Design of Ctenophore Ca2+-Regulated Photoprotein Berovin Capable of Being Converted into Active Protein Under Physiological Conditions: Computational and Experimental Approaches
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Molecular Modeling
2.3. Molecular Biology
2.4. Protein Expression and Purification
2.5. Determination of Apparent Dissociation Constant of the Apophotoprotein–Coelenterazine Complex
2.6. Bioluminescence Assay
2.7. Spectral Measurements
2.8. Photo- and Thermoinactivation
3. Results and Discussion
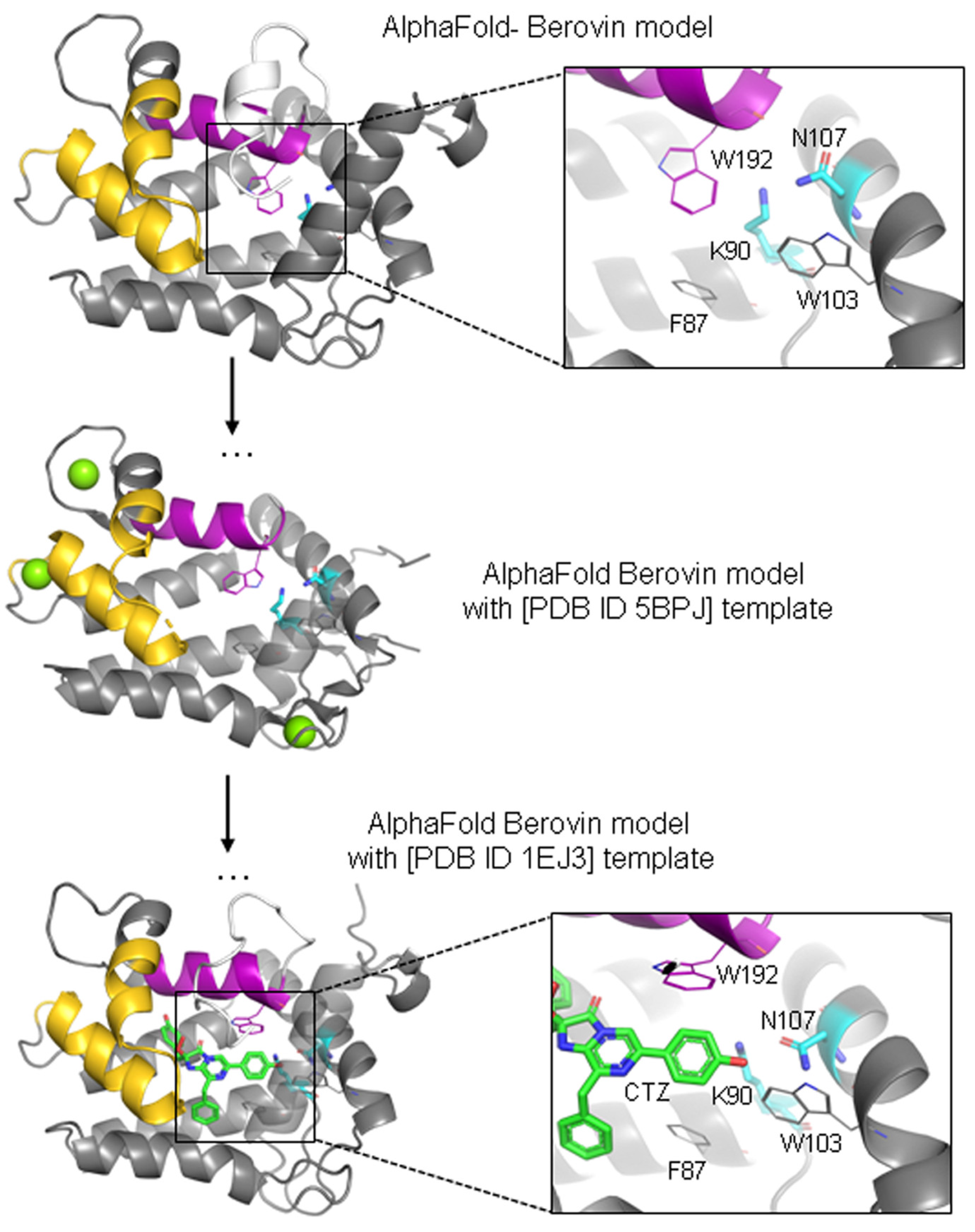
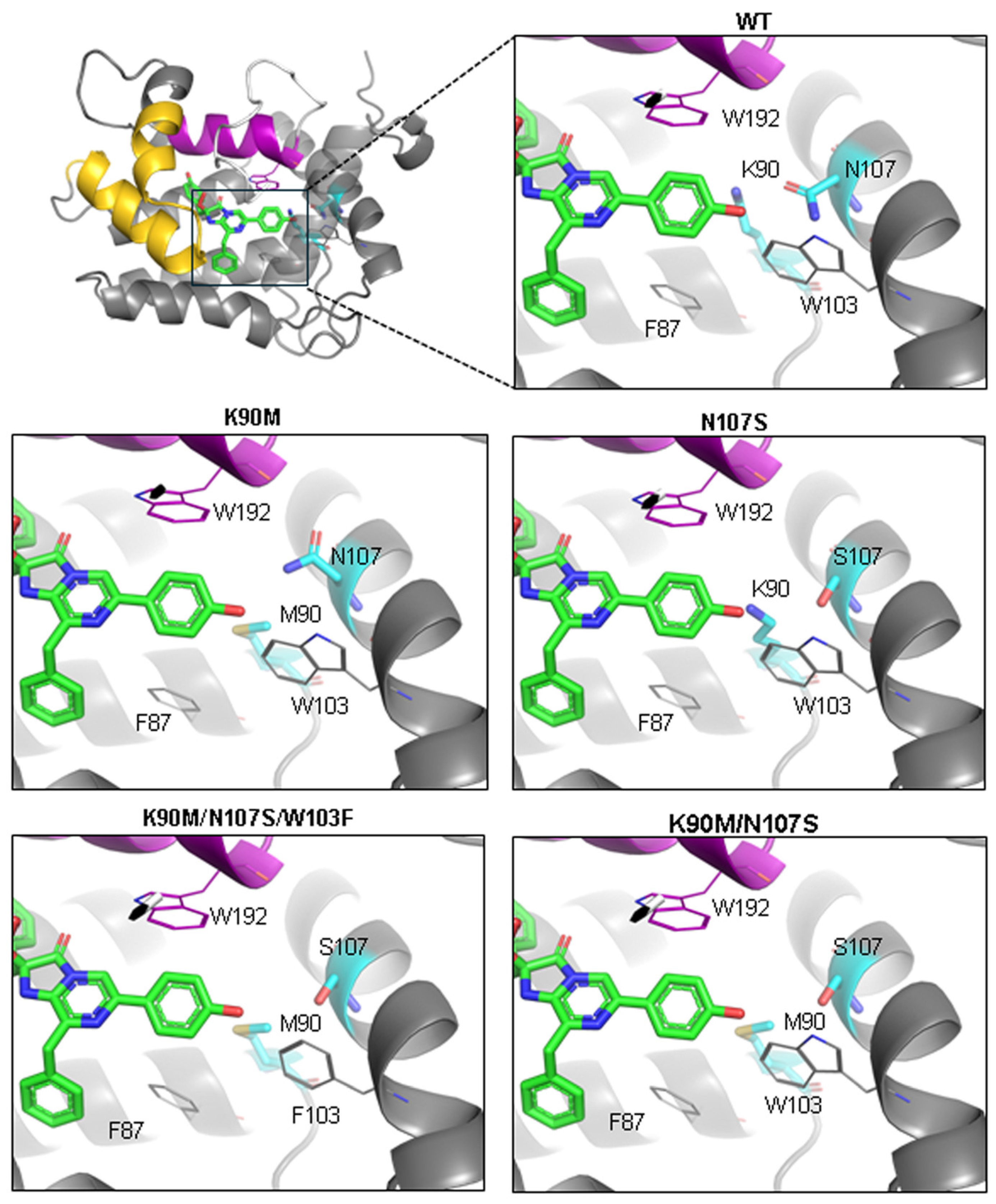
3.1. Molecular Modeling
3.2. Properties of Berovin Mutants
