Genetic Profiling of MRSA and MSSA from Food Contact Surfaces: Antibiotic, Heavy Metal and Benzalkonium Chloride Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Detection of Genes Involved in Antibiotic Resistance and Staphylococcal Chromosomal Cassette mec (SCCmec) Typing
2.3. Antimicrobial Susceptibility Testing
2.4. Disinfectant and Heavy Metal Sensitivity
2.5. Treatment with BC with or Without Milk Residues for Removal of Biofilm Methicillin-Resistant S. aureus (MRSA) and Methicillin-Susceptible S. aureus (MSSA)
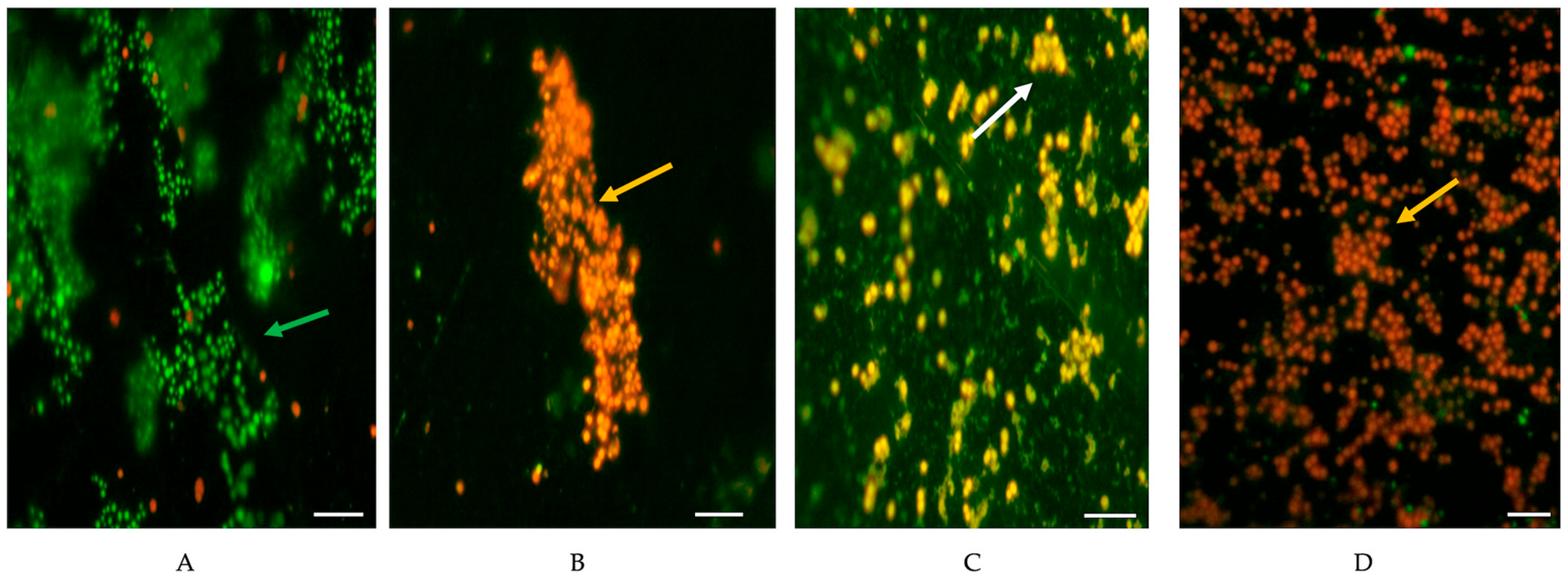
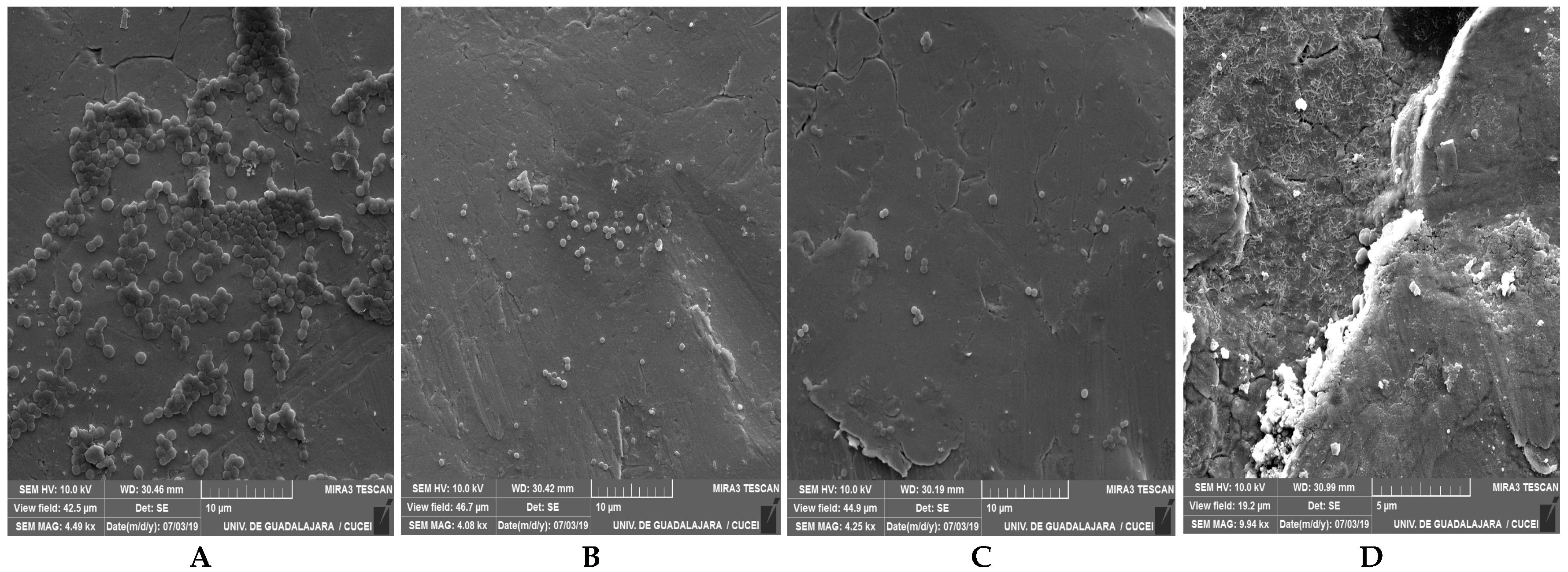
2.6. Evaluation of Cell Viability and Scanning Electron Microscopy (SEM)
2.7. Statistical Analysis
3. Results
3.1. Antimicrobial Resistance and Sensitivity to Disinfectants and Heavy Metals of MRSA and MSSA
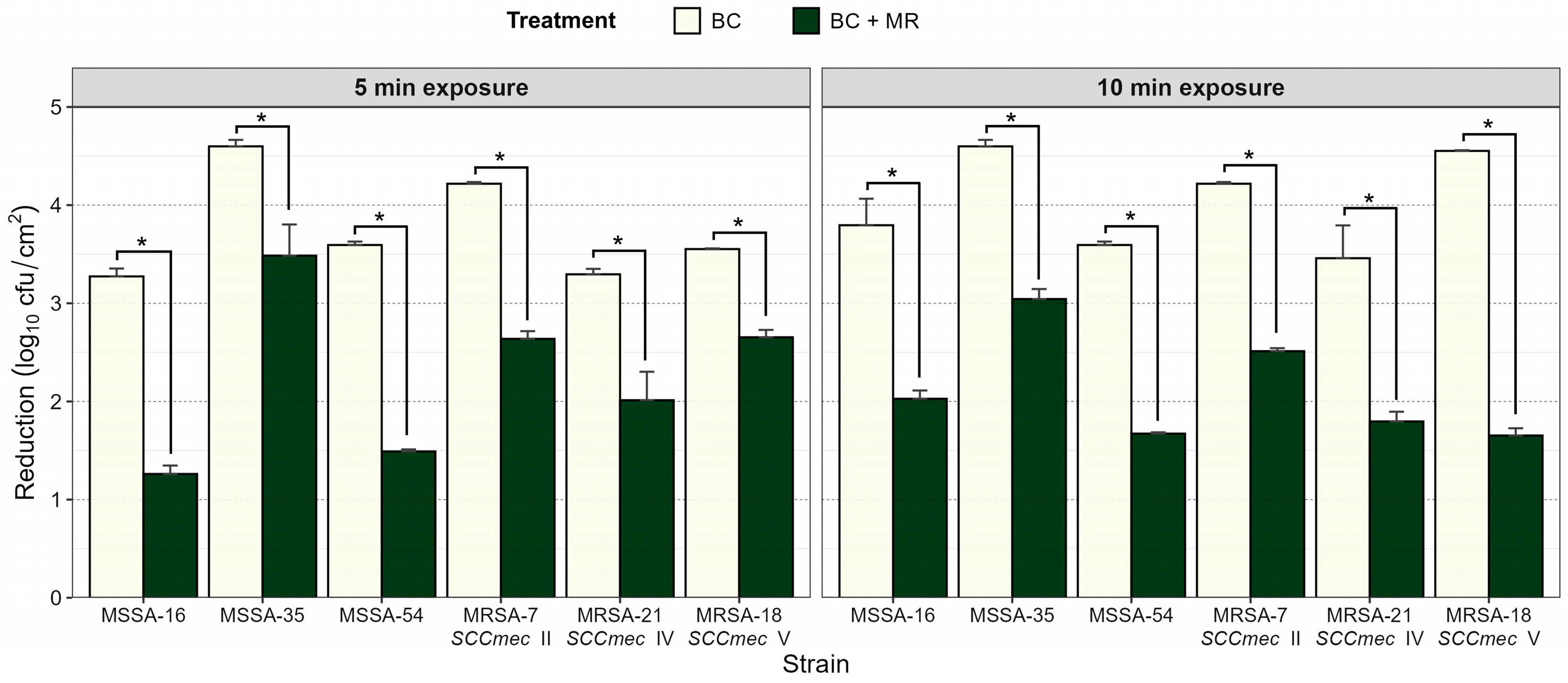
3.2. Reduction in Biofilms of MSSA and MRSA with BC with or Without Milk Residues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.E.; George, S.E.; Wolz, C.; Olson, M.E. Biofilms: A developmental niche for vancomycin-intermediate Staphylococcus aureus. Infect. Genet. Evol. 2024, 117, 105545. [Google Scholar] [CrossRef] [PubMed]
- Abril, A.G.; Villa, T.G.; Barros-Velázquez, J.; Cañas, B.; Sánchez-Pérez, A.; Calo-Mata, P.; Carrera, M. Staphylococcus aureus Exotoxins and Their Detection in the Dairy Industry and Mastitis. Toxins 2020, 12, 537. [Google Scholar] [CrossRef] [PubMed]
- Kotzamanidis, C.; Vafeas, G.; Giantzi, V.; Anastasiadou, S.; Mygdalias, S.; Malousi, A.; Loukia, E.; Daniel, S.; Zdragas, A. Staphylococcus aureus Isolated from Ruminants with Mastitis in Northern Greece Dairy Herds: Genetic Relatedness and Phenotypic and Genotypic Characterization. Toxins 2021, 13, 176. [Google Scholar] [CrossRef]
- Annamanedi, M.; Sheela, P.; Sundareshan, S.; Isloor, S.; Gupta, P.; Jasmeen, P.; Gargi, M.; Mallick, S.; Hegde, N.R. Molecular fingerprinting of bovine mastitis-associated Staphylococcus aureus isolates from India. Sci. Rep. 2021, 11, 15228. [Google Scholar] [CrossRef]
- León-Galván, M.F.; Barboza-Corona, J.E.; Lechuga-Arana, A.A.; Valencia-Posadas, M.; Aguayo, D.D.; Cedillo-Pelaez, C.; Martínez-Ortega, E.A.; Gutierrez-Chavez, A.J. Molecular detection and sensitivity to antibiotics and bacteriocins of pathogens isolated from bovine mastitis in family dairy herds of central Mexico. Biomed. Res. Int. 2015, 2015, 615153. [Google Scholar] [CrossRef]
- Vázquez, H.C.; Jäger, S.; Wolter, W.; Zschöck, M.; Vazquez, M.A.C.; El-Sayed, A. Isolation and identification of main mastitis pathogens in Mexico. Arq. Bras. Med. Vet. Zootec. 2013, 65, 377–382. [Google Scholar] [CrossRef]
- Ahmad, S.; Raqeeb, A.; Ali, F.; Anwar, M. Characterization of novel antibiotic resistance genes in Staphylococcal aureus. J. Bacteriol. Mycol. 2018, 6, 8–10. [Google Scholar] [CrossRef]
