Bridging Breeds: Transcriptomic Insights into Immune Traits of Yili, Thoroughbred, and Kazakh Horses
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals Used in Study, Study Locations, Whole Blood Collection and Storage
2.2. Measurement of Blood Physiological and Biochemical Parameters
2.3. Blood Immune Index Measurement
2.4. Statistical Analysis
2.5. RNA Extraction, Library Construction, and Quality Control
2.6. Sequencing Data Quality Control and Alignment
2.7. Differential Gene Analysis
2.8. Clustering and Enrichment Analysis of Differential Genes
2.9. Real-Time Quantitative PCR (qRT-PCR) Validation
2.10. Correlation Analysis
3. Results
3.1. Blood Physiological and Biochemical Parameters
3.2. Immune Indicators
3.3. RNA Extraction and Quality Control
3.4. Mapping Reads to the Reference Genome
3.5. Analysis of DEGs Among Thoroughbred, Yili, and Kazakh Horses
3.6. Gene Ontology (GO) Enrichment Analysis of Differentially Expressed Genes
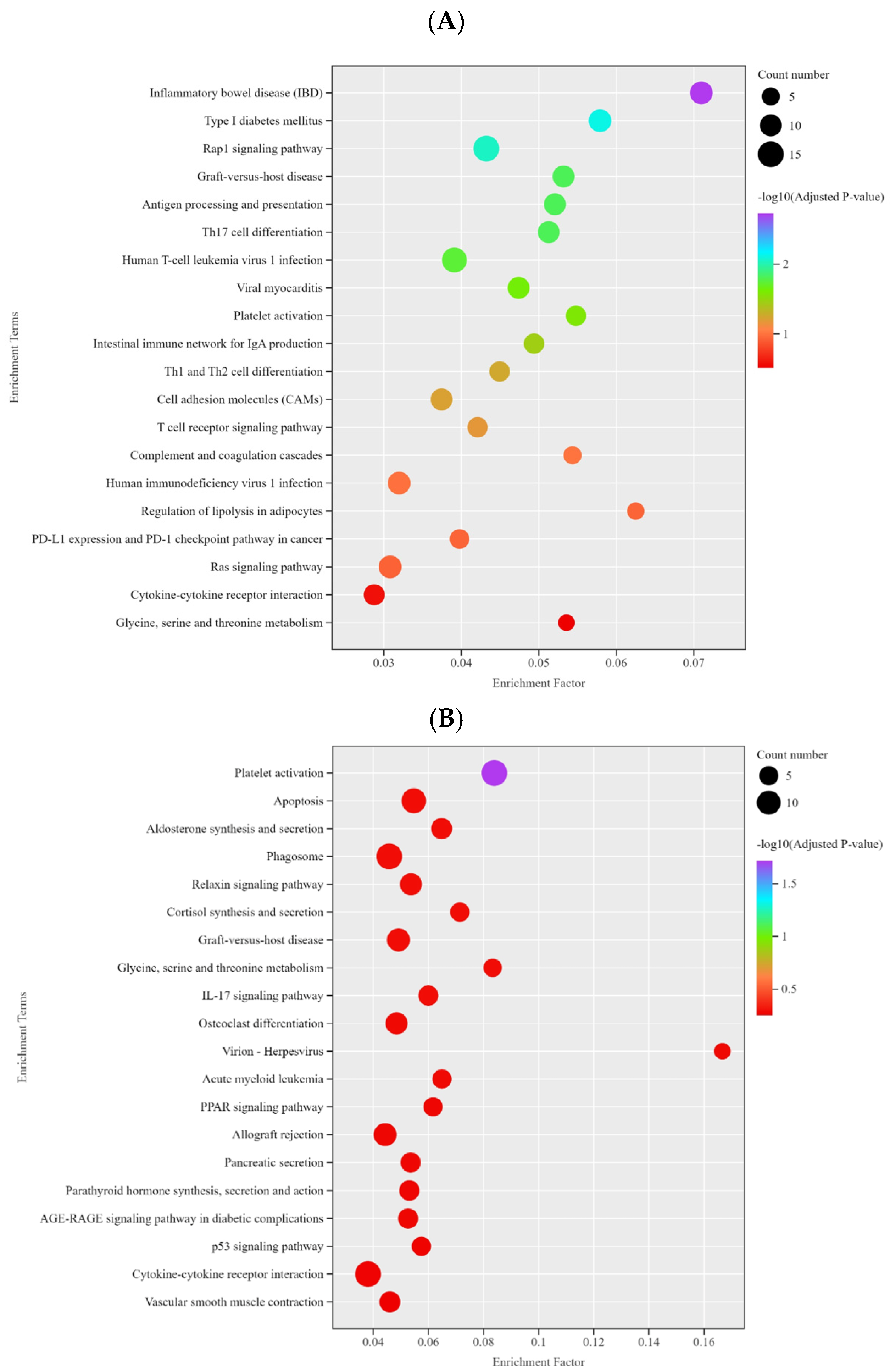
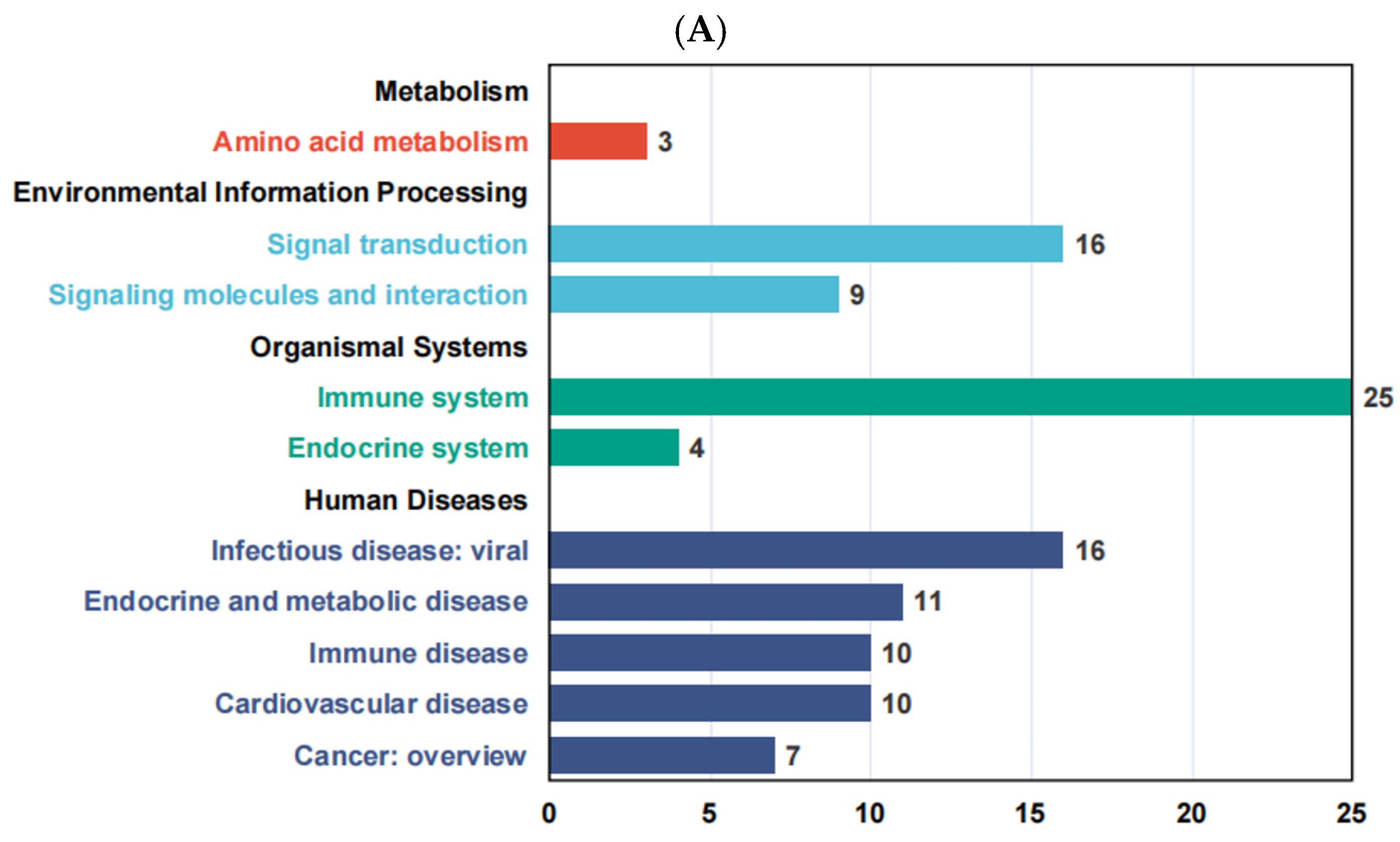
