Unusual and Unconsidered Mechanisms of Bacterial Resilience and Resistance to Quinolones
Abstract
1. Introduction
2. Altered Production of Quinolone Targets
3. Amoeba Protection
3.1. Barrier Effect?
3.2. Effects on Bacterial Transcriptome
4. Bacterial Benthonic Lifestyle
4.1. Biofilm Access
4.2. Altered Bacterial Metabolic Activity
5. Nutrient-Independent Slow Growth
6. Stringent Response and Toxin/Antitoxin Systems
6.1. Sporulation and Quiescence
7. Quinolone Inactivation/Modification
7.1. Fungi
7.2. Bacteria
8. Unconsidered Chromosomal Mutations
8.1. Toxic Metabolites Accumulation
8.2. RNA Polymerase
8.3. Aminoacyl-tRNA Synthetase Genes
8.4. Mutations in Other Targets
8.5. Small Colony Variants
8.6. Atypical Amino Acid Substitutions in GyrA
9. Unknown Mechanisms
10. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruiz, J. Transferable mechanisms of quinolone resistance from 1998 onward. Clin. Microbiol. Rev. 2019, 32, e00007-19. [Google Scholar] [CrossRef]
- Correia, S.; Poeta, P.; Hébraud, M.; Capelo, J.L.; Igrejas, G. Mechanisms of quinolone action and resistance: Where do we stand? J. Med. Microbiol. 2017, 66, 551–559. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Topoisomerase Inhibitors: Fluoroquinolone mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025320. [Google Scholar] [CrossRef]
- Pohlhaus, J.R.; Kreuzer, K.N. Norfloxacin-induced DNA gyrase cleavage complexes block Escherichia coli replication forks, causing double-stranded breaks in vivo. Mol. Microbiol. 2005, 56, 1416–1429. [Google Scholar] [CrossRef]
- Barton, N.; Crowther, A.F.; Hepworth, W.; Richardson, N.D.; Driver, G.W. New Quinolones and Therapeutic Compositions Containing Them. British Patent 830832, 23 March 1960. [Google Scholar]
- Bisacchi, G.S. Origins of the quinolone class of antibacterials: An expanded “discovery story”. J. Med. Chem. 2015, 58, 4874–4882. [Google Scholar] [CrossRef]
- Hepworth, W. Manufacture of 1-Methyl-6-nitro-4-quinolone-3-carboxylic Acid. British Patent 820295, 16 September 1959. [Google Scholar]
- Lesher, G.Y.; Froelich, E.J.; Gruett, M.D.; Bailey, J.H.; Brundage, R.P. 1,8-Naphthyridine derivatives: A new class of chemotherapy agents. J. Med. Pharm. Chem. 1962, 5, 1063–1068. [Google Scholar] [CrossRef]
- Mandomando, I.M.; Macete, E.V.; Ruiz, J.; Sanz, S.; Abacassamo, F.; Vallés, X.; Sacarlal, J.; Vila, J.; Navia, M.M.; Alonso, P.L.; et al. Etiology of diarrhea in children younger than 5 years of age admitted in a rural hospital of southern Mozambique. Am. J. Trop. Med. Hyg. 2007, 76, 522–527. [Google Scholar] [CrossRef]
- Parry, H.E. Nalidixic acid for shigellosis. Lancet 1983, 322, 1206. [Google Scholar] [CrossRef]
- Sen, D.; Dutta, P.; Deb, B.C.; Pal, S.C. Nalidixic-acid resistant Shigella dysenteriae type 1 in eastern India. Lancet 1988, 332, 911. [Google Scholar] [CrossRef]
- Emmerson, A.M.; Jones, A.M. The quinolones: Decades of development and use. J. Antimicrob. Chemother. 2003, 51 (Suppl. S1), 13–20. [Google Scholar] [CrossRef]
- AlMahmoud, T.; Elhanan, M.; Elshamsy, M.H.; Alshamsi, H.N.; Abu-Zidan, F.M. Management of infective corneal ulcers in a high-income developing country. Medicine 2019, 98, e18243. [Google Scholar] [CrossRef]
- Alonso, D.; Muñoz, J.; Ruiz, J.; Carmona, F.; Nadal, A.; Gascón, J. Salmonella ovarian abscess following travel diarrhoea episode. Arch. Gynecol. Obstet. 2007, 276, 551–553. [Google Scholar] [CrossRef]
- Bassetti, M.; Cadeo, B.; Villa, G.; Sartor, A.; Cainero, V.; Causero, A. Current antibiotic management of prosthetic joint infections in Italy: The ‘Udine strategy’. J. Antimicrob. Chemother. 2014, 69 (Suppl. S1), i41–i45. [Google Scholar] [CrossRef]
- Gentry, L.O. Review of quinolones in the treatment of infections of the skin and skin structure. J. Antimicrob. Chemother. 1991, 28 (Suppl. SC), 97–110. [Google Scholar] [CrossRef]
- Koulenti, D.; Xu, E.; Mok, I.Y.S.; Song, A.; Karageorgopoulos, D.E.; Armaganidis, A.; Lipman, J.; Tsiodras, S. Novel antibiotics for multidrug-resistant gram-positive microorganisms. Microorganisms 2019, 7, 270. [Google Scholar] [CrossRef]
- Leonov, Y.; Schlaeffer, F.; Karpuch, J.; Bourvin, A.; Shemesh, Y.; Lewinson, G. Ciprofloxacin in the treatment of nosocomial multiply resistant Acinetobacter calcoaceticus bacteremia. Infection 1990, 18, 234–236. [Google Scholar] [CrossRef]
- Okubo, Y.; Michihata, N.; Morisaki, N.; Uda, K.; Miyairi, I.; Ogawa, Y.; Matsui, H.; Fushimi, K.; Yasunaga, H. Recent trends in practice patterns and impact of corticosteroid use on pediatric Mycoplasma pneumoniae-related respiratory infections. Respir. Investig. 2018, 56, 158–165. [Google Scholar] [CrossRef]
- Ruiz, J.; Marco, F.; Oliveira, I.; Vila, J.; Gascón, J. Trends in antimicrobial resistance in Campylobacter spp. causing traveler’s diarrhea. APMIS 2007, 115, 218–224. [Google Scholar] [CrossRef]
- Vila, J.; Ruiz, J.; Sanchez, F.; Navarro, F.; Mirelis, B.; Jiménez de Anta, M.T.; Prats, G. Increase in quinolone resistance in a Haemophilus influenzae strain isolated from a patient with recurrent respiratory infections treated with ofloxacin. Antimicrob. Agents Chemother. 1999, 43, 161–162. [Google Scholar] [CrossRef][Green Version]
- Davino, G.; D’Alvano, T.; Esposito, S. The use of ozenoxacin in pediatric patients: Clinical evidence, efficacy and safety. Front. Pharmacol. 2020, 11, 559708. [Google Scholar] [CrossRef]
- Liapikou, A.; Cilloniz, C.; Palomeque, A.; Torres, T. Emerging antibiotics for community-acquired pneumonia. Expert Opin. Emerg. Drugs 2019, 24, 221–231. [Google Scholar] [CrossRef]
- Copeland, V.; McLaughlin, M.; Trifilio, S. Ciprofloxacin vs levofloxacin for prophylaxis during hematopoietic stem-cell transplantation. Clin. Transplant. 2018, 32, e13145. [Google Scholar] [CrossRef]
- Córdova-González, D.; Alfonseca-Silva, E.; Gutiérrez, L.; Tapia-Pérez, G.; Sumano, H. Intramammary preparation of enrofloxacin hydrochloride-dihydrate for bovine mastitis (biofilm-forming Staphylococcus aureus). J. Vet. Sci. 2024, 25, e6. [Google Scholar] [CrossRef]
- Nelson, J.M.; Chiller, T.M.; Powers, J.H.; Angulo, F.J. Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: A public health success story. Clin. Infect. Dis. 2007, 44, 977–980. [Google Scholar] [CrossRef]
- Trouchon, T.; Lefebvre, S. A review of enrofloxacin for veterinary use. Open J. Vet. Med. 2016, 6, 40–58. [Google Scholar] [CrossRef]
- World Organisation for Animal Health. OIE Annual Report on Antimicrobial Agents Intended for Use in Animals, 3rd ed.; World Organisation for Animal Health (OIE): Paris, France, 2018; pp. 1–129. Available online: https://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/AMR/Annual_Report_AMR_3.pdf (accessed on 13 March 2024).