- Nwobi, O.C.; Anyanwu, M.U.; Jaja, I.F.; Nwankwo, I.O.; Okolo, C.C.; Nwobi, C.A.; Ezenduka, E.V.; Oguttu, J.W. Staphylococcus aureus in Horses in Nigeria: Occurrence, Antimicrobial, Methicillin and Heavy Metal Resistance and Virulence Potentials. Antibiotics 2023, 12, 242. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Methicillin-Resistant Staphylococcus aureus. Available online: https://www.cdc.gov/antimicrobial-resistance/media/pdfs/MRSA-508.pdf (accessed on 2 August 2025).
- World Health Organization (WHO). WHO Bacterial Priority Pathogens List, 2024. Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance. Available online: https://data.who.int/indicators/i/918081E/5DD9606 (accessed on 21 September 2025).
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 5 August 2025).
- Guo, H.; Tong, Y.; Cheng, J.; Abbas, Z.; Li, Z.; Wang, J.; Zhou, Y.; Si, D.; Zhang, R. Biofilm and Small Colony Variants—An Update on Staphylococcus aureus Strategies toward Drug Resistance. Int. J. Mol. Sci. 2022, 23, 1241. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Amábile-Cuevas, C.F. Antibiotic usage and resistance in Mexico: An update after a decade of change. J. Infect. Dev. Ctries. 2021, 15, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Larsson, D.G.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Dweba, C.C.; Zishiri, O.T.; El Zowalaty, M.E. Isolation and molecular identification of virulence, antimicrobial and heavy metal resistance genes in livestock-associated methicillin-resistant Staphylococcus aureus. Pathogens 2019, 8, 79. [Google Scholar] [CrossRef]
- Di Ciccio, P.; Vergara, A.; Festino, A.R.; Paludi, D.; Zanardi, E.; Ghidini, S.; Ianieri, A. Biofilm formation by Staphylococcus aureus on food contact surfaces: Relationship with temperature and cell surface hydrophobicity. Food Control 2015, 50, 930–936. [Google Scholar] [CrossRef]
- Avila-Novoa, M.G.; Iñíguez-Moreno, M.; Solís-Velázquez, O.A.; González-Gómez, J.P.; Guerrero-Medina, P.J.; Gutiérrez-Lomelí, M. Biofilm Formation by Staphylococcus aureus Isolated from Food Contact Surfaces in the Dairy Industry of Jalisco, Mexico. J. Food Qual. 2018, 2018, 72–78. [Google Scholar] [CrossRef]
- Bridier, A.; Sanchez-Vizuete, P.; Guilbaud, M.; Piard, J.C.; Naïtali, M.; Briandet, R. Biofilm-associated persistence of food-borne pathogens. Food Microbiol. 2015, 45, 167–178. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Wassenaar, T.; Ussery, D.; Nielsen, L.; Ingmer, H. Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur. J. Microbiol. Immunol. 2015, 5, 44–61. [Google Scholar] [CrossRef]
- Avila-Novoa, M.G.; Solis-Velazquez, O.A.; Guerrero-Medina, P.J.; González-Gómez, J.P.; González-Torres, B.; Velázquez-Suárez, N.Y.; Martínez-Chávez, L.; Martínez-Gonzáles, N.E.; De la Cruz-Color, L.; Ibarra-Velázquez, L.M.; et al. Genetic and compositional analysis of biofilm formed by Staphylococcus aureus isolated from food contact surfaces. Front. Microbiol. 2022, 13, 1001700. [Google Scholar] [CrossRef] [PubMed]