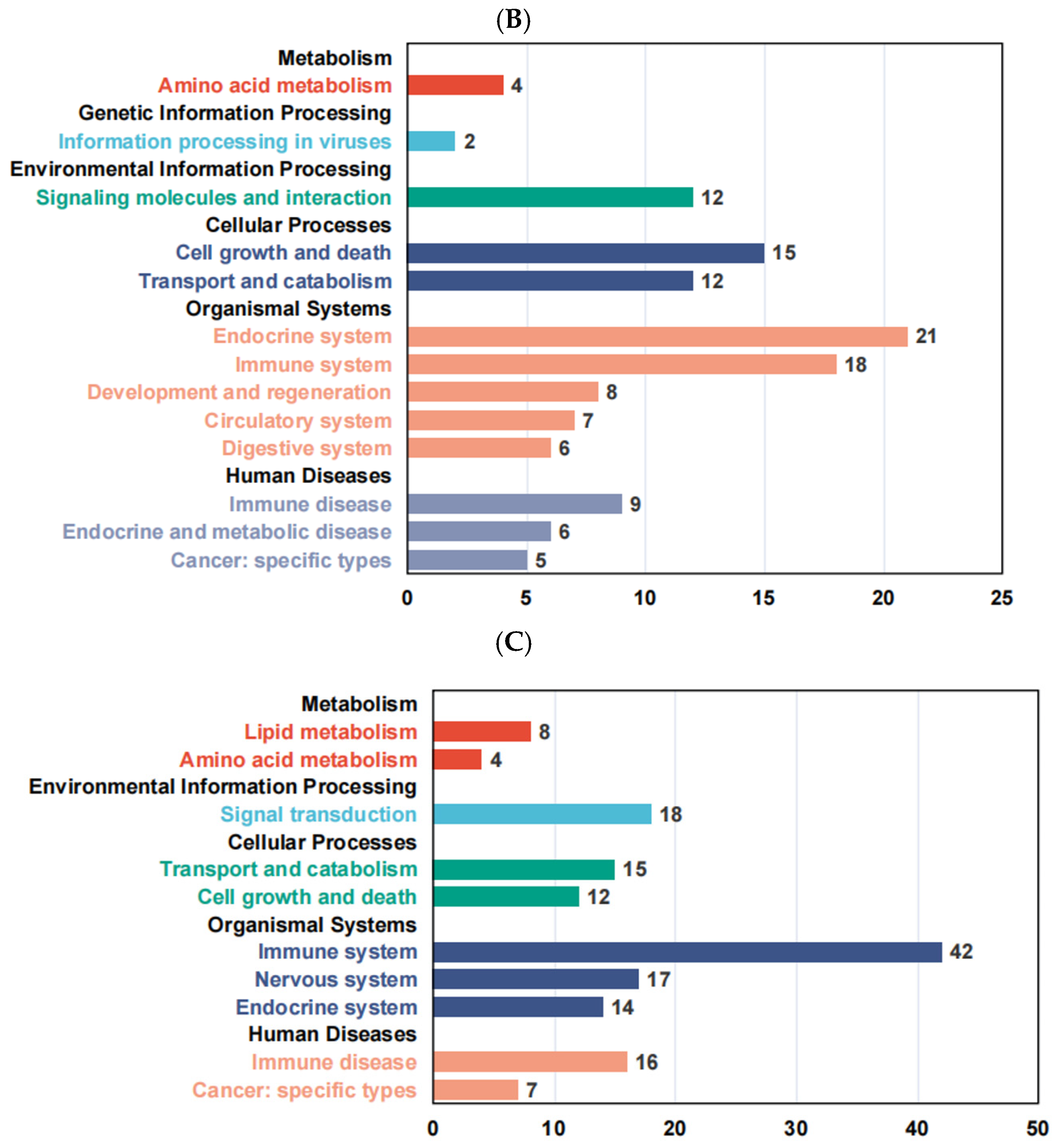
3.7. KEGG Enrichment Analysis of Differentially Expressed Genes
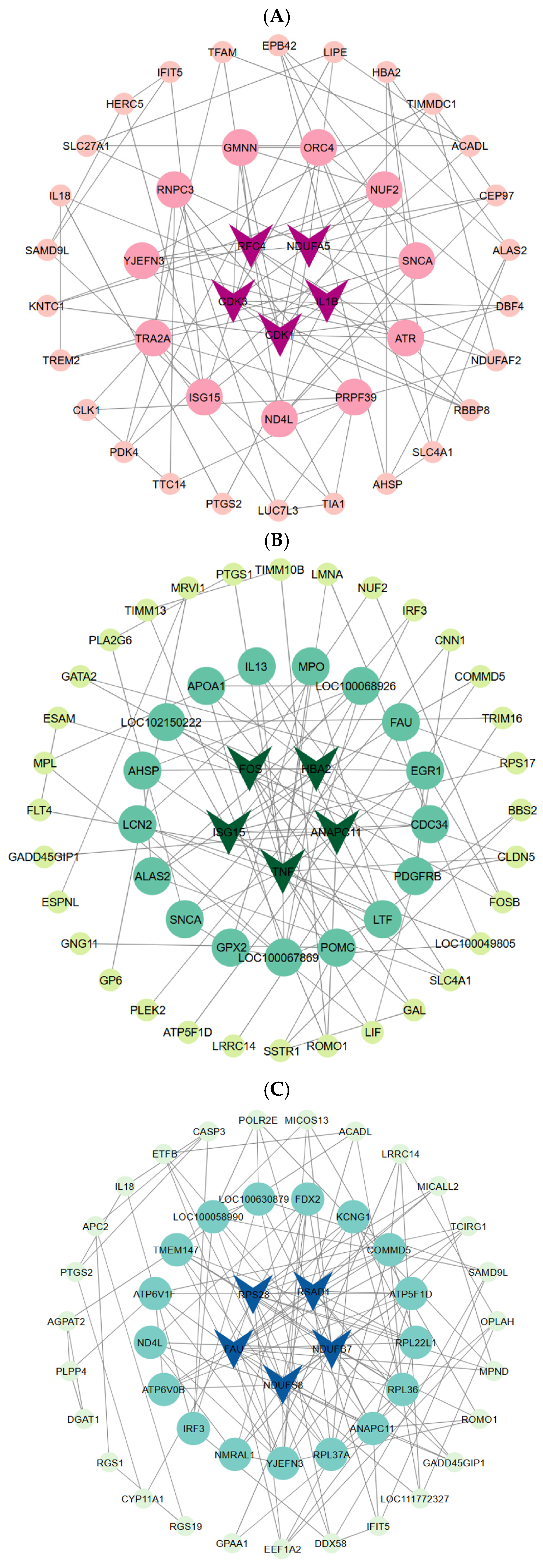
3.8. Protein–Protein Interaction (PPI) Analysis
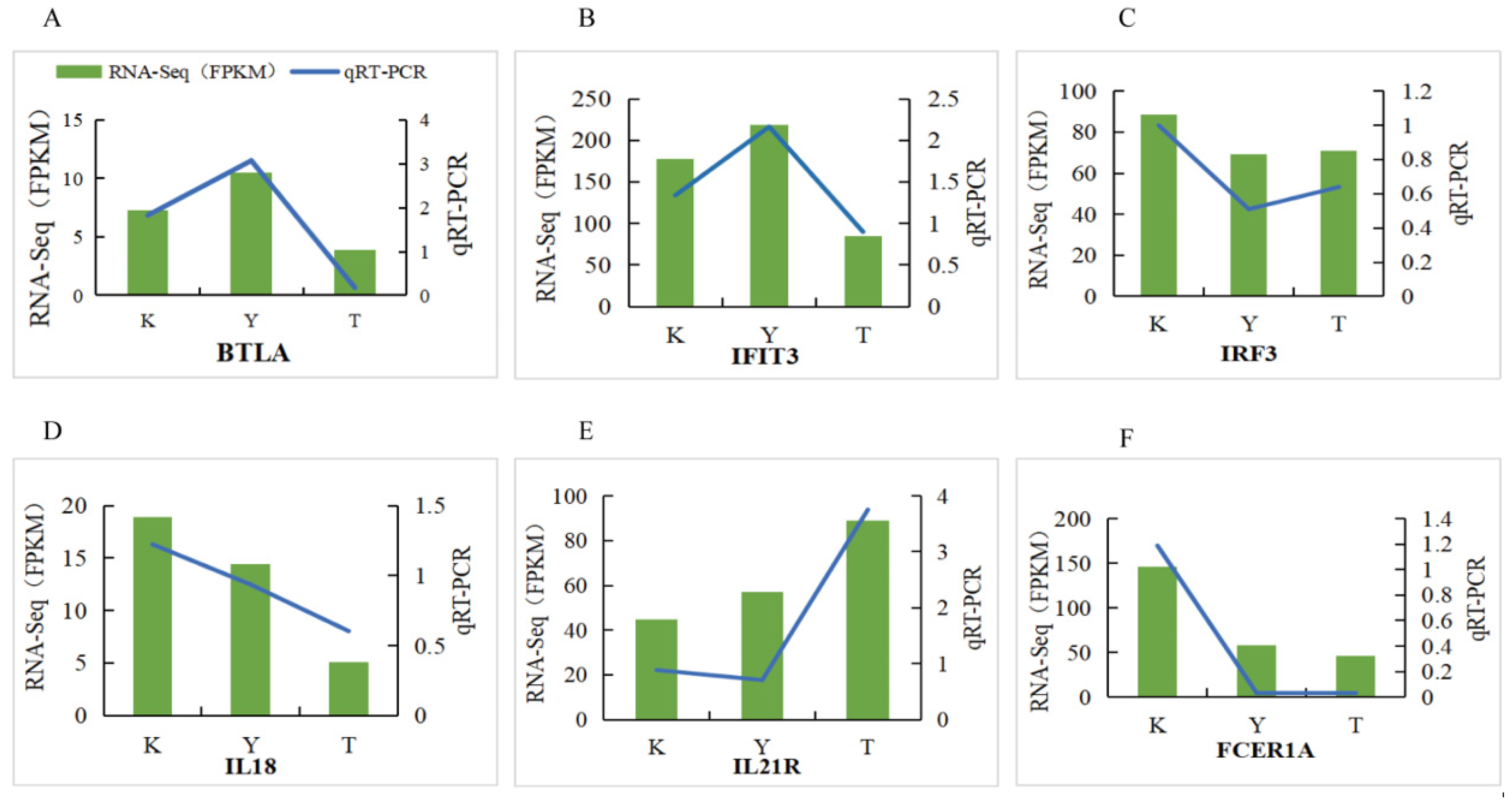
3.9. RT-qPCR Validation
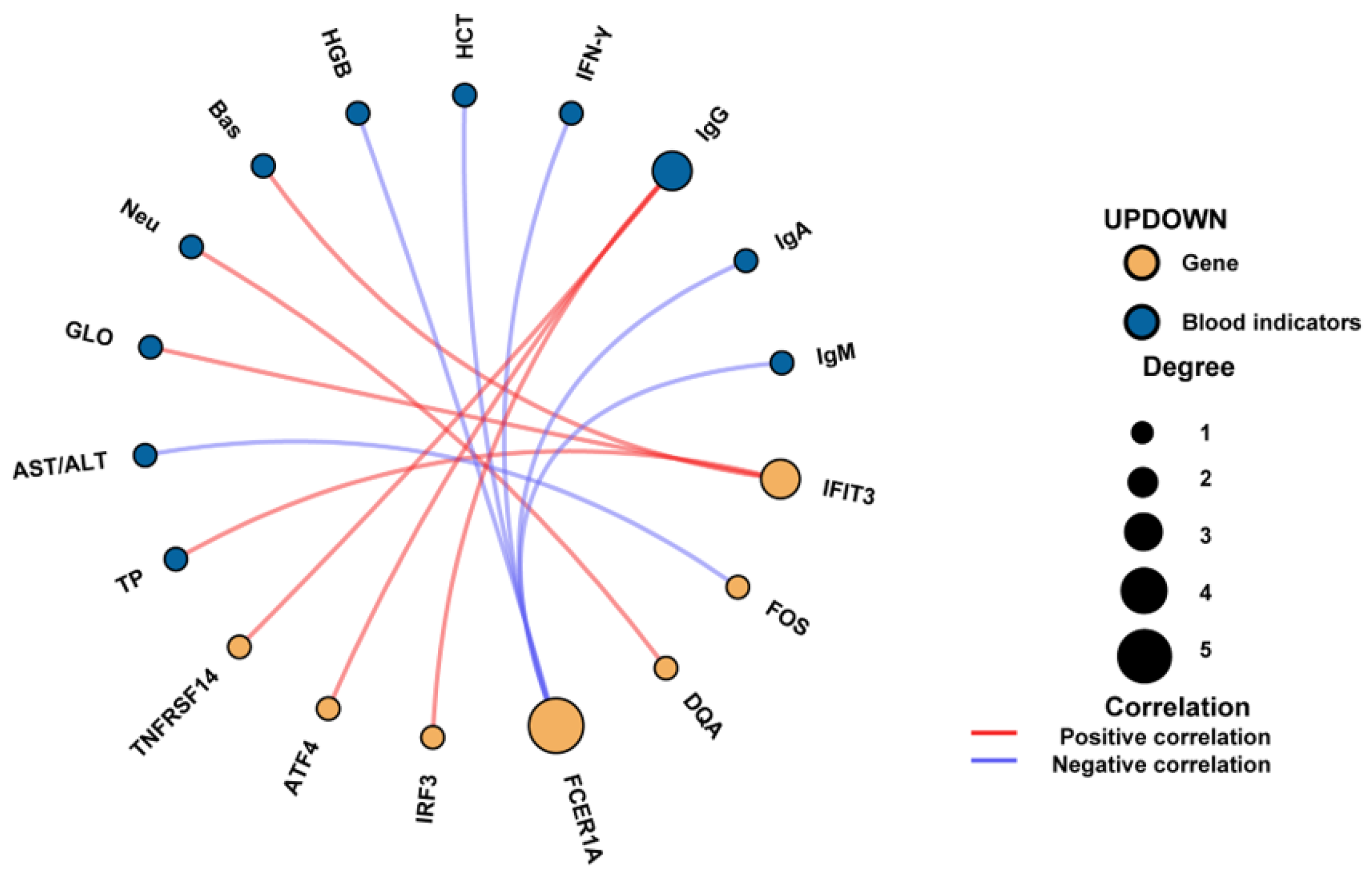
3.10. Correlation Analysis
4. Discussion
- 1.