- Dalhoff, A. Antiviral, antifungal, and antiparasitic activities of fluoroquinolones optimized for treatment of bacterial infections: A puzzling paradox or a logical consequence of their mode of action? Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 661–668. [Google Scholar] [CrossRef]
- Romero, I.C.; Saravia, N.G.; Walker, J. Selective action of fluoroquinolones against intracellular amastigotes of Leishmania (Viannia) panamensis in vitro. J. Parasitol. 2005, 91, 1474–1479. [Google Scholar] [CrossRef]
- Stergiopoulou, T.; Meletiadis, J.; Sein, T.; Papaioannidou, P.; Tsiouris, I.; Roilides, E.; Walsh, T.J. Comparative pharmacodynamic interaction analysis between ciprofloxacin, moxifloxacin and levofloxacin and antifungal agents against Candida albicans and Aspergillus fumigatus. J. Antimicrob. Chemother. 2009, 63, 343–348. [Google Scholar] [CrossRef]
- Yamaya, M.; Nishimura, H.; Hatachi, Y.; Yasuda, H.; Deng, X.; Sasaki, T.; Mizuta, K.; Kubo, H.; Nagatomi, R. Levofloxacin inhibits rhinovirus infection in primary cultures of human tracheal epithelial cells. Antimicrob. Agents Chemother. 2012, 56, 4052–4061. [Google Scholar] [CrossRef]
- Carroll, G. Neggram (nalidixic acid): A new antimicrobial chemotherapeutic agent. J. Urol. 1963, 90, 476–478. [Google Scholar] [CrossRef]
- Cullen, M.E.; Wyke, A.W.; Kuroda, R.; Fisher, L.M. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob. Agents Chemother. 1989, 33, 886–894. [Google Scholar] [CrossRef]
- Guibert, F.; Espinoza, K.; Taboada-Blanco, C.; Alonso, C.A.; Oporto, R.; Castillo, A.K.; Rojo-Bezares, B.; López, M.; Sáenz, Y.; Pons, M.J.; et al. Traditional marketed meats as a reservoir of multidrug-resistant Escherichia coli. Int. Microbiol. 2024; in press. [Google Scholar] [CrossRef]
- Hooper, D.C.; Wolfson, J.S.; Ng, E.Y.; Swartz, M.N. Mechanisms of action of and resistance to ciprofloxacin. Am. J. Med. 1987, 82, 12–20. [Google Scholar]
- Jonsson, M. Antibiotic resistance and R factors in gram-negative bacteria isolated in a hospital for infectious diseases. III. The effect of antibacterial treatment on the incidence of R factor-mediated antibiotic resistance. Scand. J. Infect. Dis. 1973, 5, 41–47. [Google Scholar] [CrossRef]
- Lishman, I.V.; Swinney, J. Studies of a new antibacterial agent, nalidixic acid. Br. J. Urol. 1963, 35, 116–121. [Google Scholar] [CrossRef]
- Martínez-Martínez, L.; Pascual, A.; Jacoby, G.A. Quinolone resistance from a transferable plasmid. Lancet 1998, 351, 797–799. [Google Scholar] [CrossRef]
- Medina, A.; Rivera, F.P.; Ochoa, T.J.; Riveros, M.; Pons, M.J.; Ruiz, J. Transferable mechanisms of quinolone resistance are more frequent among enterotoxigenic Escherichia coli isolates displaying low-level quinolone resistance. Trop. Biomed. 2023, 40, 183–187. [Google Scholar] [CrossRef]
- Pitout, J.D.; Wei, Y.; Church, D.L.; Gregson, D.B. Surveillance for plasmid-mediated quinolone resistance determinants in Enterobacteriaceae within the Calgary Health Region, Canada: The emergence of aac(6′)-Ib-cr. J. Antimicrob. Chemother. 2008, 61, 999–1002. [Google Scholar] [CrossRef]
- Pons, M.J.; Mosquito, S.G.; Gomes, C.; del Valle, L.J.; Ochoa, T.J.; Ruiz, J. Analysis of quinolone-resistance in commensal and diarrheagenic Escherichia coli isolates from infants in Lima, Peru. Trans. R. Soc. Trop. Med. Hyg. 2014, 108, 22–28. [Google Scholar] [CrossRef]
- Ricci, V.; Tzakas, P.; Buckley, A.; Piddock, L.J. Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob. Agents Chemother. 2006, 50, 38–42. [Google Scholar] [CrossRef]
- Ruiz, E.; Sáenz, Y.; Zarazaga, M.; Rocha-Gracia, R.; Martínez-Martínez, L.; Arlet, G.; Torres, C. qnr, aac(6′)-Ib-cr and qepA genes in Escherichia coli and Klebsiella spp.: Genetic environments and plasmid and chromosomal location. J. Antimicrob. Chemother. 2012, 67, 886–897. [Google Scholar] [CrossRef]
- Tavío, M.M.; Vila, J.; Ruiz, J.; Ruiz, J.; Martin-Sanchez, A.M.; Jiménez de Anta, M.T. Mechanisms involved in the development of resistance to fluoroquinolones in Escherichia coli isolates. J. Antimicrob. Chemother. 1999, 44, 735–742. [Google Scholar] [CrossRef]
- Vila, J.; Ruiz, J.; Marco, F.; Barceló, A.; Goñi, P.; Giralt, E.; Jiménez de Anta, M.T. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and minimal inhibitory concentration. Antimicrob. Agents Chemother. 1994, 38, 2477–2479. [Google Scholar] [CrossRef]
- Vinué, L.; Hooper, D.C.; Jacoby, G.A. Chromosomal mutations that accompany qnr in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents 2018, 51, 479–483. [Google Scholar] [CrossRef]
- Gellert, M.; Mizuuchi, K.; O’Dea, M.H.; Itoh, T.; Tomizawa, J.I. Nalidixic acid resistance: A second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 1977, 74, 4772–4776. [Google Scholar] [CrossRef]
- Everett, M.J.; Jin, Y.F.; Ricci, V.; Piddock, L.J. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother. 1996, 40, 2380–2386. [Google Scholar] [CrossRef]
- Guillemin, I.; Jarlier, V.; Cambau, E. Correlation between quinolone susceptibility patterns and sequences in the A and B subunits of DNA gyrase in Mycobacteria. Antimicrob. Agents Chemother. 1998, 42, 2084–2088. [Google Scholar] [CrossRef]
- Gomes, C.; Martínez-Puchol, S.; Ruiz-Roldán, L.; Pons, M.J.; del Valle Mendoza, J.; Ruiz, J. Development and characterisation of highly antibiotic resistant Bartonella bacilliformis mutants. Sci. Rep. 2016, 6, 33584. [Google Scholar] [CrossRef]
- Madurga, S.; Sánchez-Céspedes, J.; Belda, I.; Vila, J.; Giralt, E. Mechanism of binding of fluoroquinolones to the quinolone resistance-determining region of DNA gyrase: Towards an understanding of the molecular basis of quinolone resistance. ChemBioChem 2008, 9, 2081–2086. [Google Scholar] [CrossRef]
- Mendoza-Mujica, G.; Flores-Leon, D.; Ruiz, J. Molecular characterization of fluoroquinolone-resistant Bartonella bacilliformis. Pathogens 2021, 10, 876. [Google Scholar] [CrossRef]
- Palumbo, M.; Gatto, B.; Zagotto, G.; Palù, G. On the mechanism of action of quinolone drugs. Trends Microbiol. 1993, 1, 232–235. [Google Scholar] [CrossRef]
- Ruiz, J. Mechanisms of resistance to quinolones: Target alterations, decreased accumulation and DNA Gyrase protection. J. Antimicrob. Chemother. 2003, 51, 1109–1117. [Google Scholar] [CrossRef]
