL3, and CXCL8 as Potential Prognosis Markers in Cervical Cancer: Evidence of E6/E7 from HPV16 and 18 in Chemokines Regulation. Biomedicines 2023, 11, 2655. [Google Scholar] [CrossRef] [PubMed]
- Osiagwu, D.D.; Azenabor, A.E.; Osijirin, A.A.; Awopetu, P.I.; Oyegbami, F.R. Evaluation of Interleukin 8 and Interleukin 10 Cytokines in Liquid Based Cervical Cytology Samples. Pan Afr. Med. J. 2019, 32, 148. [Google Scholar] [CrossRef]
- Tjiong, M.Y.; van der Vange, N.; ten Kate, F.J.; Tjong-A-Hung, S.P.; ter Schegget, J.; Burger, M.P.; Out, T.A. Increased IL-6 and IL-8 Levels in Cervicovaginal Secretions of Patients with Cervical Cancer. Gynecol. Oncol. 1999, 73, 285–291. [Google Scholar] [CrossRef]
- Cotrim, A.C.d.M.; França, E.L.; Martins, J.S.; Silva, K.P.G.; Fujimori, M.; Ghalfi, Y.C.; Machado, I.T.; Honorio-França, A.C.; Tozetti, I.A. Correlation between Melatonin Concentration and Cytokines in Cervical Mucus in Positive Samples for the Presence of Human Papillomavirus. Cancer Immunol. Immunother. 2021, 70, 2721–2726. [Google Scholar] [CrossRef]
- Kumar, A.; Cherukumilli, M.; Mahmoudpour, S.H.; Brand, K.; Bandapalli, O.R. ShRNA-Mediated Knock-down of CXCL8 Inhibits Tumor Growth in Colorectal Liver Metastasis. Biochem. Biophys. Res. Commun. 2018, 500, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.-J.; Chen, S.-U.; Chou, C.-H.; Lin, M.-C. Lysophosphatidic Acid Receptor 2/3-Mediated IL-8-Dependent Angiogenesis in Cervical Cancer Cells. Int. J. Cancer 2012, 131, 789–802. [Google Scholar] [CrossRef]
- Wu, S.; Shang, H.; Cui, L.; Zhang, Z.; Zhang, Y.; Li, Y.; Wu, J.; Li, R.-K.; Xie, J. Targeted Blockade of Interleukin-8 Abrogates Its Promotion of Cervical Cancer Growth and Metastasis. Mol. Cell Biochem. 2013, 375, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Shuai, H.; Luo, X.; Wang, X.; Guan, B. The Clinical and Prognostic Value of CXCL8 in Cervical Carcinoma Patients: Immunohistochemical Analysis. Biosci. Rep. 2017, 37, BSR20171021. [Google Scholar] [CrossRef]
- Olukomogbon, T.; Akpobome, B.; Omole, A.; ACCME Research Group; Adebamowo, C.A.; Adebamowo, S.N. Association Between Cervical Inflammatory Mediators and Prevalent Cervical Human Papillomavirus Infection. JCO Glob. Oncol. 2024, 10, e2300380. [Google Scholar] [CrossRef] [PubMed]
- Grimm, C.; Watrowski, R.; Polterauer, S.; Baumühlner, K.; Natter, C.; Rahhal, J.; Heinze, G.; Schuster, E.; Hefler, L.; Reinthaller, A. Vascular Endothelial Growth Factor Gene Polymorphisms and Risk of Cervical Intraepithelial Neoplasia. Int. J. Gynecol. Cancer 2011, 21, 597–601. [Google Scholar] [CrossRef]
- Tabangin, M.E.; Woo, J.G.; Martin, L.J. The Effect of Minor Allele Frequency on the Likelihood of Obtaining False Positives. BMC Proc. 2009, 3 (Suppl. S7), S41. [Google Scholar] [CrossRef]
- Politi, C.; Roumeliotis, S.; Tripepi, G.; Spoto, B. Sample Size Calculation in Genetic Association Studies: A Practical Approach. Life 2023, 13, 235. [Google Scholar] [CrossRef]
- Park, S.L.; Cheng, I.; Haiman, C.A. Genome-Wide Association Studies of Cancer in Diverse Populations. Cancer Epidemiol. Biomark. Prev. 2018, 27, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Watrowski, R.; Castillo-Tong, D.C.; Fabjani, G.; Schuster, E.; Fischer, M.; Zeillinger, R. The 811 C/T Polymorphism in the 3′ Untranslated Region of the Selenoprotein 15-kDa (Sep15) Gene and Breast Cancer in Caucasian Women. Tumour Biol. 2016, 37, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Yang, X.; Liu, Y.; Song, Q.; Pan, X.; Chen, W.; Jiang, W.; Xu, D.; Song, Y.; Chen, R. Genetic Polymorphisms in DNA Repair Genes and Their Association with Risk of Cervical Cancer: A Systematic Review and Meta-Analysis. J. Obstet. Gynaecol. Res. 2022, 48, 2405–2418. [Google Scholar] [CrossRef] [PubMed]
- Lao, O.; Lu, T.T.; Nothnagel, M.; Junge, O.; Freitag-Wolf, S.; Caliebe, A.; Balascakova, M.; Bertranpetit, J.; Bindoff, L.A.; Comas, D.; et al. Correlation between Genetic and Geographic Structure in Europe. Curr. Biol. 2008, 18, 1241–1248. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, M.; Sironi, M.; Pozzoli, U.; Ferrer-Admetlla, A.; Pattini, L.; Nielsen, R. Signatures of Environmental Genetic Adaptation Pinpoint Pathogens as the Main Selective Pressure through Human Evolution. PLoS Genet. 2011, 7, e1002355. [Google Scholar] [CrossRef]