- Avila-Novoa, M.G.; Iñiguez-Moreno, M.; González-Gómez, J.P.; Zacarías-Castillo, E.; Guerrero-Medina, P.J.; Padilla-Frausto, J.J.; Navarro-Villarruel, C.L.; Gutiérrez-Lomelí, M. Detection of enterotoxin genes of Staphylococcus aureus isolates from food contact surfaces in the dairy industry of Jalisco, Mexico. Biotecnia 2018, 20, 72–78. [Google Scholar] [CrossRef]
- Gan, T.; Shu, G.; Fu, H.; Yan, Q.; Zhang, W.; Tang, H.; Yin, L.; Zhao, L.; Lin, J. Antimicrobial resistance and genotyping of Staphylococcus aureus obtained from food animals in Sichuan Province, China. BMC Vet. Res. 2021, 17, 177. [Google Scholar] [CrossRef] [PubMed]
- Deepak, S.J.; Kannan, P.; Savariraj, W.R.; Ayyasamy, E.; Tuticorin Maragatham Alagesan, S.K.; Ravindran, N.B.; Sundaram, S.; Mohanadasse, N.Q.; Kang, Q.; Cull, C.A.; et al. Characterization of Staphylococcus aureus isolated from milk samples for their virulence, biofilm, and antimicrobial resistance. Sci. Rep. 2024, 14, 25635. [Google Scholar] [CrossRef]
- Shah, M.S.; Qureshi, S.; Kashoo, Z.; Farooq, S.; Wani, S.A.; Hussain, M.I.; Banday, M.S.; Khan, A.A.; Gull, B.; Habib, A.; et al. Methicillin resistance genes and in vitro biofilm formation among Staphylococcus aureus isolates from bovine mastitis in India. Comp. Immunol. Microbiol. Infect. Dis. 2019, 64, 117–124. [Google Scholar] [CrossRef]
- CLSI M100-ED35:2025; Performance Standards for Antimicrobial Susceptibility Testing. 35th ed. CLSI Supplement M100S. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2025.
- Mekhloufi, O.A.; Chieffi, D.; Hammoudi, A.; Bensefia, S.A.; Fanelli, F.; Fusco, V. Prevalence, enterotoxigenic potential and antimicrobial resistance of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA) isolated from Algerian ready to eat foods. Toxins 2021, 13, 835. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Blasco, M.D.; Esteve, C.; Alcaide, E. Multiresistant waterborne pathogens isolated from water reservoirs and cooling systems. J. Appl. Microbiol. 2008, 105, 469–475. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Hemati, M.; Shabanpour, Z.; Dehkordi, S.H.; Bahadoran, S.; Lotalian, S.; Khoibani, S. Effects of benzalkonium chloride on planktonic growth and biofilm formation by animal bacterial pathogens. Jundishapur J. Microbiol. 2015, 8, e16058. [Google Scholar] [CrossRef]
- Avila-Novoa, M.G.; González-Gómez, J.P.; Guerrero-Medina, P.J.; Cardona-López, M.A.; Ibarra-Velazquez, L.M.; Velazquez-Suarez, N.Y.; Morales-del Río, J.A.; Gutiérrez-Lomelí, M. Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) strains isolated from dairy products: Relationship of ica-dependent/independent and components of biofilms produced in vitro. Int. Dairy J. 2021, 119, 105066. [Google Scholar] [CrossRef]
- Borucki, M.K.; Peppin, J.D.; White, D.; Loge, F.; Call, D.R. Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7336–7342. [Google Scholar] [CrossRef] [PubMed]
- Fratesi, S.E.; Lynch, F.L.; Kirkland, B.L.; Brown, L.R. Effects of SEM preparation techniques on the appearance of bacteria and biofilms in the Carter Sandstone. J. Sediment. Res. 2004, 74, 858–867. [Google Scholar] [CrossRef]