- Sierra, J.M.; Marco, F.; Ruiz, J.; Jiménez de Anta, M.T.; Vila, J. Correlation between the activity of different fluoroquinolones and the presence of mechanisms of quinolone resistance in epidemiologically related and unrelated strains of methicillin-susceptible and -resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2002, 8, 781–790. [Google Scholar] [CrossRef]
- Han, X.Y.; Andrade, R.A. Brevundimonas diminuta infections and its resistance to fluoroquinolones. J. Antimicrob. Chemother. 2005, 55, 853–859. [Google Scholar] [CrossRef]
- del Valle, L.J.; Flores, L.; Vargas, M.; García-de-la-Guarda, R.; Quispe, R.L.; Ibañez, Z.B.; Alvarado, D.; Ramírez, P.; Ruiz, J. Bartonella bacilliformis, endemic pathogen of the Andean region, is intrinsically resistant to quinolones. Int. J. Infect. Dis. 2010, 14, e506–e510. [Google Scholar] [CrossRef][Green Version]
- Hirai, K.; Aoyama, H.; Suzue, S.; Irikura, T.; Iyobe, S.; Mitsuhashi, S. Isolation and characterization of norfloxacin-resistant mutants of Escherichia coli K-12. Antimicrob. Agents Chemother. 1986, 30, 248–253. [Google Scholar] [CrossRef]
- Palma, N.; Pons, M.J.; Gomes, C.; Mateu, J.; Riveros, M.; García, W.; Jacobs, J.; García, C.; Ochoa, T.J.; Ruiz, J. Resistance to quinolones, cephalosporins and macrolides in Escherichia coli causing bacteraemia in Peruvian children. J. Glob. Antimicrob. Resist. 2017, 11, 28–33. [Google Scholar] [CrossRef]
- Redgrave, L.S.; Sutton, S.B.; Webber, M.A.; Piddock, L.J. Fluoroquinolone resistance: Mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014, 22, 438–445. [Google Scholar] [CrossRef]
- Sáenz, Y.; Ruiz, J.; Zarazaga, M.; Teixidó, M.; Torres, C.; Vila, J. Effect of the efflux pump inhibitor Phe-Arg-beta-naphthylamide on the MIC values of the quinolones, tetracycline and chloramphenicol, in Escherichia coli isolates of different origin. J. Antimicrob. Chemother. 2004, 53, 544–545. [Google Scholar] [CrossRef]
- García-León, G.; Ruiz de Alegría Puig, C.; García de la Fuente, C.; Martínez-Martínez, L.; Martínez, J.L.; Sánchez, M.B. High-level quinolone resistance is associated with the overexpression of smeVWX in Stenotrophomonas maltophilia clinical isolates. Clin. Microbiol. Infect. 2015, 21, 464–467. [Google Scholar] [CrossRef]
- Valdezate, S.; Vindel, A.; Saéz-Nieto, J.A.; Baquero, F.; Cantón, R. Preservation of topoisomerase genetic sequences during in vivo and in vitro development of high-level resistance to ciprofloxacin in isogenic Stenotrophomonas maltophilia strains. J. Antimicrob. Chemother. 2005, 56, 220–223. [Google Scholar] [CrossRef]
- Ribera, A.; Doménech-Sánchez, A.; Ruiz, J.; Benedi, V.J.; de Anta, M.T.J.; Vila, J. Mutations in gyrA and parC QRDRs are not relevant for quinolone resistance in epidemiological unrelated Stenotrophomonas maltophilia clinical isolates. Microb. Drug Resist. 2002, 8, 245–252. [Google Scholar] [CrossRef]
- Ribera, A.; Jurado, A.; Ruiz, J.; Marco, F.; Del Valle, O.; Mensa, J.; Chaves, J.; Hernández, G.; de Anta, M.J.; Vila, J. In vitro activity of clinafloxacin in comparison with other quinolones against Stenotrophomonas maltophilia clinical isolates in the presence and absence of reserpine. Diagn. Microbiol. Infect. Dis. 2002, 42, 123–128. [Google Scholar] [CrossRef]
- Babcock, G.F.; Berryhill, D.L.; Marsh, D.H. R-factors of Escherichia coli from dressed beef and humans. Appl. Microbiol. 1973, 25, 21–23. [Google Scholar] [CrossRef]
- Courvalin, P. Plasmid-mediated 4-quinolone resistance: A real or apparent absence? Antimicrob. Agents Chemother. 1990, 34, 681–684. [Google Scholar] [CrossRef]
- Lebek, G. Medizinische aspekte der infektiösen antibiotika-resistenz gram-negativer darmbakterien. Pathol. Microbiol. 1967, 30, 1015–1036. [Google Scholar] [CrossRef]
- Munshi, M.H.; Sack, D.A.; Haider, K.; Ahmed, Z.U.; Rahaman, M.M.; Morshed, M.G. Plasmid-mediated resistance to nalidixic acid in Shigella dysenteriae type 1. Lancet 1987, 330, 419–421. [Google Scholar] [CrossRef]
- Panhotra, B.R.; Desai, B.; Sharma, P.L. Nalidixic-acid-resistant Shigella dysenteriae I. Lancet 1985, 325, 763. [Google Scholar] [CrossRef]
- Pons, M.J.; Gomes, C.; Ruiz, J. QnrVC, a new transferable Qnr-like family. Enferm. Infecc. Microbiol. Clin. 2013, 31, 191–192. [Google Scholar] [CrossRef]
- Robicsek, A.; Strahilevitz, J.; Jacoby, G.A.; Macielag, M.; Abbanat, D.; Park, C.H.; Bush, K.; Hooper, D.C. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006, 12, 83–88. [Google Scholar] [CrossRef]
- Ruiz, J. Analysis of the presence of erroneous Qnr sequences in GenBank. J. Antimicrob. Chemother. 2018, 73, 1213–1216. [Google Scholar] [CrossRef]
- Ruiz, J. In silico analysis of transferable QepA variants and related chromosomal efflux pumps. Rev. Esp. Quimioterap. 2018, 31, 537–541. [Google Scholar]
- Ruiz, J. CrpP, a passenger or a hidden stowaway in the Pseudomonas aeruginosa genome? J. Antimicrob. Chemother. 2019, 74, 3397–3399. [Google Scholar] [CrossRef]
- Zubyk, H.L.; Wright, G.D. CrpP is not a fluoroquinolone-inactivating enzyme. Antimicrob. Agents Chemother. 2021, 65, e0077321. [Google Scholar] [CrossRef]
- Pérez-Moreno, M.O.; Estepa, V.; Sáenz, Y.; Cortell-Ortolá, M.; Fort-Gallifa, I.; Ruiz, J.; Torres, C. Intrahospitalary dissemination of Klebsiella pneumoniae carrying blaDHA-1 and qnrB4 genes within a novel complex class 1 integron. Diagn. Microbiol. Infect. Dis. 2012, 73, 210–211. [Google Scholar] [CrossRef]
- Quiroga, M.P.; Arduino, S.M.; Merkier, A.K.; Quiroga, C.; Petroni, A.; Argentinian Integron Study Group; Roy, P.H.; Centrón, D. Distribution and functional identification of complex class 1 integrons. Infect. Genet. Evol. 2013, 19, 88–96. [Google Scholar] [CrossRef]
- Ruiz, J.; Jurado, A.; Garcia-Méndez, E.; Marco, F.; Aguilar, L.; Jiménez de Anta, M.T.; Vila, J. Frequency of selection of fluoroquinolone-resistant mutants of Neisseria gonorrhoeae exposed to gemifloxacin and four other quinolones. J. Antimicrob. Chemother. 2001, 48, 545–548. [Google Scholar] [CrossRef][Green Version]
- Ruiz, J.; Sierra, J.M.; Jiménez de Anta, M.T.; Vila, J. Characterization of sparfloxacin-resistant mutants of Staphylococcus aureus obtained in vitro. Int. J. Antimicrob. Agents 2001, 18, 107–112. [Google Scholar] [CrossRef]
- Soto, S.M.; Ruíz, J.; Mendoza, M.C.; Vila, J. In vitro fluoroquinolone-resistant mutants of Salmonella enterica serotype Enteritidis: Analysis of mechanisms involved in resistance. Int. J. Antimicrob. Agents 2003, 22, 537–540. [Google Scholar] [CrossRef]