- Shepherd, R.; Cheung, A.S.; Pang, K.; Saffery, R.; Novakovic, B. Sexual Dimorphism in Innate Immunity: The Role of Sex Hormones and Epigenetics. Front. Immunol. 2020, 11, 604000. [Google Scholar] [CrossRef]
- Ambros, V. MicroRNA-Mediated Gene Regulation and the Resilience of Multicellular Animals. Postepy Biochem. 2024, 70, 62–70. [Google Scholar] [CrossRef]
- Zhao, X.; Fan, X.; Lin, X.; Guo, B.; Yu, Y. Deciphering Age-Specific Molecular Features in Cervical Cancer and Constructing an Angio-Immune Prognostic Model. Medicine 2024, 103, e37717. [Google Scholar] [CrossRef] [PubMed]
- Voutsadakis, I.A. PI3KCA Mutations in Uterine Cervix Carcinoma. J. Clin. Med. 2021, 10, 220. [Google Scholar] [CrossRef]
- McKenna, G.; O’Flatharta, C.; Ranera, B.; Shaw, G.; Barron, V.; Barry, F.; Murphy, M. Investigation of the Role of Interleukin 16 in Chondrogenesis of Mesenchymal Stem Cells and in Osteoarthritis. Osteoarthr. Cartil. 2013, 21, S239–S240. [Google Scholar] [CrossRef][Green Version]
- Kostov, S.; Kornovski, Y.; Yordanov, A.; Watrowski, R.; Slavchev, S.; Ivanova, Y.; Ganev, T.; Yalçın, H.; Selçuk, I. Surgical Anatomy and Dissection of the Hypogastric Plexus in Nerve-Sparing Radical Hysterectomy. Diagnostics 2023, 14, 83. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Sun, H.; Chen, Y.; Wang, L.; Song, O.; Zhang, J.; Li, D.; Liu, X.; Feng, L. Perineural Invasion in Cervical Cancer: A Hidden Trail for Metastasis. Diagnostics 2024, 14, 1517. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Wang, Y.; Wang, Z.; Cui, Y.; Sun, X.; Wang, Y. Weighted Gene Co-Expression Network Analysis Identified Cancer Cell Proliferation as a Common Phenomenon During Perineural Invasion. OncoTargets Ther. 2019, 12, 10361–10374. [Google Scholar] [CrossRef]
- Sirico, A.; Simonelli, S.; Pignatiello, S.; Fulgione, C.; Sarno, L.; Chiuso, F.; Maruotti, G.M.; Sansone, M.; Guida, M.; Insabato, L. BDNF and NGF Expression in Preneoplastic Cervical Disease According to HIV Status. Int. J. Mol. Sci. 2023, 24, 10729. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, J.; Wang, R.; Chen, H.; Wang, R.; Wang, W.; Yang, X. NGF Signaling Interacts with the Hippo/YAP Pathway to Regulate Cervical Cancer Progression. Front. Oncol. 2021, 11, 688794. [Google Scholar] [CrossRef]
- Long, Y.; Yao, D.-S.; Wei, Y.-S.; Wu, G.-T. Effects of Nerve Growth Factor Expression on Perineural Invasion and Worse Prognosis in Early-Stage Cervical Cancer. Chin. Med. J. 2018, 131, 2360–2363. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, Q.; Huang, T.; Xu, C.; Yang, X.; Zhang, L.; Wang, J.; Yang, L.; Zheng, X.; Fan, Q.; et al. Cervical Cancer-Produced Neuromedin-B Reprograms Schwann Cells to Initiate Perineural Invasion. Cell Death Dis. 2024, 15, 636. [Google Scholar] [CrossRef]
- Heer, E.; Harper, A.; Escandor, N.; Sung, H.; McCormack, V.; Fidler-Benaoudia, M.M. Global Burden and Trends in Premenopausal and Postmenopausal Breast Cancer: A Population-Based Study. Lancet Glob. Health 2020, 8, e1027–e1037. [Google Scholar] [CrossRef] [PubMed]
- Phipps, A.I.; Ichikawa, L.; Bowles, E.J.A.; Carney, P.A.; Kerlikowske, K.; Miglioretti, D.L.; Buist, D.S.M. Defining Menopausal Status in Epidemiologic Studies: A Comparison of Multiple Approaches and Their Effects on Breast Cancer Rates. Maturitas 2010, 67, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Montégut, L.; López-Otín, C.; Kroemer, G. Aging and Cancer. Mol. Cancer 2024, 23, 106. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Zhang, X.; Zheng, S.; Khanabdali, R.; Kalionis, B.; Wu, J.; Wan, W.; Tai, X. An Update on Inflamm-Aging: Mechanisms, Prevention, and Treatment. J. Immunol. Res. 2016, 2016, 8426874. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.; Anderson, C.; Lippman, S.M. Physical Activity, Sedentary Behaviour, Diet, and Cancer: An Update and Emerging New Evidence. Lancet Oncol. 2017, 18, e457–e471. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).