- Scarcity and access restrictions of specific breeds:
- 2.
- Strict inclusion criteria and resource limitations:
- 3.
- Context-specific geographic and historical focus:
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Librado, P.; Fages, A.; Gaunitz, C.; Leonardi, M.; Wagner, S.; Khan, N.; Hanghøj, K.; Alquraishi, S.A.; Alfarhan, A.H.; Al-Rasheid, K.A.; et al. The Evolutionary Origin and Genetic Makeup of Domestic Horses. Genetics 2016, 204, 423–434. [Google Scholar] [CrossRef]
- Robert, L.; Fulop, T. Longevity and its regulation: Centenarians and beyond. Aging 2014, 39, 198–211. [Google Scholar]
- Farhud, D.D. Hypothetical Strategies of Gene and Environmental Influence on Life Expectancy: A Brief Review. Iran. J. Public Health 2022, 51, 2382. [Google Scholar] [CrossRef] [PubMed]
- Klecel, W.; Martyniuk, E. From the Eurasian steppes to the Roman circuses: A review of early development of horse breeding and management. Animals 2021, 11, 1859. [Google Scholar] [CrossRef]
- Dronca, D. Attempts for Optimization the Genetic Improvement Actions in Horse Populations of Nonius Variety from the Izvin Stud, Timiș County; CABI: Oxford, UK, 2007. [Google Scholar]
- Ramzan, P.; Palmer, L.; Dallas, R.; Shepherd, M. Subclinical ultrasonographic abnormalities of the suspensory ligament branch of the athletic horse: A survey of 60 Thoroughbred racehorses. Equine Vet. J. 2013, 45, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Dulati, K.; Aidierhan, S.; Gemingguli, M. Characteristics of Genetic Resources of Kazakh Horse. Agric. Biotechnol. (2164-4993) 2018, 7, 88–91. [Google Scholar]
- Liu, L.-L.; Fang, C.; Meng, J.; Detilleux, J.; Liu, W.-J.; Yao, X.-K. Genome-wide analysis reveals signatures of selection for gait traits in Yili horse. bioRxiv 2018, 471797. [Google Scholar] [CrossRef]
- Abbas, A.; Lichtman, A.; Pober, J. Cellular and Molecular Immunology; Peking University Medica: Beijing, China, 1991. [Google Scholar]
- Marcoux, G.; Laroche, A.; Espinoza Romero, J.; Boilard, E. Role of platelets and megakaryocytes in adaptive immunity. Platelets 2021, 32, 340–351. [Google Scholar] [CrossRef]
- Ye, X.; Zhu, Y.; Wang, Z. Evaluation of the Detection Value of T Lymphocytes and Immunoglobulins in Leukemia Patients. Contemp. Med. 2021, 27, 24–27. [Google Scholar]
- Ono, T.; Yamada, Y.; Hata, A.; Shimokawa Miyama, T.; Shibano, K.; Iwata, E.; Ohzawa, E.; Kitagawa, H. Reference values of hematological and blood biochemical parameters for the Noma horse. J. Equine Sci. 2019, 30, 69–73. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Z.; Sun, F.; Cao, Y.; Zhang, Q.; Ding, D.; Jin, X.; Jin, H.; Zhang, S. Comparative analysis of blood routine and immunological indicators among Changbai Mountain wild hybrid pigs, Min pigs, and Large White pigs. Chin. J. Vet. Med. 2018, 38, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Study on the Expression of Immune-Related Genes and Spleen Expression Profiling in Mongolian Horses. Ph.D. Thesis, Inner Mongolia Agricultural University, Hohhot, China, 2013. [Google Scholar]
- Mach, N.; Gao, Y.; Lemonnier, G.; Lecardonnel, J.; Oswald, I.P.; Estellé, J.; Rogel-Gaillard, C. The peripheral blood transcriptome reflects variations in immunity traits in swine: Towards the identification of biomarkers. BMC Genom. 2013, 14, 894. [Google Scholar] [CrossRef]
- Khodadoust, M.S.; Olsson, N.; Wagar, L.E.; Haabeth, O.A.; Chen, B.; Swaminathan, K.; Rawson, K.; Liu, C.L.; Steiner, D.; Lund, P. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature 2017, 543, 723–727. [Google Scholar] [CrossRef]
- Anthoney, N.; Foldi, I.; Hidalgo, A. Toll and Toll-like receptor signalling in development. Development 2018, 145, dev156018. [Google Scholar] [CrossRef]
- Zhang, X.; Li, X.; Hua, R.; Fang, Y.; Yue, T.; Li, J.; Lu, Y.; Yue, W.; Gao, Z.; Liu, S.; et al. Identification of stable reference genes for qRT-PCR in Stropharia rugosoannulata using mRNA-sequencing data. PLoS ONE 2025, 20, e0323272. [Google Scholar] [CrossRef]