- Ruíz, J.; Navia, M.M.; Marco, F.; Vila, J. Mecanismos de resistencia a betalactámicos y ácido nalidíxico en aislados clínicos de Salmonella enterica serotipo hadar y bsilla. Enferm. Infecc. Microbiol. Clin. 2004, 22, 251–256. [Google Scholar] [CrossRef]
- Stapleton, P.; Wu, P.J.; King, A.; Shannon, K.; French, G.; Philips, I. Incidence and mechanisms of resistance to the combination of amoxicillin and clavulanate in Escherichia coli. Antimicrob. Agents Chemother. 1995, 39, 2478–2483. [Google Scholar] [CrossRef]
- Wu, P.J.; Shannon, K.; Phillips, I. Effect of hyperproduction of TEM-1 β-lactamase on in vitro susceptibility of Escherichia coli to β-lactam antibiotics. Antimicrob. Agents Chemother. 1994, 38, 494–498. [Google Scholar] [CrossRef][Green Version]
- Ince, D.; Hooper, D.C. Quinolone resistance due to reduced target enzyme expression. J. Bacteriol. 2003, 185, 6883–6892. [Google Scholar] [CrossRef]
- Straney, R.; Krah, R.; Menzel, R. Mutations in the -10 TATAAT sequence of the gyrA promoter affect both promoter strength and sensitivity to DNA supercoiling. J. Bacteriol. 1994, 176, 5999–6006. [Google Scholar] [CrossRef]
- Aleixandre, V.; Herrera, G.; Urios, A.; Blanco, M. Effects of ciprofloxacin on plasmid DNA supercoiling of Escherichia coli topoisomerase I and gyrase mutants. Antimicrob. Agents Chemother. 1991, 35, 20–23. [Google Scholar] [CrossRef]
- Dages, S.; Dages, K.; Zhi, X.; Leng, F. Inhibition of the gyrA promoter by transcription-coupled DNA supercoiling in Escherichia coli. Sci. Rep. 2018, 8, 14759. [Google Scholar] [CrossRef]
- Pan, X.S.; Hamlyn, P.J.; Talens-Visconti, R.; Alovero, F.L.; Manzo, R.H.; Fisher, L.M. Small-colony mutants of Staphylococcus aureus allow selection of gyrase-mediated resistance to dual-target fluoroquinolones. Antimicrob. Agents Chemother. 2002, 46, 2498–2506. [Google Scholar] [CrossRef]
- Hecht, A.; Glasgow, J.; Jaschke, P.R.; Bawazer, L.A.; Munson, M.S.; Cochran, J.R.; Endy, D.; Salit, M. Measurements of translation initiation from all 64 codons in E. coli. Nucleic Acids Res. 2017, 45, 3615–3626. [Google Scholar] [CrossRef]
- Samba-Louaka, A.; Delafont, V.; Rodier, M.H.; Cateau, E.; Héchard, Y. Free-living amoebae and squatters in the wild: Ecological and molecular features. FEMS Microbiol. Rev. 2019, 43, 415–434. [Google Scholar] [CrossRef]
- Cirillo, J.D.; Falkow, S.; Tompkins, L.S. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 1994, 62, 3254–3261. [Google Scholar] [CrossRef]
- Molmeret, M.; Horn, M.; Wagner, M.; Santic, M.; Abu Kwaik, Y. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 2005, 71, 20–28. [Google Scholar] [CrossRef]
- Saisongkorh, W.; Robert, C.; La Scola, B.; Raoult, D.; Rolain, J.M. Evidence of transfer by conjugation of type IV secretion system genes between Bartonella species and Rhizobium radiobacter in amoeba. PLoS ONE 2010, 5, e12666. [Google Scholar] [CrossRef]
- Maschio, V.J.; Corção, G.; Rott, M.B. identification of Pseudomonas spp. as amoeba-resistant microorganisms in isolates of Acanthamoeba. Rev. Inst. Med. Trop. Sao Paulo 2015, 57, 81–83. [Google Scholar] [CrossRef]
- Judy, B.M.; Whitlock, G.C.; Torres, A.G.; Estes, D.M. Comparison of the in vitro and in vivo susceptibilities of Burkholderia mallei to ceftazidime and levofloxacin. BMC Microbiol. 2009, 9, 88. [Google Scholar] [CrossRef]
- Kaldestad, M.; Haugland, G.T.; Rønneseth, A.; Wergeland, H.I.; Samuelsen, O.B. Antibiotic uptake by cultured Atlantic cod leucocytes and effect on intracellular Francisella noatunensis subsp. noatunensis replication. Dis. Aquat. Organ. 2014, 108, 11–21. [Google Scholar] [CrossRef][Green Version]
- Ortillés, A.; Belloc, J.; Rubio, E.; Fernández, M.T.; Benito, M.; Cristóbal, J.A.; Calvo, B.; Goñi, P. In-vitro development of an effective treatment for Acanthamoeba keratitis. Int. J. Antimicrob. Agents 2017, 50, 325–333. [Google Scholar] [CrossRef]
- Michot, J.M.; Seral, C.; Van Bambeke, F.; Mingeot-Leclercq, M.P.; Tulkens, P.M. Influence of efflux transporters on the accumulation and efflux of four quinolones (ciprofloxacin, levofloxacin, garenoxacin, and moxifloxacin) in J774 macrophages. Antimicrob. Agents Chemother. 2005, 49, 2429–2437. [Google Scholar] [CrossRef]
- Lismond, A.; Tulkens, P.M.; Mingeot-Leclercq, M.P.; Courvalin, P.; Van Bambeke, F. Cooperation between prokaryotic (Lde) and eukaryotic (MRP) efflux transporters in J774 macrophages infected with Listeria monocytogenes: Studies with ciprofloxacin and moxifloxacin. Antimicrob. Agents Chemother. 2008, 52, 3040–3046. [Google Scholar] [CrossRef]
- Behera, H.S.; Satpathy, G. Characterisation and expression analysis of trophozoite and cyst proteins of Acanthamoeba spp. isolated from Acanthamoeba keratitis (AK) patient. Mol. Biochem. Parasitol. 2016, 205, 29–34. [Google Scholar] [CrossRef]
- Fouque, E.; Trouilhé, M.C.; Thomas, V.; Hartemann, P.; Rodier, M.H.; Héchard, Y. Cellular, biochemical, and molecular changes during encystment of free-living amoebae. Eukaryot. Cell 2012, 11, 382–387. [Google Scholar] [CrossRef]
- Lee, X.; Reimmann, C.; Greub, G.; Sufrin, J.; Croxatto, A. The Pseudomonas aeruginosa toxin L-2-amino-4-methoxy-trans-3-butenoic acid inhibits growth and induces encystment in Acanthamoeba castellanii. Microbes Infect. 2012, 14, 268–272. [Google Scholar] [CrossRef]
- Maal-Bared, R.; Dixon, B.; Axelsson-Olsson, D. Fate of internalized Campylobacter jejuni and Mycobacterium avium from encysted and excysted Acanthamoeba polyphaga. Exp. Parasitol. 2019, 199, 104–110. [Google Scholar] [CrossRef]
- Barker, J.; Scaife, H.; Brown, M.R. Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob. Agents Chemother. 1995, 39, 2684–2688. [Google Scholar] [CrossRef]
- Dubois, V.; Pawlik, A.; Bories, A.; Le Moigne, V.; Sismeiro, O.; Legendre, R.; Varet, H.; Rodríguez-Ordóñez, M.D.P.; Gaillard, J.L.; Coppée, J.Y.; et al. Mycobacterium abscessus virulence traits unraveled by transcriptomic profiling in amoeba and macrophages. PLoS Pathog. 2019, 15, e1008069. [Google Scholar] [CrossRef]
- Winiecka-Krusnell, J.; Linder, E. Bacterial infections of free-living amoebae. Res. Microbiol. 2001, 152, 613–619. [Google Scholar] [CrossRef]
- Vieira, A.; Ramesh, A.; Seddon, A.M.; Karlyshev, A.V. CmeABC Multidrug efflux pump contributes to antibiotic resistance and promotes Campylobacter jejuni survival and multiplication in Acanthamoeba polyphaga. Appl. Environ. Microbiol. 2017, 83, e01600-17. [Google Scholar] [CrossRef]