- Kou, X.; Cai, H.; Huang, S.; Ni, Y.; Luo, B.; Qian, H.; Ji, H.; Wang, X. Prevalence and Characteristics of Staphylococcus aureus Isolated from Retail Raw Milk in Northern Xinjiang, China. Front. Microbiol. 2021, 12, 705947. [Google Scholar] [CrossRef] [PubMed]
- Eidaroos, N.H.; Algammal, A.M.; Mohamaden, W.I.; Alenzi, A.M.; Alghamdi, S.; Kabrah, A.; El-Mahallawy, H.S.; Eid, H.M.; Algwad, A.A.; Asfor, S.A.; et al. Virulence traits, agr typing, multidrug resistance patterns, and biofilm ability of MDR Staphylococcus aureus recovered from clinical and subclinical mastitis in dairy cows. BMC Microbiol. 2025, 25, 155. [Google Scholar] [CrossRef]
- Caruso, M.; Latorre, L.; Santagada, G.; Fraccalvieri, R.; Miccolupo, A.; Sottili, R.; Palazzo, L.; Parisi, A. Methicillin-resistant Staphylococcus aureus (MRSA) in sheep and goat bulk tank milk from Southern Italy. Small Rumin. Res. 2016, 135, 26–31. [Google Scholar] [CrossRef]
- Chen, C.; Sun, C.; Li, J.; Ji, X.; Wang, Y.; Song, C.; Wang, G. Characterisation of Staphylococcus aureus isolates from bovine mastitis in Ningxia, Western China. J. Glob. Antimicrob. Resist. 2021, 25, 232–237. [Google Scholar] [CrossRef]
- Szczuka, E.; Porada, K.; Wesołowska, M.; Łęska, B. Occurrence and characteristics of Staphylococcus aureus isolated from dairy products. Molecules 2022, 27, 4649. [Google Scholar] [CrossRef]
- Biswas, S.; Islam, M.A.; Islam, J.; Khatun, M.M.; Rahman, M.Z. Detection of multidrug-resistant methicillin-resistant Staphylococcus aureus from healthy black Bengal goat in Bangladesh. J. Infect. Dev. Ctries. 2024, 18, 1891–1898. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Delgado, S.; Vázquez-Sánchez, D.; Martínez, B.; Cabo, M.L.; Rodríguez, A.; Herrera, J.J.; García, P. Incidence of Staphylococcus aureus and analysis of associated bacterial communities on food industry surfaces. Appl. Environ. Microbiol. 2012, 78, 8547–8554. [Google Scholar] [CrossRef]
- Takata, T.; Shirakawa, T.; Ito, J.; Okamoto, A.; Massi, M.N.; Kinoshita, S.; Hatta, M.; Kawabata, M. SCCmec typing and detection of VISA-related genes in methicillin-resistant Staphylococcus aureus clinical strains from Kobe University Hospital, Japan. Southeast Asian J. Trop. Med. Public Health 2006, 37, 1149–1155. [Google Scholar] [PubMed]
- Sağlam, M.; Kılıç, İ.H.; Zer, Y. Investigation of SCCmec types using the real time PCR method in cefoxitin-resistant Staphylococcus aureus isolates. Indian J. Med. Microbiol. 2024, 50, 100649. [Google Scholar] [CrossRef] [PubMed]
- Cuny, C.; Strommenger, B.; Witte, W.; Stanek, C. Clusters of infections in horses with MRSA ST1, ST254, and ST398 in a veterinary hospital. Microb. Drug Resist. 2008, 14, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Otter, J.A.; Havill, N.L.; Boyce, J.M.; French, G.L. Comparison of community-associated methicillin-resistant Staphylococcus aureus from teaching hospitals in London and the USA, 2004–2006: Where is USA300 in the UK? Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 835–839. [Google Scholar] [CrossRef]