- Gaunitz, C.; Fages, A.; Hanghøj, K.; Albrechtsen, A.; Khan, N.; Schubert, M.; Seguin-Orlando, A.; Owens, I.J.; Felkel, S.; Bignon-Lau, O. Ancient genomes revisit the ancestry of domestic and Przewalski’s horses. Science 2018, 360, 111–114. [Google Scholar] [CrossRef]
- Orlando, L. The evolutionary and historical foundation of the modern horse: Lessons from ancient genomics. Annu. Rev. Genet. 2020, 54, 563–581. [Google Scholar] [CrossRef]
- Banday, M.T.; Wani, M.A.; Othman, S.I.; Rudayni, H.A.; Allam, A.A.; Alshahrani, M.Y.; Ibrahim, E.H.; Nabi, S.; Adil, S. Impact of Rumex nepalensis on Performance, Blood Markers, Immunity, Intestinal Microbiology and Histomorphology in Broiler Chicken. Vet. Sci. 2024, 11, 463. [Google Scholar] [CrossRef]
- Li, Y.; Han, G. Horse Veterinary Handbook; China Agricultural Press: Beijing, China, 2016. [Google Scholar]
- Frank, G.; Tim, J.; Mark, H. Equine Veterinary Diagnostic Techniques; China Agricultural Press: Beijing, China, 2018. [Google Scholar]
- Slanzon, G.S.; Toledo, A.F.; Silva, A.P.; Coelho, M.G.; da Silva, M.D.; Cezar, A.M.; Bittar, C.M.M. Red propolis as an additive for preweaned dairy calves: Effect on growth performance, health, and selected blood parameters. J. Dairy Sci. 2019, 102, 8952–8962. [Google Scholar] [CrossRef] [PubMed]
- Toğaçar, M.; Ergen, B.; Cömert, Z. Classification of white blood cells using deep features obtained from Convolutional Neural Network models based on the combination of feature selection methods. Appl. Soft Comput. 2020, 97, 106810. [Google Scholar] [CrossRef]
- Underhill, D.M.; Ozinsky, A. Phagocytosis of microbes: Complexity in action. Annu. Rev. Immunol. 2002, 20, 825–852. [Google Scholar] [CrossRef]
- Clapperton, M.; Bishop, S.C.; Glass, E.J. Innate immune traits differ between Meishan and Large White pigs. Vet. Immunol. Immunopathol. 2005, 104, 131–144. [Google Scholar] [CrossRef]
- Renoux, C.; Faivre, M.; Bessaa, A.; Da Costa, L.; Joly, P.; Gauthier, A.; Connes, P. Impact of surface-area-to-volume ratio, internal viscosity and membrane viscoelasticity on red blood cell deformability measured in isotonic condition. Sci. Rep. 2019, 9, 6771. [Google Scholar] [CrossRef] [PubMed]
- Khummuang, S.; Lee, H.G.; Joo, S.S.; Park, J.W.; Choi, J.Y.; Oh, J.H.; Kim, K.H.; Youn, H.H.; Kim, M.; Cho, B.W. Comparison for immunophysiological responses of Jeju and Thoroughbred horses after exercise. Asian-Australas J. Anim. Sci. 2020, 33, 424–435. [Google Scholar] [CrossRef]
- Tan, X.; Li, H.; Deng, H.; Dong, H. Discussion on Yili horse improvement and specialization in new meat horse breeds. Grass-Fed Livest. 2014, 3, 7–10. [Google Scholar]
- Chebbo, M.; Assou, S.; Pantesco, V.; Duez, C.; Alessi, M.C.; Chanez, P.; Gras, D. Platelets Purification Is a Crucial Step for Transcriptomic Analysis. Int. J. Mol. Sci. 2022, 23, 3100. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.J.C.; Morgan, P.K.; Man, K.; Lancaster, G.I.; Murphy, A.J. The influence of metabolic disorders on adaptive immunity. Cell. Mol. Immunol. 2024, 21, 1109–1119. [Google Scholar] [CrossRef] [PubMed]
- Ivy, J.L. Role of carbohydrate in physical activity. Clin. Sports Med. 1999, 18, 469–484. [Google Scholar] [CrossRef]
- Durham, A.E.; Frank, N.; McGowan, C.M.; Menzies-Gow, N.J.; Roelfsema, E.; Vervuert, I.; Feige, K.; Fey, K. ECEIM consensus statement on equine metabolic syndrome. J. Vet. Intern. Med. 2019, 33, 335–349. [Google Scholar] [CrossRef]
- Horton, R.E.; Vidarsson, G. Antibodies and their receptors: Different potential roles in mucosal defense. Front. Immunol. 2013, 4, 200. [Google Scholar] [CrossRef] [PubMed]