- Grinnage-Pulley, T.; Zhang, Q. Genetic basis and functional consequences of differential expression of the CmeABC efflux pump in Campylobacter jejuni isolates. PLoS ONE 2015, 10, 0131534. [Google Scholar] [CrossRef]
- Yan, M.; Sahin, O.; Lin, J.; Zhang, Q. Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J. Antimicrob. Chemother. 2006, 58, 1154–1159. [Google Scholar] [CrossRef]
- Colvin, K.M.; Irie, Y.; Tart, C.S.; Urbano, R.; Whitney, J.C.; Ryder, C.; Howell, P.L.; Wozniak, D.J.; Parsek, M.R. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 2012, 14, 1913–1928. [Google Scholar] [CrossRef]
- Oliveira, F.; Lima, C.A.; Brás, S.; França, Â.; Cerca, N. Evidence for inter- and intraspecies biofilm formation variability among a small group of coagulase-negative staphylococci. FEMS Microbiol. Lett. 2015, 362, fnv175. [Google Scholar] [CrossRef][Green Version]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Petrova, O.E.; Sauer, K. Escaping the biofilm in more than one way: Desorption, detachment or dispersion. Curr. Opin. Microbiol. 2016, 30, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Akers, K.S.; Cardile, A.; Wenke, J.C.; Murray, C.K. Biofilm formation by clinical isolates and its relevance to clinical infections. Adv. Exp. Med. Biol. 2015, 830, 1–28. [Google Scholar] [CrossRef] [PubMed]
- del Pozo, J.L. Biofilm-related disease. Expert Rev. Anti Infect. Ther. 2018, 16, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Trautner, B.W.; Darouiche, R.O. Role of biofilm in catheter-associated urinary tract infection. Am. J. Infect. Control 2004, 32, 177–183. [Google Scholar] [CrossRef]
- Steenackers, H.P.; Parijs, I.; Dubey, A.; Foster, K.R.; Vanderleyden, J. Experimental evolution in biofilm populations. FEMS Microbiol. Rev. 2016, 40, 373–397. [Google Scholar] [CrossRef] [PubMed]
- Taghizadeh, L.; Karimi, A.; Presterl, E.; Heitzinger, C. Bayesian inversion for a biofilm model including quorum sensing. Comput. Biol. Med. 2020, 117, 103582. [Google Scholar] [CrossRef]
- Li, Z.; Ding, Z.; Liu, Y.; Jin, X.; Xie, J.; Li, T.; Zeng, Z.; Wang, Z.; Liu, J. Phenotypic and genotypic characteristics of biofilm formation in clinical isolates of Acinetobacter baumannii. Infect. Drug Resist. 2021, 14, 2613–2624. [Google Scholar] [CrossRef]
- Park, H.R.; Hong, M.K.; Hwang, S.; Park, Y.; Kwon, K.H.; Yoon, J.W.; Shin, S.; Kim, J.H.; Park, Y.H. Characterisation of Pseudomonas aeruginosa related to bovine mastitis. Acta Vet. Hung. 2014, 62, 1–12. [Google Scholar] [CrossRef]
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-How P. aeruginosa can escape antibiotics. Front. Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef]
- Anderl, J.N.; Franklin, M.J.; Stewart, P.S. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemoter. 2000, 44, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Suci, P.A.; Mittelman, M.W.; Yu, F.P.; Geesey, G.G. Investigation of ciprofloxacin penetration into Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 1994, 38, 2125–2133. [Google Scholar] [CrossRef] [PubMed]
- Billings, N.; Millan, M.; Caldara, M.; Rusconi, R.; Tarasova, Y.; Stocker, R.; Ribbeck, K. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2013, 9, e1003526. [Google Scholar] [CrossRef] [PubMed]
- Colvin, K.M.; Gordon, V.D.; Murakami, K.; Borlee, B.R.; Wozniak, D.J.; Wong, G.C.; Parsek, M.R. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011, 7, e1001264. [Google Scholar] [CrossRef] [PubMed]
- Becker, P.; Hufnagle, W.; Peters, G.; Herrmann, M. Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis. Appl. Environ. Microbiol. 2001, 67, 2958–2965. [Google Scholar] [CrossRef] [PubMed]
- Booth, S.C.; Workentine, M.L.; Wen, J.; Shaykhutdinov, R.; Vogel, H.J.; Ceri, H.; Turner, R.J.; Weljie, A.M. Differences in metabolism between the biofilm and planktonic response to metal stress. J. Proteome Res. 2011, 10, 3190–3199. [Google Scholar] [CrossRef]
- Crumplin, G.C.; Smith, J.T. Nalidixic acid: An antibacterial paradox. Antimicrob. Agents Chemother. 1975, 8, 251–261. [Google Scholar] [CrossRef]
- Uruén, C.; Chopo-Escuin, G.; Tommassen, J.; Mainar-Jaime, R.C.; Arenas, J. Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics 2020, 10, 3. [Google Scholar] [CrossRef]
- Kolpen, M.; Lerche, C.J.; Kragh, K.N.; Sams, T.; Koren, K.; Jensen, A.S.; Line, L.; Bjarnsholt, T.; Ciofu, O.; Moser, C.; et al. Hyperbaric oxygen sensitizes anoxic Pseudomonas aeruginosa biofilm to ciprofloxacin. Antimicrob. Agents Chemother. 2017, 61, e01024-17. [Google Scholar] [CrossRef]
- Soares, A.; Roussel, V.; Pestel-Caron, M.; Barreau, M.; Caron, F.; Bouffartigues, E.; Chevalier, S.; Etienne, M. Understanding ciprofloxacin failure in Pseudomonas aeruginosa biofilm: Persister cells survive matrix disruption. Front. Microbiol. 2019, 10, 2603. [Google Scholar] [CrossRef]
- Clerch, B.; Rivera, E.; Llagostera, M. Identification of a pKM101 region which confers a slow growth rate and interferes with susceptibility to quinolone in Escherichia coli AB1157. J. Bacteriol. 1996, 178, 5568–5572. [Google Scholar] [CrossRef][Green Version]
- Cheng, G.; Hao, H.; Dai, M.; Liu, Z.; Yuan, Z. Antibacterial action of quinolones: From target to network. Eur. J. Med. Chem. 2013, 66, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Drlica, K.; Malik, M.; Kerns, R.J.; Zhao, X. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 2008, 52, 385–392. [Google Scholar] [CrossRef]
- Pacios, O.; Blasco, L.; Bleriot, I.; Fernandez-Garcia, L.; Ambroa, A.; López, M.; Bou, G.; Cantón, R.; Garcia-Contreras, R.; Wood, T.K.; et al. (p)ppGpp and its role in bacterial persistence: New challenges. Antimicrob. Agents Chemother. 2020, 64, e01283-20. [Google Scholar] [CrossRef] [PubMed]
- Sonika, S.; Singh, S.; Mishra, S.; Verma, S. Toxin-antitoxin systems in bacterial pathogenesis. Heliyon 2023, 9, e14220. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; King, K.; Alqahtani, M.; Worden, M.; Muthuraman, P.; Cioffi, C.L.; Bakshi, C.S.; Malik, M. Stringent response governs the oxidative stress resistance and virulence of Francisella tularensis. PLoS ONE 2019, 14, e0224094. [Google Scholar] [CrossRef] [PubMed]