- Dastmalchi Saei, H.; Panahi, M. Genotyping and antimicrobial resistance of Staphylococcus aureus isolates from dairy ruminants: Differences in the distribution of clonal types between cattle and small ruminants. Arch. Microbiol. 2020, 202, 115–125. [Google Scholar] [CrossRef]
- Ren, Q.; Liao, G.; Wu, Z.; Lv, J.; Chen, W. Prevalence and characterization of Staphylococcus aureus isolates from subclinical bovine mastitis in southern Xinjiang, China. J. Dairy Sci. 2020, 103, 3368–3380. [Google Scholar] [CrossRef]
- Oniciuc, E.A.; Nicolau, A.I.; Hernández, M.; Rodríguez-Lázaro, D. Presence of methicillin-resistant Staphylococcus aureus in the food chain. Trends Food Sci. Technol. 2017, 61, 49–59. [Google Scholar] [CrossRef]
- Tartor, Y.H.; Enany, M.E.; Ismail, N.I.; El-Demerdash, A.S.; Eidaroos, N.H.; Algendy, R.M.; Mahmmod, Y.; Elsohaby, I. Vancomycin-resistant Staphylococcus aureus endangers Egyptian dairy herds. Sci. Rep. 2024, 14, 30606. [Google Scholar] [CrossRef]
- Wendlandt, S.; Feßler, A.T.; Monecke, S.; Ehricht, R.; Schwarz, S.; Kadlec, K. The diversity of antimicrobial resistance genes among staphylococci of animal origin. Int. J. Med. Microbiol. 2013, 303, 338–349. [Google Scholar] [CrossRef]
- El-Banna, T.E.S.; Sonbol, F.I.; Kamer, A.M.A.; Badr, S.A.M.M. Genetic diversity of macrolides resistant Staphylococcus aureus clinical isolates and the potential synergistic effect of vitamins, C and K3. BMC Microbiol. 2024, 24, 30. [Google Scholar] [CrossRef]
- Page, S.W.; Gautier, P. Use of antimicrobial agents in livestock. Rev. Sci. Tech. 2012, 31, 145–188. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, A.; Sobhani, K.; Mahmoudi, P. A systematic review and meta-analysis revealed a high-level antibiotic resistance of bovine mastitis Staphylococcus aureus in Iran. Res. Vet. Sci. 2023, 161, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Painuli, S.; Semwal, P.; Sharma, R.; Akash, S. Superbugs or multidrug resistant microbes: A new threat to the society. Health Sci. Rep. 2023, 6, e1480. [Google Scholar] [CrossRef] [PubMed]
- Chudobova, D.; Dostalova, S.; Blazkova, I.; Michalek, P.; Ruttkay-Nedecky, B.; Sklenar, M.; Nejdl, L.; Kudr, J.; Gumulec, J.; Tmejova, K.; et al. Effect of ampicillin, streptomycin, penicillin and tetracycline on metal resistant and non-resistant Staphylococcus aureus. Int. J. Environ. Res. Public Health 2014, 11, 3233–3255. [Google Scholar] [CrossRef]
- Aonghusa, C.N.; Gray, N.F. Laundry detergents as a source of heavy metals in Irish domestic wastewater. J. Environ. Sci. Health A Toxic Hazard. Subst. Environ. Eng. 2002, 37, 1–6. [Google Scholar] [CrossRef]
- Eggers, S.; Safdar, N.; Malecki, K.M. Heavy metal exposure and nasal Staphylococcus aureus colonization: Analysis of the National Health and Nutrition Examination Survey (NHANES). Environ. Health 2018, 17, 2. [Google Scholar] [CrossRef]
- Agency for Toxic Substances and Disease Registry (ATSDR). Cadmium ToxGuide. (Listeriosis). Available online: https://www.atsdr.cdc.gov/toxguides/toxguide-5.pdf (accessed on 8 September 2025).