- Pahud, J.J.; Mach, J.P. Equine secretory IgA and secretory component. Int. Arch. Allergy Appl. Immunol. 1972, 42, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Blunden, A.S.; Mackintosh, M.E. The microflora of the lower respiratory tract of the horse: An autopsy study. Br. Vet. J. 1991, 147, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Casanova, J.-L.; MacMicking, J.D.; Nathan, C.F. Interferon-γ and infectious diseases: Lessons and prospects. Science 2024, 384, eadl2016. [Google Scholar] [PubMed]
- Wang, Y.; Chu, H. The effect of moxibustion on the expression of TNF-α in the hippocampus and colon tissue of rats with an IBS-D model. Shanghai J. Acupunct. Moxibustion 2020, 39, 1449–1459. [Google Scholar]
- Liu, Y.; Mai, Z.; Zhou, Y.; Cui, Y.; Zhang, C.; Zhang, T.; Lu, Y.; Kuang, L.; Wu Muerbek, B.; Liu, J. Changes in TNF-α and IL-6 content in different breeds of horses in three regions of Xinjiang. Heilongjiang Anim. Husb. Vet. Med. 2016, 12, 91–96. [Google Scholar]
- Zhao, X.; Zhu, L.; Yin, Q.; Xu, Z.; Jia, Q.; Yang, R.; He, K. F2RL3 methylation in the peripheral blood as a potential marker for the detection of coronary heart disease: A case-control study. Front. Genet. 2022, 13, 833923. [Google Scholar] [CrossRef]
- Cheng, M.; Luo, J.; Duan, Y.; Yang, Y.; Shi, C.; Sun, Y.; Lu, Y.; Wang, J.; Li, X.; Wang, J. African swine fever virus MGF505-3R inhibits cGAS-STING-mediated IFN-β pathway activation by degrading TBK1. Anim. Dis. 2022, 2, 13. [Google Scholar] [CrossRef]
- Chikhalya, A.; Dittmann, M.; Zheng, Y.; Sohn, S.-Y.; Rice, C.M.; Hearing, P. Human IFIT3 protein induces interferon signaling and inhibits adenovirus immediate early gene expression. Mbio 2021, 12, e02829-21. [Google Scholar]
- Tidball, J.G.; Villalta, S.A. Regulatory interactions between muscle and the immune system during muscle regeneration. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2010, 298, R1173–R1187. [Google Scholar] [CrossRef]
- Bosurgi, L.; Manfredi, A.A.; Rovere-Querini, P. Macrophages in injured skeletal muscle: A perpetuum mobile causing and limiting fibrosis, prompting or restricting resolution and regeneration. Front. Immunol. 2011, 2, 62. [Google Scholar] [CrossRef]
- Campbell, A.L.; Smith, N.C.; Reilly, J.H.; Kerr, S.C.; Leach, W.J.; Fazzi, U.G.; Rooney, B.P.; Murrell, G.A.; Millar, N.L. IL-21 receptor expression in human tendinopathy. Mediat. Inflamm. 2014, 2014, 481206. [Google Scholar] [CrossRef]
- Bradley, P.; Thomas, P.G. Using T cell receptor repertoires to understand the principles of adaptive immune recognition. Annu. Rev. Immunol. 2019, 37, 547–570. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Q.; Mi, Z.; Liu, H.; Zhang, F. Adverse reactions of long-term use of traditional immunosuppressants in the treatment of skin diseases. China J. Lepr. Ski. Dis. 2024, 40, 311–316. [Google Scholar]
- Li, H.; Li, Y.; Luo, S.; Zhang, Y.; Feng, Z.; Li, S. The roles and mechanisms of the NF-κB signaling pathway in tendon disorders. Front. Vet. Sci. 2024, 11, 1382239. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, M.W.; Cheung, T.C.; Ware, C.F. The signaling networks of the herpesvirus entry mediator (TNFRSF14) in immune regulation. Immunol. Rev. 2011, 244, 169–187. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, I.; Suryawanshi, A.; Hong, Y.; Ranganathan, P.; Shanmugam, A.; Ahmad, S.; Swafford, D.; Manicassamy, B.; Ramesh, G.; Koni, P.A. Homeostatic PPARα signaling limits inflammatory responses to commensal microbiota in the intestine. J. Immunol. 2016, 196, 4739–4749. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Luo, S.; Zhan, Y.; Lu, Q. The roles of PPARγ and its agonists in autoimmune diseases: A comprehensive review. J. Autoimmun. 2020, 113, 102510. [Google Scholar] [CrossRef] [PubMed]



| Item | Content (%) |
|---|---|
| Ingredients | |
| Corn | 17.28 |
| Wheat bran | 5.26 |
| Soybean meal | 9.26 |
| Dicalcium phosphate | 1.15 |
| Salt | 0.63 |
| Premix | 0.31 |
| Methionine | 0.19 |
| Dried hay | 65.92 |
| Total | 100 |
| Nutrient levels | |
| Dry matter | 95.31 |
| Crude protein | 12.69 |
| Crude fat | 1.77 |