- Potrykus, K.; Cashel, M. (p)ppGpp: Still magical? Annu. Rev. Microbiol. 2008, 62, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Liu, W.; Du, Q.; Zhang, H.; Han, D. Recent advances in bacterial persistence mechanisms. Int. J. Mol. Sci. 2023, 24, 14311. [Google Scholar] [CrossRef]
- Das, B.; Bhadra, R.K. (p)ppGpp Metabolism and antimicrobial resistance in bacterial pathogens. Front. Microbiol. 2020, 11, 563944. [Google Scholar] [CrossRef]
- Viducic, D.; Ono, T.; Murakami, K.; Susilowati, H.; Kayama, S.; Hirota, K.; Miyake, Y. Functional analysis of spoT, relA and dksA genes on quinolone tolerance in Pseudomonas aeruginosa under nongrowing condition. Microbiol. Immunol. 2006, 50, 349–357. [Google Scholar] [CrossRef]
- Dörr, T.; Vulić, M.; Lewis, K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010, 8, e1000317. [Google Scholar] [CrossRef]
- Muthuramalingam, M.; White, J.C.; Murphy, T.; Ames, J.R.; Bourne, C.R. The toxin from a ParDE toxin-antitoxin system found in Pseudomonas aeruginosa offers protection to cells challenged with anti-gyrase antibiotics. Mol. Microbiol. 2019, 111, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Kamruzzaman, M.; Iredell, J. A ParDE-family toxin antitoxin system in major resistance plasmids of Enterobacteriaceae confers antibiotic and heat tolerance. Sci. Rep. 2019, 9, 9872. [Google Scholar] [CrossRef]
- Kang, S.M.; Kim, D.H.; Jin, C.; Lee, B.J. A systematic overview of type II and III toxin-antitoxin systems with a focus on druggability. Toxins 2018, 10, 515. [Google Scholar] [CrossRef]
- Rittershaus, E.S.; Baek, S.H.; Sassetti, C.M. The normalcy of dormancy: Common themes in microbial quiescence. Cell Host Microbe 2013, 13, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Bush, N.G.; Evans-Roberts, K.; Maxwell, A. DNA Topoisomerases. EcoSal Plus 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Ocampo, P.S.; Lázár, V.; Papp, B.; Arnoldini, M.; Abel zur Wiesch, P.; Busa-Fekete, R.; Fekete, G.; Pál, C.; Ackermann, M.; Bonhoeffer, S. Antagonism between bacteriostatic and bactericidal antibiotics is prevalent. Antimicrob. Agents Chemother. 2014, 58, 4573–4582. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Thawng, C.N.; Choi, J.H.; Lee, K.; Cha, C.J. Polymorphism of antibiotic-inactivating enzyme driven by ecology expands the environmental resistome. ISME J. 2018, 12, 267–276. [Google Scholar] [CrossRef]
- Chávez-Jacobo, V.M.; Hernández-Ramírez, K.C.; Romo-Rodríguez, P.; Pérez-Gallardo, R.V.; Campos-García, J.; Gutiérrez-Corona, J.F.; García-Merinos, J.P.; Meza-Carmen, V.; Silva-Sánchez, J.; Ramírez-Díaz, M.I. CrpP is a novel ciprofloxacin-modifying enzyme encoded by the Pseudomonas aeruginosa pUM505 plasmid. Antimicrob. Agents Chemother. 2018, 62, e02629-17. [Google Scholar] [CrossRef]
- Martens, R.; Wetzstein, H.G.; Zadrazil, F.; Capelari, M.; Hoffmann, P.; Schmeer, N. Degradation of the fluoroquinolone enrofloxacin by wood-rotting fungi. Appl. Environ. Microbiol. 1996, 62, 4206–4209. [Google Scholar] [CrossRef]
- Ben Ayed, A.; Akrout, I.; Albert, Q.; Greff, S.; Simmler, C.; Armengaud, J.; Kielbasa, M.; Turbé-Doan, A.; Chaduli, D.; Navarro, D.; et al. Biotransformation of the fluoroquinolone, levofloxacin, by the white-rot fungus Coriolopsis gallica. J. Fungi 2022, 8, 965. [Google Scholar] [CrossRef]
- Čvančarová, M.; Moeder, M.; Filipová, A.; Cajthaml, T. Biotransformation of fluoroquinolone antibiotics by ligninolytic fungi—Metabolites, enzymes and residual antibacterial activity. Chemosphere 2015, 136, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Rusch, M.; Kauschat, A.; Spielmeyer, A.; Römpp, A.; Hausmann, H.; Zorn, H.; Hamscher, G. Biotransformation of the antibiotic danofloxacin by Xylaria longipes leads to an efficient reduction of its antibacterial activity. J. Agric. Food Chem. 2015, 63, 6897–6904. [Google Scholar] [CrossRef] [PubMed]
- Wetzstein, H.G.; Stadler, M.; Tichy, H.V.; Dalhoff, A.; Karl, W. Degradation of ciprofloxacin by basidiomycetes and identification of metabolites generated by the brown rot fungus Gloeophyllum striatum. Appl. Environ. Microbiol. 1999, 65, 1556–1563. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Liu, C.X.; Xu, Q.M.; Cheng, J.S.; Yuan, Y.J. Simultaneous removal of ciprofloxacin, norfloxacin, sulfamethoxazole by co-producing oxidative enzymes system of Phanerochaete chrysosporium and Pycnoporus sanguineus. Chemosphere 2018, 195, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Suryadi, H.; Judono, J.J.; Putri, M.R.; Eclessia, A.D.; Ulhaq, J.M.; Agustina, D.N.; Sumiati, T. Biodelignification of lignocellulose using ligninolytic enzymes from white-rot fungi. Heliyon 2022, 8, e08865. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Li, X.; Hu, M.; Li, S.; Zhai, Q.; Jiang, Y. Efficient enzymatic degradation used as pre-stage treatment for norfloxacin removal by activated sludge. Bioprocess. Biosyst. Eng. 2017, 40, 1261–1270. [Google Scholar] [CrossRef]
- Chen, Z.; Li, N.; Lan, Q.; Zhang, X.; Wu, L.; Liu, J.; Yang, R. Laccase inducer Mn2+ inhibited the intracellular degradation of norfloxacin by Phanerochaete chrysosporium. Int. Biodeterior. Biodegrad. 2021, 164, 105300. [Google Scholar] [CrossRef]
- Adjei, M.D.; Heinze, T.M.; Deck, J.; Freeman, J.P.; Williams, A.J.; Sutherland, J.B. Transformation of the antibacterial agent norfloxacin by environmental mycobacteria. Appl. Environ. Microbiol. 2006, 72, 5790–5793. [Google Scholar] [CrossRef]
- Amorim, C.L.; Moreira, I.S.; Maia, A.S.; Tiritan, M.E.; Castro, P.M. Biodegradation of ofloxacin, norfloxacin, and ciprofloxacin as single and mixed substrates by Labrys portucalensis F11. Appl. Microbiol. Biotechnol. 2014, 98, 3181–3190. [Google Scholar] [CrossRef]
- Kim, D.W.; Heinze, T.M.; Kim, B.S.; Schnackenberg, L.K.; Woodling, K.A.; Sutherland, J.B. Modification of norfloxacin by a Microbacterium sp. strain isolated from a wastewater treatment plant. Appl. Environ. Microbiol. 2011, 77, 6100–6108. [Google Scholar] [CrossRef]
- Pan, L.J.; Li, J.; Li, C.X.; Tang, X.D.; Yu, G.W.; Wang, Y. Study of ciprofloxacin biodegradation by a Thermus sp. isolated from pharmaceutical sludge. J. Hazard. Mater. 2018, 343, 59–67. [Google Scholar] [CrossRef]
- Chen, Y.; Rosazza, J.P.; Reese, C.P.; Chang, H.Y.; Nowakowski, M.A.; Kiplinger, J.P. Microbial models of soil metabolism: Biotransformations of danofloxacin. J. Ind. Microbiol. Biotechnol. 1997, 19, 378–384. [Google Scholar] [CrossRef]