- Bondurant, S.; Mckinney, T.; Bondurant, L.; Fitzpatrick, L. Evaluation of a benzalkonium chloride hand sanitizer in reducing transient Staphylococcus aureus bacterial skin contamination in health care workers. Am. J. Infect. Control 2020, 48, 522–526. [Google Scholar] [CrossRef]
- Chojnacki, M.; Dobrotka, C.; Osborn, R.; Johnson, W.; Young, M.; Meyer, B.; Laskey, E.; Wozniak, R.A.F.; Dewhurst, S.; Dunman, P.M. Evaluating the Antimicrobial Properties of Commercial Hand Sanitizers. mSphere 2021, 6, e00062-21. [Google Scholar] [CrossRef]
- Tahir, S.; Emanuel, S.; Inglis, D.W.; Vickery, K.; Deva, A.K.; Hu, H. Mild Positive Pressure Improves the Efficacy of Benzalkonium Chloride against Staphylococcus aureus Biofilm. Bioengineering 2022, 9, 461. [Google Scholar] [CrossRef]
- Houari, A.; Di Martino, P. Effect of chlorhexidine and benzalkonium chloride on bacterial biofilm formation. Lett. Appl. Microbiol. 2007, 45, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.C.; Hugie, C.N.; Kile, M.L.; Navab-Daneshmand, T. Association between heavy metals and antibiotic-resistant human pathogens in environmental reservoirs: A review. Front. Environ. Sci. Eng. 2019, 13, 46. [Google Scholar] [CrossRef]
- Komijani, M.; Shamabadi, N.S.; Shahin, K.; Eghbalpour, F.; Tahsili, M.R.; Bahram, M. Heavy metal pollution promotes antibiotic resistance potential in the aquatic environment. Environ. Pollut. 2021, 274, 116569. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Wang, C.; Zhao, Q.; Wang, Y.; Huo, M.; Wang, J.; Wang, S. Prevalence and dissemination of antibiotic resistance genes and coselection of heavy metals in Chinese dairy farms. J. Hazard. Mater. 2016, 320, 10–17. [Google Scholar] [CrossRef]
- Azmi, K.; Qrei, W.; Abdeen, Z. Screening of genes encoding adhesion factors and biofilm production in methicillin resistant strains of Staphylococcus aureus isolated from Palestinian patients. BMC Genom. 2019, 20, 578. [Google Scholar] [CrossRef]
- Chmielewski, R.A.N.; Frank, J.F. Biofilm Formation and Control in Food Processing Facilities. Compr. Rev. Food Sci. Food Saf. 2003, 2, 22–32. [Google Scholar] [CrossRef]
- Lorite, G.S.; Rodrigues, C.M.; Souza, A.A.; De Kranz, C.; Mizaikoff, B.; Cotta, M.A. The role of conditioning film formation and surface chemical changes on Xylella fastidiosa adhesion and biofilm evolution. J. Colloid Interface Sci. 2011, 359, 289–295. [Google Scholar] [CrossRef]
- Sugimoto, S.; Sato, F.; Miyakawa, R.; Chiba, A.; Onodera, S.; Hori, S.; Mizunoe, Y. Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and -sensitive strains of Staphylococcus aureus. Sci. Rep. 2018, 8, 2254. [Google Scholar] [CrossRef]
| Antimicrobial Class According to the WHO | Antibiotic | No. (%) of MRSA (n = 28) and MSSA (n = 39) Strains | ||||||
|---|---|---|---|---|---|---|---|---|
| Resistant | Susceptible | Intermediate | ||||||
| MRSA | MSSA | MRSA | MSSA | MRSA | MSSA | |||
| Highly important | Lincosamides | CLM | 19 (28.3) | 25 (37.3) | 9 (13.4) | 14 (20.8) | - | - |