| Neutral detergent fiber (NDF) | 42.23 |
| Acid detergent fiber (ADF) | 37.98 |
| Crude ash | 8.16 |
| Calcium (Ca) | 0.86 |
| Phosphorus (P) | 0.41 |
| Digestible energy (MJ/kg) | 9.03 |
| Label | Gene | Primer Sequences | Product Size (bp) | Annealing Temp (°C) |
|---|---|---|---|---|
| A | BTLA | TCAGCACCATTGTCTGAGCC | 86 | 62.46 |
| AGCACCAACATGAACCTCCT | ||||
| B | IFIT3 | TGGCCATCGCGACATACTG | 118 | 59.9 |
| AGGGCAAGGAGAACTTTGACA | ||||
| C | IRF3 | AACCGGAAGGAAGTGTTGCG | 87 | 59.71 |
| CGCTGGGGTCAGAAACTCAT | ||||
| D | IL18 | GAGCAGTGAGGAGTTCCGTT | 184 | 60.32 |
| CCAGCGGCCATCTTTATTCC | ||||
| E | IL21R | CGTGACCTTCTCGGGACTTT | 104 | 59.9 |
| CTCCCCGCATCTTGTACTGC | ||||
| F | FCER1A | GGATTGGGACGTCTTCAAGGT | 125 | 58.82 |
| CCTCGCAGTAATAGGTGCCA | ||||
| GAPDH | CTAAGTCGTCTCTGCTGAG | 113 | 59.81 |
| Index | Kazakh Horses | Thoroughbreds | Yili Horses |
|---|---|---|---|
| WBC (109/L) | 9.22 ± 0.47 | 9.69 ± 1.34 | 10.48 ± 0.68 |
| Neu # (109/L) | 4.01 ± 0.32 b | 5.80 ± 1.24 a | 5.44 ± 0.16 a |
| Lym # (109/L) | 4.23 ± 0.15 | 3.28 ± 0.39 | 4.26 ± 1.00 |
| Mon # (109/L) | 0.34 ± 0.06 a | 0.18 ± 0.01 b | 0.19 ± 0.05 b |
| Eos # (109/L) | 0.52 ± 0.09 | 0.38 ± 0.20 | 0.58 ± 0.17 |
| Bas # (109/L) | 0.08 ± 0.03 | 0.04 ± 0.02 | 0.08 ± 0.02 |
| RBC (1012/L) | 6.35 ± 1.03 b | 8.86 ± 0.87 a | 8.40 ± 1.22 ab |
| HGB (g/L) | 107.33 ± 15.18 b | 152.00 ± 13.00 a | 143.33 ± 18.15 a |
| HCT (%) | 30.80 ± 4.71 b | 41.73 ± 0.71 a | 39.33 ± 3.94 a |
| MCV (fL) | 48.63 ± 2.29 | 47.17 ± 2.31 | 47.07 ± 2.30 |
| MCH (pg) | 16.97 ± 0.85 | 17.20 ± 0.78 | 17.07 ± 0.38 |
| MCHC (g/L) | 349.00 ± 4.58 b | 364.33 ± 2.31 a | 363.33 ± 10.02 a |
| RDW-CV (%) | 22.50 ± 1.37 | 21.37 ± 1.17 | 22.20 ± 0.44 |
| RDW-SD (fL) | 37.83 ± 2.74 | 35.57 ± 0.25 | 36.80 ± 1.40 |
| PLT (109/L) | 104.00 ± 5.57 b | 227.67 ± 21.35 a | 184.67 ± 23.80 a |
| Index | Kazakh Horses | Thoroughbreds | Yili Horses |
|---|---|---|---|
| Glu (mmol/L) | 3.02 ± 0.3 b | 3.71 ± 0.05 a | 3.01 ± 0.19 b |
| ALT (U/L) | 5.87 ± 1.10 | 5.13 ± 0.32 | 5.5 ± 0.46 |
| AST (U/L) | 241.87 ± 36.23 | 255.37 ± 5.27 | 247.47 ± 32.10 |
| ALP (U/L) | 215.93 ± 14.45 | 251.43 ± 8.56 | 240.70 ± 32.92 |
| TP (g/L) | 57.93 ± 5.33 | 50.07 ± 2.49 | 56.30 ± 13.87 |
| TC (mmol/L) | 2.33 ± 0.62 | 1.92 ± 0.28 | 2.22 ± 0.57 |
| UREA (mmol/L) | 8.51 ± 2.03 | 7.28 ± 1.73 | 7.40 ± 3.65 |
| ALB (g/L) | 22.33 ± 4.52 | 25.30 ± 2.56 | 21.80 ± 6.58 |
| Index | Kazakh Horses | Thoroughbreds | Yili Horses |
|---|---|---|---|
| IFN-γ (pg/mL) | 57.93 ± 27.62 b | 83.79 ± 36.51 a | 73.28 ± 19.31 ab |
| TNF-α (pg/mL) | 23.62 ± 13.14 b | 37.56 ± 15.12 a | 45.5 ± 22.96 a |
| IL-4 (pg/mL) | 20.38 ± 11.87 b | 39.73 ± 17.82 a | 32.49 ± 19.03 ab |
| IgA (g/L) | 2.13 ± 0.16 b | 3.19 ± 0.23 a | 3.09 ± 0.13 a |
| IgG (g/L) | 19.67 ± 0.81 b | 21.96 ± 0.82 a | 21.59 ± 1.18 a |
| IgM (g/L) | 1.27 ± 0.09 b | 1.79 ± 0.13 a | 1.78 ± 0.11 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Yang, X.; Wang, C.; Wang, J.; Meng, J.; Yao, X.; Zeng, Y.; Ren, W. Bridging Breeds: Transcriptomic Insights into Immune Traits of Yili, Thoroughbred, and Kazakh Horses. Life 2025, 15, 1496. https://doi.org/10.3390/life15101496
Wang T, Yang X, Wang C, Wang J, Meng J, Yao X, Zeng Y, Ren W. Bridging Breeds: Transcriptomic Insights into Immune Traits of Yili, Thoroughbred, and Kazakh Horses. Life. 2025; 15(10):1496. https://doi.org/10.3390/life15101496
Chicago/Turabian StyleWang, Tongliang, Xixi Yang, Chuankun Wang, Jianwen Wang, Jun Meng, Xinkui Yao, Yaqi Zeng, and Wanlu Ren. 2025. "Bridging Breeds: Transcriptomic Insights into Immune Traits of Yili, Thoroughbred, and Kazakh Horses" Life 15, no. 10: 1496. https://doi.org/10.3390/life15101496
APA StyleWang, T., Yang, X., Wang, C., Wang, J., Meng, J., Yao, X., Zeng, Y., & Ren, W. (2025). Bridging Breeds: Transcriptomic Insights into Immune Traits of Yili, Thoroughbred, and Kazakh Horses. Life, 15(10), 1496. https://doi.org/10.3390/life15101496