- Kim, D.W.; Feng, J.; Chen, H.; Kweon, O.; Gao, Y.; Yu, L.R.; Burrowes, V.J.; Sutherland, J.B. Identification of the enzyme responsible for N-acetylation of norfloxacin by Microbacterium sp. strain 4N2-2. Appl. Environ. Microbiol. 2013, 79, 314–321. [Google Scholar] [CrossRef]
- Millanao, A.R.; Mora, A.Y.; Saavedra, C.; Villagra, N.A.; Mora, G.C.; Hidalgo, A.A. Inactivation of glutamine synthetase-coding gene glnA increases susceptibility to quinolones through increasing outer membrane protein F in Salmonella enterica Serovar Typhi. Front. Microbiol. 2020, 11, 428. [Google Scholar] [CrossRef]
- Blánquez, A.; Guillén, F.; Rodríguez, J.; Arias, M.E.; Hernández, M. The degradation of two fluoroquinolone based antimicrobials by SilA, an alkaline laccase from Streptomyces ipomoeae. World J. Microbiol. Biotechnol. 2016, 32, 52. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Choudhary, P.; Chatterjee, S.; Akhter, Y. Comparative analysis of SilA-laccase mediated degradation of ciprofloxacin, norfloxacin and ofloxacin and interpretation of the possible catalytic mechanism. J. Biomol. Struct. Dyn. 2024, 42, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A.; Griffin, C.M.; Hooper, D.C. Citrobacter spp. as a source of qnrB alleles. Antimicrob. Agents Chemother. 2011, 55, 4979–4984. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Xu, X.; Guo, Q.; Zhao, X.; Ye, X.; Guo, Y.; Wang, M. Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J. Antimicrob. Chemother. 2012, 67, 1655–1659. [Google Scholar] [CrossRef]
- Tavío, M.M.; Vila, J.; Ruiz, J.; Amicosante, G.; Franceschini, N.; Martín-Sánchez, A.M.; Jiménez de Anta, M.T. In vitro selected fluoroquinolone-resistant mutants of Citrobacter freundii: Analysis of the quinolone resistance acquisition. J. Antimicrob. Chemother. 2000, 45, 521–524. [Google Scholar] [CrossRef]
- Turkmani, A.; Psaroulaki, A.; Christidou, A.; Chochlakis, D.; Tabaa, D.; Tselentis, Y. In vitro-selected resistance to fluoroquinolones in two Brucella strains associated with mutational changes in gyrA. Int. J. Antimicrob. Agents 2008, 32, 227–232. [Google Scholar] [CrossRef]
- Tavío, M.M.; Vila, J.; Ruiz, J.; Martín Sánchez, A.M.; Jiménez de Anta, M.T. Decreased permeability and enhanced proton-dependent active efflux in the development of resistance to quinolones in Morganella morganii. Int. J. Antimicrob. Agents 2000, 14, 157–160. [Google Scholar] [CrossRef]
- Vinué, L.; Corcoran, M.A.; Hooper, D.C.; Jacoby, G.A. Mutations that enhance the ciprofloxacin resistance of Escherichia coli with qnrA1. Antimicrob. Agents Chemother. 2016, 60, 537–545. [Google Scholar] [CrossRef]
- Helling, R.B.; Janes, B.K.; Kimball, H.; Tran, T.; Bundesmann, M.; Check, P.; Phelan, D.; Miller, C. Toxic waste disposal in Escherichia coli. J. Bacteriol. 2002, 184, 3699–3703. [Google Scholar] [CrossRef]
- Webber, M.A.; Whitehead, R.N.; Mount, M.; Loman, N.J.; Pallen, M.J.; Piddock, L.J. Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J. Antimicrob. Chemother. 2015, 70, 2241–2248. [Google Scholar] [CrossRef]
- Pietsch, F.; Bergman, J.M.; Brandis, G.; Marcusson, L.L.; Zorzet, A.; Huseby, D.L.; Hughes, D. Ciprofloxacin selects for RNA polymerase mutations with pleiotropic antibiotic resistance effects. J. Antimicrob. Chemother. 2017, 72, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Garoff, L.; Huseby, D.L.; Praski Alzrigat, L.; Hughes, D. Effect of aminoacyl-tRNA synthetase mutations on susceptibility to ciprofloxacin in Escherichia coli. J. Antimicrob. Chemother. 2018, 73, 3285–3292. [Google Scholar] [CrossRef] [PubMed]
- Cesaro, A.; Bettoni, R.R.; Lascols, C.; Mérens, A.; Soussy, C.J.; Cambau, E. Low selection of topoisomerase mutants from strains of Escherichia coli harbouring plasmid-borne qnr genes. J. Antimicrob. Chemother. 2008, 61, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Kawamura, K.; Arakawa, Y. Contribution of QnrA, a plasmid mediated quinolone resistance peptide, to survival of Escherichia coli exposed to a lethal ciprofloxacin concentration. Jpn. J. Infect. Dis. 2015, 68, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Helling, R.B.; Adams, B.S. Nalidixic acid-resistant auxotrophs of Escherichia coli. J. Bacteriol. 1970, 104, 1027–1029. [Google Scholar] [CrossRef]
- Kaspersen, H.; Sekse, C.; Zeyl Fiskebeck, E.; Slettemeås, J.S.; Simm, R.; Norström, M.; Urdahl, A.M.; Lagesen, K. Dissemination of quinolone-resistant Escherichia coli in the Norwegian broiler and pig production chains and possible persistence in the broiler production environment. Appl. Environ. Microbiol. 2020, 86, e02769-19. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.J.; Ma, C.J.; Kim, J.C.; Ahn, J. Proteomics-based discrimination of differentially expressed proteins in antibiotic-sensitive and antibiotic-resistant Salmonella Typhimurium, Klebsiella pneumoniae, and Staphylococcus aureus. Arch. Microbiol. 2019, 201, 1259–1275. [Google Scholar] [CrossRef]
- Moore, S.D.; Sauer, R.T. Revisiting the mechanism of macrolide-antibiotic resistance mediated by ribosomal protein L22. Proc. Natl. Acad. Sci. USA 2008, 105, 18261–18266. [Google Scholar] [CrossRef]
- Power, E.G.; Phillips, I. Correlation between umuC induction and Salmonella mutagenicity assay for quinolone antimicrobial agents. FEMS Microbiol. Lett. 1993, 112, 251–254. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ysern, P.; Clerch, B.; Castaño, M.; Gibert, I.; Barbé, J.; Llagostera, M. Induction of SOS genes in Escherichia coli and mutagenesis in Salmonella typhimurium by fluoroquinolones. Mutagenesis 1990, 5, 63–66. [Google Scholar] [CrossRef]
- Breidenstein, E.B.; Khaira, B.K.; Wiegand, I.; Overhage, J.; Hancock, R.E. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob. Agents Chemother. 2008, 52, 4486–4491. [Google Scholar] [CrossRef]
- Agnello, M.; Finkel, S.E.; Wong-Beringer, A. Fitness cost of fluoroquinolone resistance in clinical isolates of Pseudomonas aeruginosa differs by type III secretion genotype. Front. Microbiol. 2016, 7, 1591. [Google Scholar] [CrossRef]
- Agnello, M.; Wong-Beringer, A. Differentiation in quinolone resistance by virulence genotype in Pseudomonas aeruginosa. PLoS ONE 2012, 7, e42973. [Google Scholar] [CrossRef]
- Horna, G.; Amaro, C.; Palacios, A.; Guerra, H.; Ruiz, J. High frequency of the exoU+/exoS+ genotype associated with multidrug-resistant “high-risk clones” of Pseudomonas aeruginosa clinical isolates from Peruvian hospitals. Sci. Rep. 2019, 9, 10874. [Google Scholar] [CrossRef]