| Sulfonamides | STX | 5 (7.4) | 7 (10.4) | 23 (34.3) | 32 (47.7) | - | - | |
| Cyclic peptides | TE | 8 (11.9) | 3 (4.4) | 20 (29.8) | 36 (53.7) | - | - | |
| Critically important | Macrolides | E | 5 (7.4) | 4 (5.9) | 23 (34.3) | 35 (52.2) | - | - |
| Aminoglycosides | GE | 6 (8.9) | 2 (2.9) | 22 (32.8) | 37 (55.2) | - | - | |
| Fluoroquinolones | CPF | 11 (11.9) | 22 (32.8) | 12 (17.9) | 10 (14.9) | 5 (7.4) | 7 (10.4) | |
| β-Lactams | PE | 28 (41.7) | 39 (58.2) | - | - | - | - | |
| Glycopeptides | VA * | - | 5 (7.4) | 28 (41.7) | 34 (50.7) | - | - | |
| Strain No. | SCCmec | Antimicrobial Resistance Genes | Antibiotic Resistance | MAR Index | |||||
|---|---|---|---|---|---|---|---|---|---|
| ermB | ermC | ermA | tetM | blaZ | aacA-aphD | ||||
| MSRA-2 | SCCmec II | - | - | - | - | + | + | PE + GE | 0.25 |
| MSRA-6 | SCCmec II | - | - | - | - | + | - | PE + CLM | 0.25 |
| MSRA-7 | SCCmec II | + | + | - | + | + | - | PE + CPF + CLM + E + TE | 0.62 |
| MSRA-8 | SCCmec II | - | - | - | + | + | - | PE + CLM + TE | 0.37 |
| MSRA-9 | SCCmec II | - | - | - | - | + | - | PE + CLM | 0.25 |
| MSRA-10 | SCCmec II | - | - | - | - | + | - | PE | 0.12 |
| MSRA-11 | SCCmec II | - | - | - | - | + | - | PE | 0.12 |
| MSRA-23 | SCCmec II | + | - | - | + | + | + | PE + CPF + CLM + E + TE + GE | 0.75 |
| MSRA-27 | SCCmec II | + | - | - | + | + | - | PE + CPF + CLM + E + TE + GE | 0.75 |
| MSRA-28 | SCCmec II | - | - | - | - | + | - | PE | 0.12 |
| MSRA-37 | SCCmec II | - | - | - | + | + | - | PE + CLM + TE | 0.37 |
| MSRA-40 | SCCmec II | - | - | - | - | - | - | PE + CLM | 0.25 |
| MSRA-43 | SCCmec II | - | - | - | + | + | - | PE + CLM + TE + STX | 0.50 |
| MSRA-44 | SCCmec II | - | - | - | - | + | - | PE + CLM | 0.25 |
| MSRA-3 | SCCmec IV | - | - | + | - | + | + | PE + CPF + CLM + E + GE | 0.62 |
| MSRA-13 | SCCmec IV | + | - | - | - | - | + | PE + CLM + E + GE | 0.50 |
| MSRA-17 | SCCmec IV | - | - | - | - | + | - | PE + CPF + CLM | 0.37 |
| MSRA-21 | SCCmec IV | - | - | - | + | + | - | PE + CPF + CLM + TE + STX | 0.62 |
| MSRA-56 | SCCmec IV | - | - | - | + | - | PE | 0.12 | |
| MSRA-4 | SCCmec V | - | - | - | + | + | - | PE + TE | 0.25 |
| MSRA-5 | SCCmec V | - | - | - | - | + | - | PE + CPF + CLM + STX | 0.50 |
| MSRA-12 | SCCmec V | - | - | - | + | + | - | PE + CPF + CLM + TE + STX | 0.62 |
| MSRA-15 | SCCmec V | - | - | - | - | + | - | PE + CLM + STX | 0.37 |
| MSRA-18 | SCCmec V | - | - | - | - | - | - | PE + CPF | 0.25 |
| MSRA-24 | SCCmec V | - | - | - | - | + | - | PE | 0.12 |
| MSRA-34 | SCCmec V | - | - | - | - | + | + | PE + GE | 0.25 |
| MSRA-39 | SCCmec V | - | + | - | - | + | - | PE + CPF + CLM + E | 0.50 |
| MSRA-62 | SCCmec V | - | - | - | - | - | - | PE + CPF + CLM | 0.37 |
| No. of Antimicrobial | Resistance Profile | Staphylococcus aureus | MAR Index | |
|---|---|---|---|---|
| No. (%) of MSSA (n = 39) | No. (%) of MRSA (n = 28) | |||
| 6 | PE + CPF + CLM + E + TE + GE | - | 2 (2.98) | 0.75 |
| PE + CPF + CLM + VA + GE + STX | 1 (1.49) | - | ||