- Lim, W.S.; Phang, K.K.; Tan, A.H.; Li, S.F.; Ow, D.S. Small colony variants and single nucleotide variations in Pf1 region of PB1 phage-resistant Pseudomonas aeruginosa. Front. Microbiol. 2016, 7, 282. [Google Scholar] [CrossRef]
- Ramiro, R.S.; Costa, H.; Gordo, I. Macrophage adaptation leads to parallel evolution of genetically diverse Escherichia coli small-colony variants with increased fitness in vivo and antibiotic collateral sensitivity. Evol. Appl. 2016, 9, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Curtis, T.D.; Gram, L.; Knudsen, G.M. The small colony variant of Listeria monocytogenes is more tolerant to antibiotics and has altered survival in RAW 264.7 murine macrophages. Front. Microbiol. 2016, 7, 1056. [Google Scholar] [CrossRef] [PubMed]
- Mitsuyama, J.; Yamada, H.; Maehana, J.; Fukuda, Y.; Kurose, S.; Minami, S.; Todo, Y.; Watanabe, Y.; Narita, H. Characteristics of quinolone-induced small colony variants in Staphylococcus aureus. J. Antimicrob. Chemother. 1997, 39, 697–705. [Google Scholar] [CrossRef]
- Al-Maleki, A.R.; Vellasamy, K.M.; Mariappan, V.; Venkatraman, G.; Tay, S.T.; Vadivelu, J. Transcriptome analysis of Burkholderia pseudomallei SCV reveals an association with virulence, stress resistance and intracellular persistence. Genomics 2020, 112, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Sandoz, K.M.; Popham, D.L.; Beare, P.A.; Sturdevant, D.E.; Hansen, B.; Nair, V.; Heinzen, R.A. Transcriptional profiling of Coxiella burnetii reveals extensive cell wall remodeling in the small cell variant developmental form. PLoS ONE 2016, 11, e0149957. [Google Scholar] [CrossRef] [PubMed]
- Brouillette, E.; Martinez, A.; Boyll, B.J.; Allen, N.E.; Malouin, F. Persistence of a Staphylococcus aureus small-colony variant under antibiotic pressure in vivo. FEMS Immunol. Med. Microbiol. 2004, 41, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.A.; Olsen, R.J.; Long, S.W.; Rosato, A.E.; Musser, J.M. Identification of point mutations in clinical Staphylococcus aureus strains that produce small-colony variants auxotrophic for menadione. Infect. Immun. 2014, 82, 1600–1605. [Google Scholar] [CrossRef]
- de Souza, D.C.; Cogo, L.L.; Palmeiro, J.K.; Dalla-Costa, L.M.; de Oliveira Tomaz, A.P.; Riedi, C.A.; Rosario Filho, N.A. Thymidine-auxotrophic Staphylococcus aureus small-colony variant bacteremia in a patient with cystic fibrosis. Pediatr. Pulmonol. 2020, 55, 1388–1393. [Google Scholar] [CrossRef]
- Hubbard, A.T.M.; Bulgasim, I.; Roberts, A.P. A novel hemA mutation is responsible for a small-colony-variant phenotype in Escherichia coli. Microbiology 2021, 167, 000962. [Google Scholar] [CrossRef]
- Irvine, S.; Bunk, B.; Bayes, H.K.; Spröer, C.; Connolly, J.P.R.; Six, A.; Evans, T.J.; Roe, A.J.; Overmann, J.; Walker, D. Genomic and transcriptomic characterization of Pseudomonas aeruginosa small colony variants derived from a chronic infection model. Microb. Genom. 2019, 5, e000262. [Google Scholar] [CrossRef]
- Precit, M.R.; Wolter, D.J.; Griffith, A.; Emerson, J.; Burns, J.L.; Hoffman, L.R. Optimized in vitro antibiotic susceptibility testing method for small-colony variant Staphylococcus aureus. Antimicrob. Agents Chemother. 2016, 60, 1725–1735. [Google Scholar] [CrossRef] [PubMed]
- Eggertsson, G.; Söll, D. Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol. Rev. 1988, 52, 354–374. [Google Scholar] [CrossRef] [PubMed]
- Minarini, L.A.R.; Darini, A.L.C. Mutations in the quinolone resistance-determining regions of gyrA and parC in Enterobacteriaceae isolates from Brazil. Braz. J. Microbiol. 2012, 43, 1309–1314. [Google Scholar] [CrossRef] [PubMed]
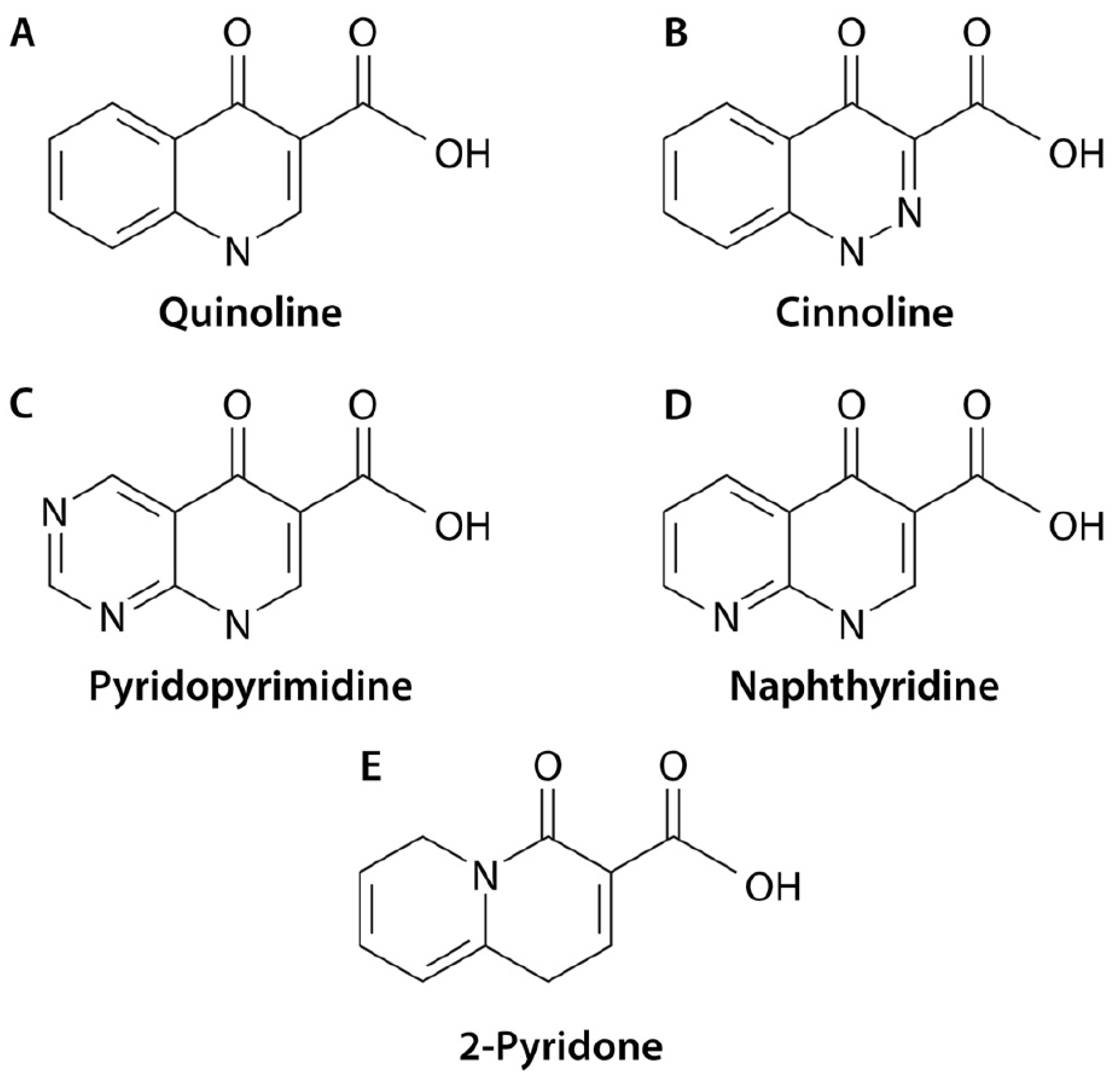
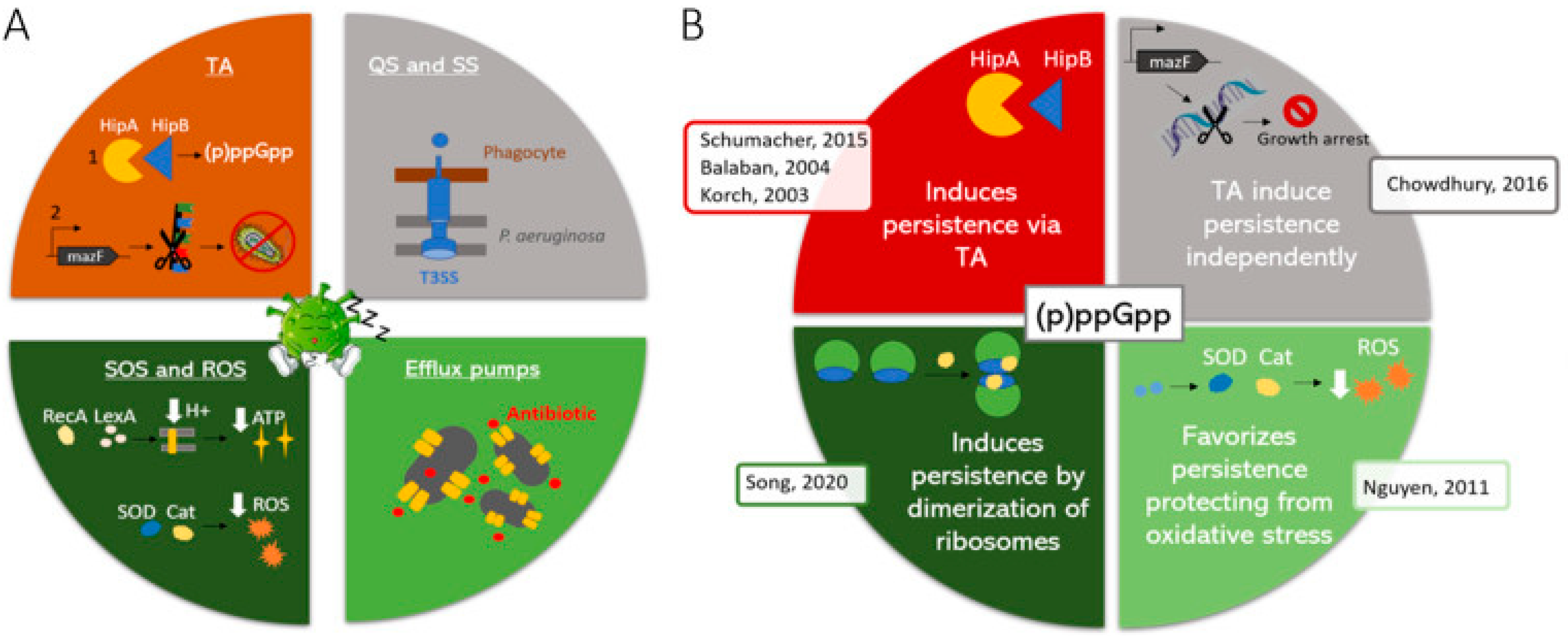

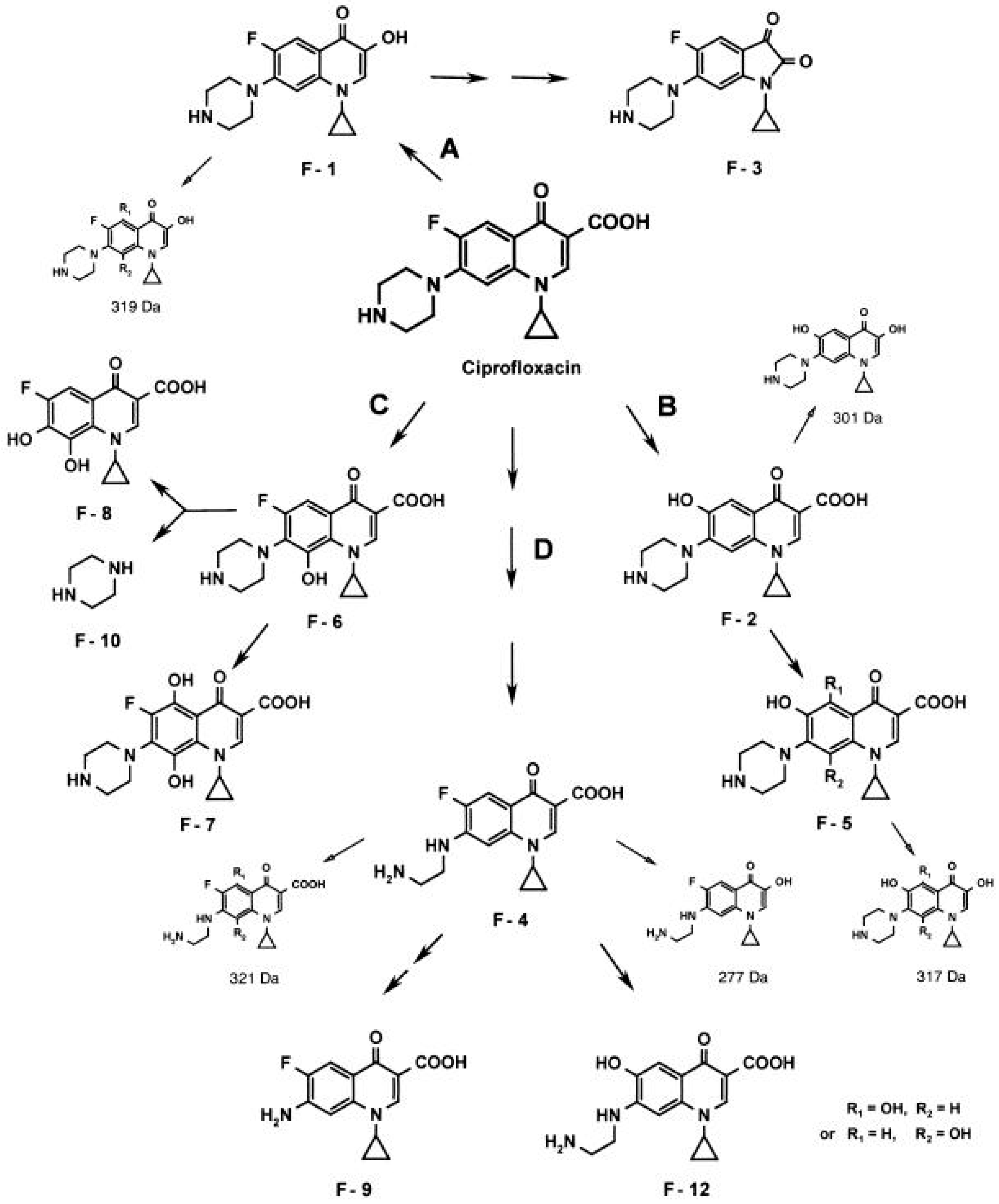
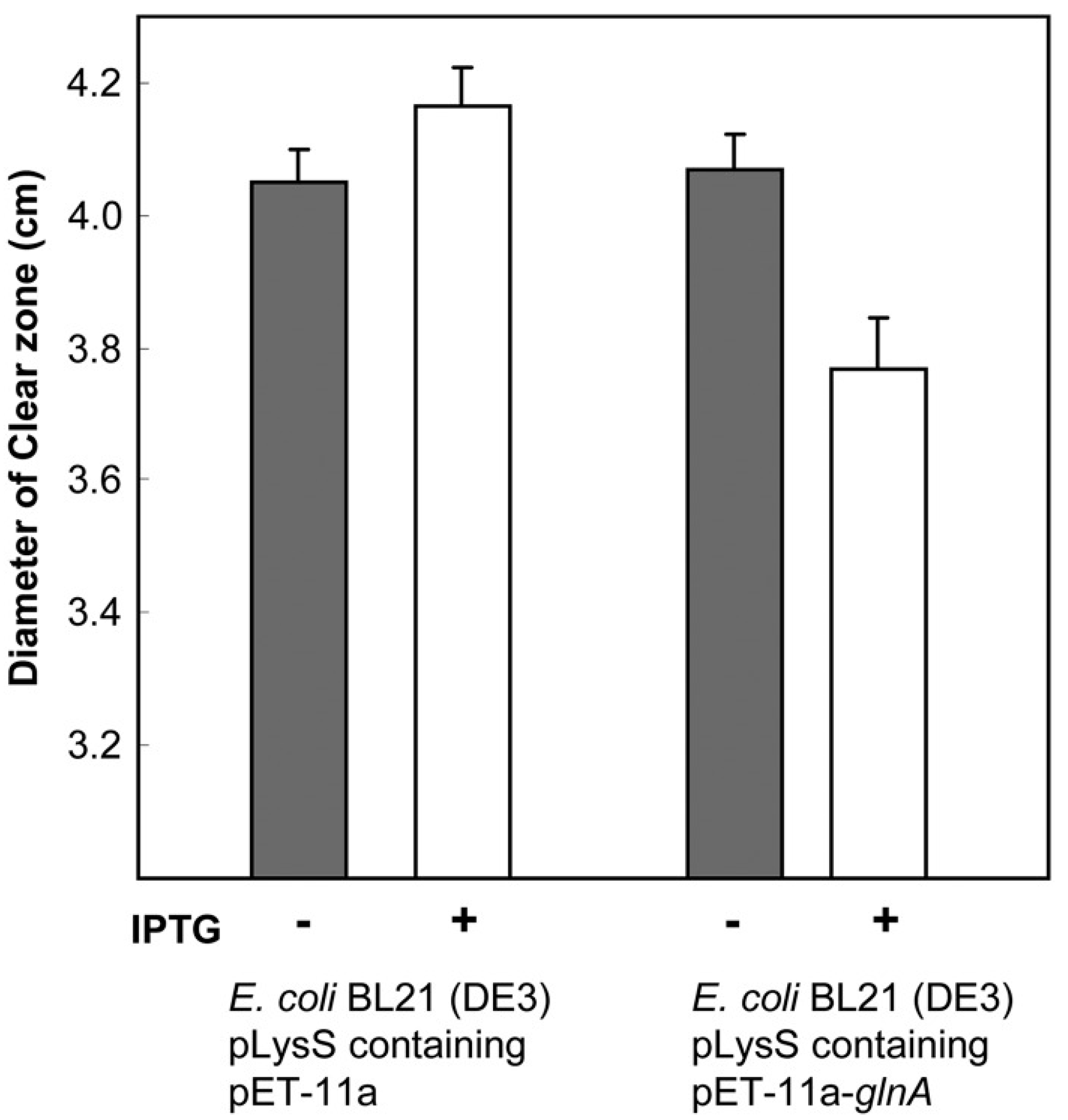
| Cause of Resistance/Tolerance | ||||||
|---|---|---|---|---|---|---|
| Low Levels of Targets | Decreased Antibiotic Access | Transcriptomic/Metabolic Alterations | Quinolones Inactivation/Degradation | Resistance | Tolerance | |
| Expression levels of targets | Y | NA | NC | -- | Y | -- |
| Amoeba protection | NC | Y | Y | NA | Y | Y |
| Biofilm | NC | V | Y | -- | -- | Y |
| Nutrient-Independent Slow Growth | NC | NC | NC | -- | -- | Y |
| SR and T/A Systems 1 | NC | NC | Y | NA | -- | Y |
| Quinolones’ modification 2 | -- | -- | -- | Y | Y | -- |
| Chromosomal mutations 3 | Y | Y | Y | NA | Y | -- |
| Mutation | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Microorg. | D 1 | S 2 | Protein Encoded | Alt. 3 | Effect 4 | Impact 5 | Reference |
| icdA | E. coli | Tn | IS10 | Isocitrate dehydrogenase | LF | MA | ↑ acrAB-tolC 6 | [177,178] |
| purB | E. coli | Tn | -- | adenylosuccinate lyase | LF | MA | ↑ acrAB-tolC | [178] |
| cysH | E. coli | Tn | -- | 3′-phosphoadenosine 5′-phosphosulfate (PAPS) sulfotransferase | LF | MA | ↑ acrAB-tolC | [178] |
| metE | E. coli | Tn | -- | homocysteine methyltransferase | LF | MA | ↑ acrAB-tolC | [178] |
| rpoA | S. enterica | -- | N294Y | RNA polymerase subunit | TBC | TBC | [179] | |
| rpoB | E. coli | -- | R146C, H1244L, E1272G, A1277V, E1279G, Δ442–445, DuS445 | RNA polymerase subunit | TBC | TBC | ↓ OmpF, ↑ MdtK | [177,180] |
| leuS | E. coli | -- | L41H, D162N, S496P | aminoacyl-tRNA synthetase (Leu) | RA | SR | ↑ MdtK; ↑ ydhIJK | [181] |
| aspS | E. coli | -- | D207A | aminoacyl-tRNA synthetase (Asp) | TBC | SR | [181] | |
| thrS | E. coli | -- | H244P, I582S | aminoacyl-tRNA synthetase (Thr) | TBC | TBC | [181] | |
| gyrA | E. coli | -- | Ser83STOP | DNA-Gyrase subunit | RE | TBC | ↓ GyrA | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz, J. Unusual and Unconsidered Mechanisms of Bacterial Resilience and Resistance to Quinolones. Life 2024, 14, 383. https://doi.org/10.3390/life14030383
Ruiz J. Unusual and Unconsidered Mechanisms of Bacterial Resilience and Resistance to Quinolones. Life. 2024; 14(3):383. https://doi.org/10.3390/life14030383
Chicago/Turabian StyleRuiz, Joaquim. 2024. "Unusual and Unconsidered Mechanisms of Bacterial Resilience and Resistance to Quinolones" Life 14, no. 3: 383. https://doi.org/10.3390/life14030383
APA StyleRuiz, J. (2024). Unusual and Unconsidered Mechanisms of Bacterial Resilience and Resistance to Quinolones. Life, 14(3), 383. https://doi.org/10.3390/life14030383