| PE + CPF + CLM + E + TE + STX | 1 (1.49) | - | ||
| 5 | PE + CPF + CLM + TE + STX | - | 2 (2.98) | 0.62 |
| PE + CPF + CLM + E + TE | - | 1 (1.49) | ||
| PE + CPF + CLM + E + GE | - | 1 (1.49) | ||
| 4 | PE + CLM + TE + STX | - | 1 (1.49) | 0.50 |
| PE + CLM + E + GE | - | 1 (1.49) | ||
| PE + CPF + CLM + STX | - | 1 (1.49) | ||
| PE + CPF + CLM + E | - | 1 (1.49) | ||
| PE + CPF + CLM + STX | 3 (4.47) | - | ||
| PE + CPF + CLM + VA | 1 (1.49) | - | ||
| PE + CPF + CLM + TE | 1 (1.49) | - | ||
| PE + CLM + E + VA | 1 (1.49) | - | ||
| PE + CLM + VA + STX | 1 (1.49) | - | ||
| 3 | PE + CPF + CLM | 9 (13.43) | 2 (2.98) | 0.37 |
| PE + CLM + TE | - | 2 (2.98) | ||
| PE + CLM + STX | - | 1 (1.49) | ||
| PE + E + TE | 1 (1.49) | - | ||
| PE + CLM + VA | 1 (1.49) | - | ||
| 2 | PE + TE | - | 1 (1.49) | 0.25 |
| PE + GE | 1 (1.49) * | 2 (2.98) | ||
| PE + CPF | 6 (8.95) * | 1 (1.49) | ||
| PE + CLM | 6 (8.95) * | 4 (5.97) | ||
| PE + STX | 1 (1.49) * | - | ||
| PE + E | 1 (1.49) * | - | ||
| 1 | PE | 1 (1.49) * | 3 (4.47) | 0.12 |
| PE | 2 (2.98) * | 2 (2.98) | ||
| PE | 1 (1.49) * | - | ||
| µg/mL | No. (%) of MRSA (n = 28) | No. (%) of MSSA (n = 39) | Total | |
|---|---|---|---|---|
| BC (MIC) | 50 | 2 (2.9) | - | 2 (2.9) |
| 25 | 8 (11.9) | 12 (17.9) | 20 (29.8) | |
| 12.5 | 2 (2.9) | 2 (2.9) | 4 (5.9) | |
| 6.25 | 6 (8.9) | 1 (1.4) | 7 (10.4) | |
| 3.12 | 2 (2.9) | 7 (10.4) | 9 (13.4) | |
| 1.56 | 8 (11.9) | 17 (25.3) | 25 (37.3) | |
| ClCd2 | Higher 400 | 23 (34.3) | 19 (28.3) | 42 (62.6) |
| 200 | - | 1 (1.4) | 1 (1.4) | |
| 100 | - | 1 (1.4) | 1 (1.4) | |
| 70 | 1 (1.4) | 7 (10.4) | 8 (11.9) | |
| 50 | 2 (2.9) | 3 (4.4) | 5 (7.4) | |
| 25 | 2 (2.9) | 8 (11.9) | 10 (14.9) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avila-Novoa, M.G.; Solis-Velazquez, O.A.; González-Gómez, J.P.; Guerrero-Medina, P.J.; Gutiérrez-Lomelí, M. Genetic Profiling of MRSA and MSSA from Food Contact Surfaces: Antibiotic, Heavy Metal and Benzalkonium Chloride Resistance. Life 2025, 15, 1811. https://doi.org/10.3390/life15121811
Avila-Novoa MG, Solis-Velazquez OA, González-Gómez JP, Guerrero-Medina PJ, Gutiérrez-Lomelí M. Genetic Profiling of MRSA and MSSA from Food Contact Surfaces: Antibiotic, Heavy Metal and Benzalkonium Chloride Resistance. Life. 2025; 15(12):1811. https://doi.org/10.3390/life15121811
Chicago/Turabian StyleAvila-Novoa, María Guadalupe, Oscar Alberto Solis-Velazquez, Jean Pierre González-Gómez, Pedro Javier Guerrero-Medina, and Melesio Gutiérrez-Lomelí. 2025. "Genetic Profiling of MRSA and MSSA from Food Contact Surfaces: Antibiotic, Heavy Metal and Benzalkonium Chloride Resistance" Life 15, no. 12: 1811. https://doi.org/10.3390/life15121811
APA StyleAvila-Novoa, M. G., Solis-Velazquez, O. A., González-Gómez, J. P., Guerrero-Medina, P. J., & Gutiérrez-Lomelí, M. (2025). Genetic Profiling of MRSA and MSSA from Food Contact Surfaces: Antibiotic, Heavy Metal and Benzalkonium Chloride Resistance. Life, 15(12), 1811. https://doi.org/10.3390/life15121811