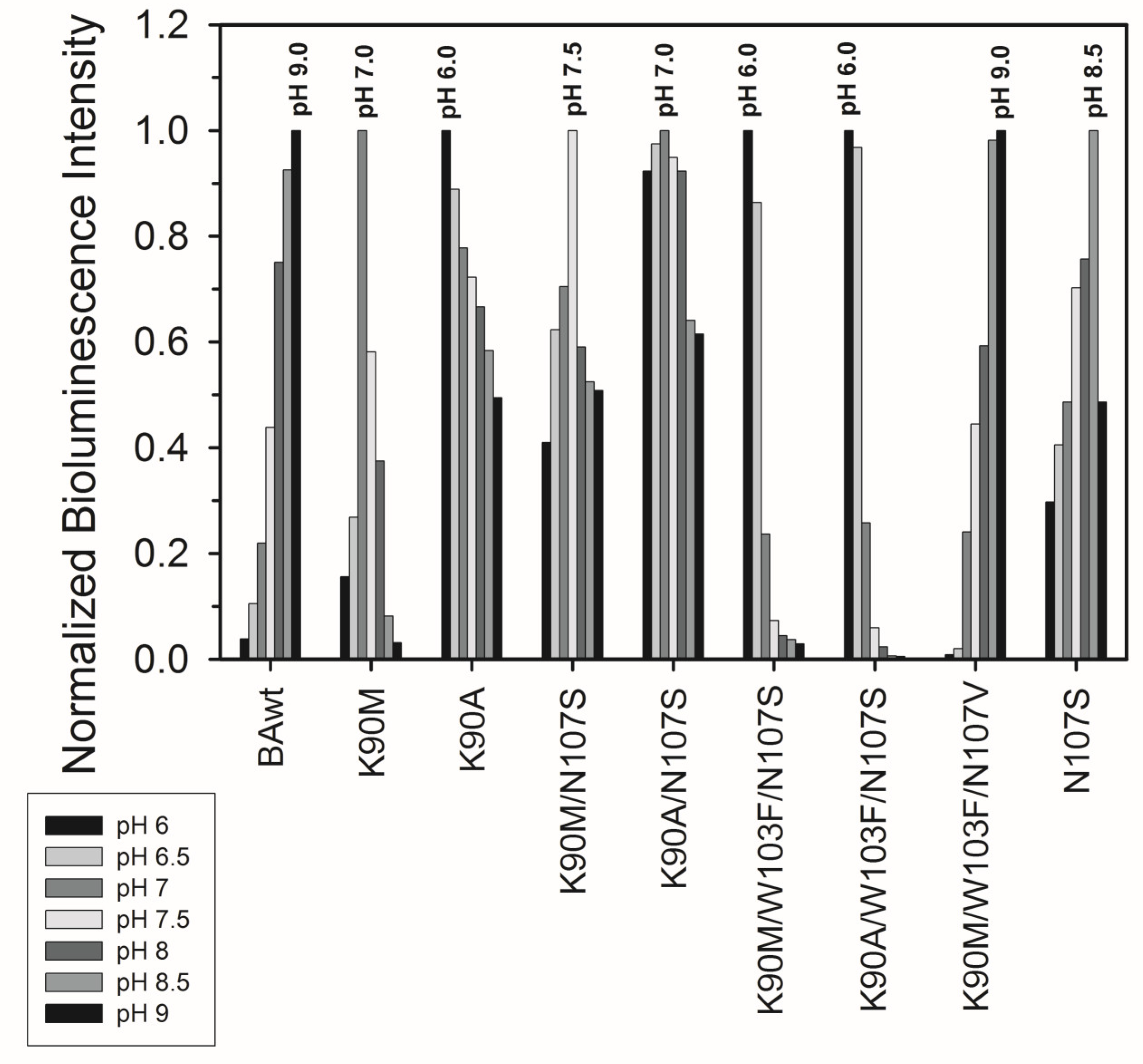
3.2.1. Conversion of Apo-Berovin Mutants into Active Photoproteins
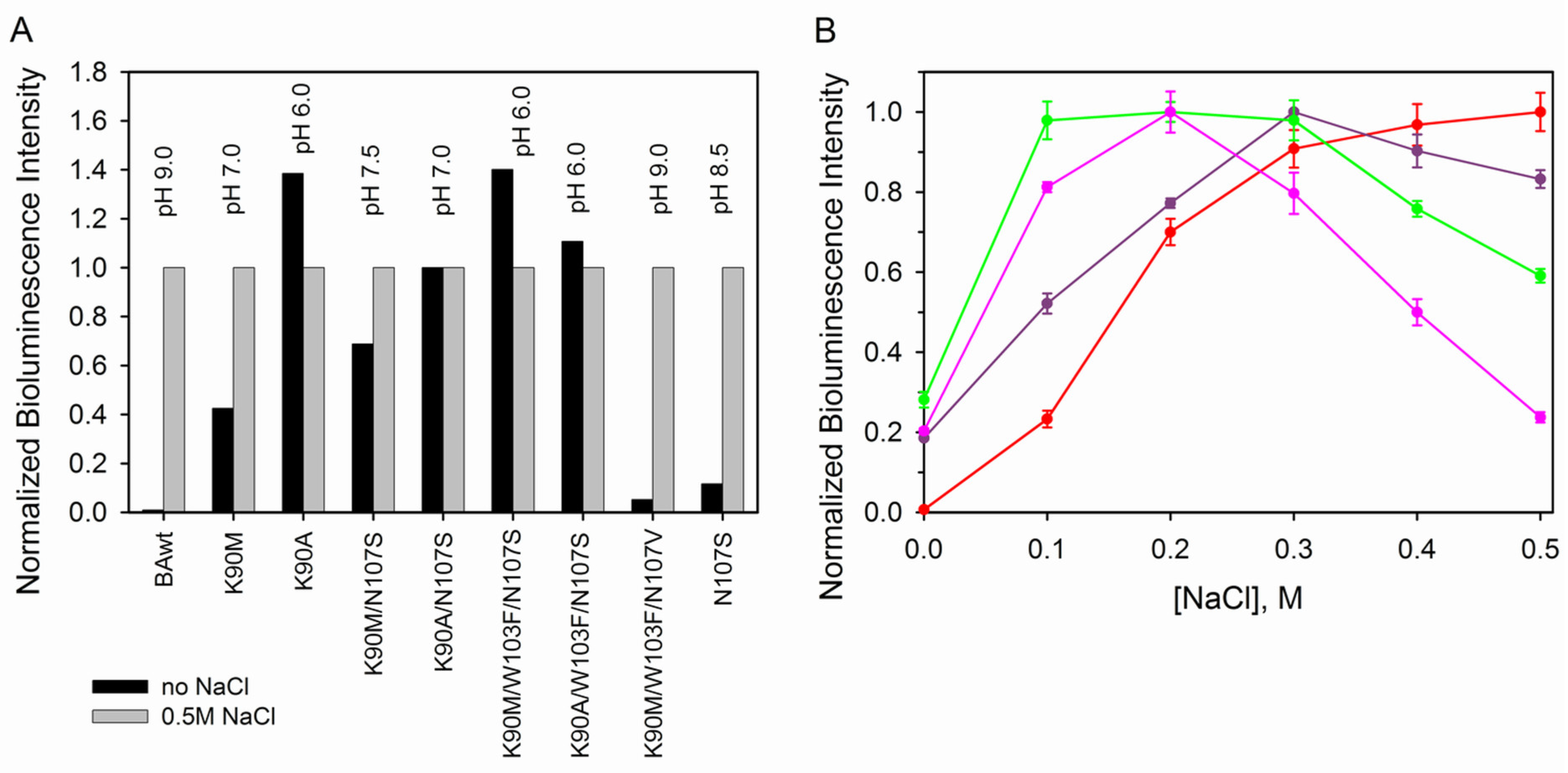
3.2.2. Bioluminescence Activity and Spectral Properties
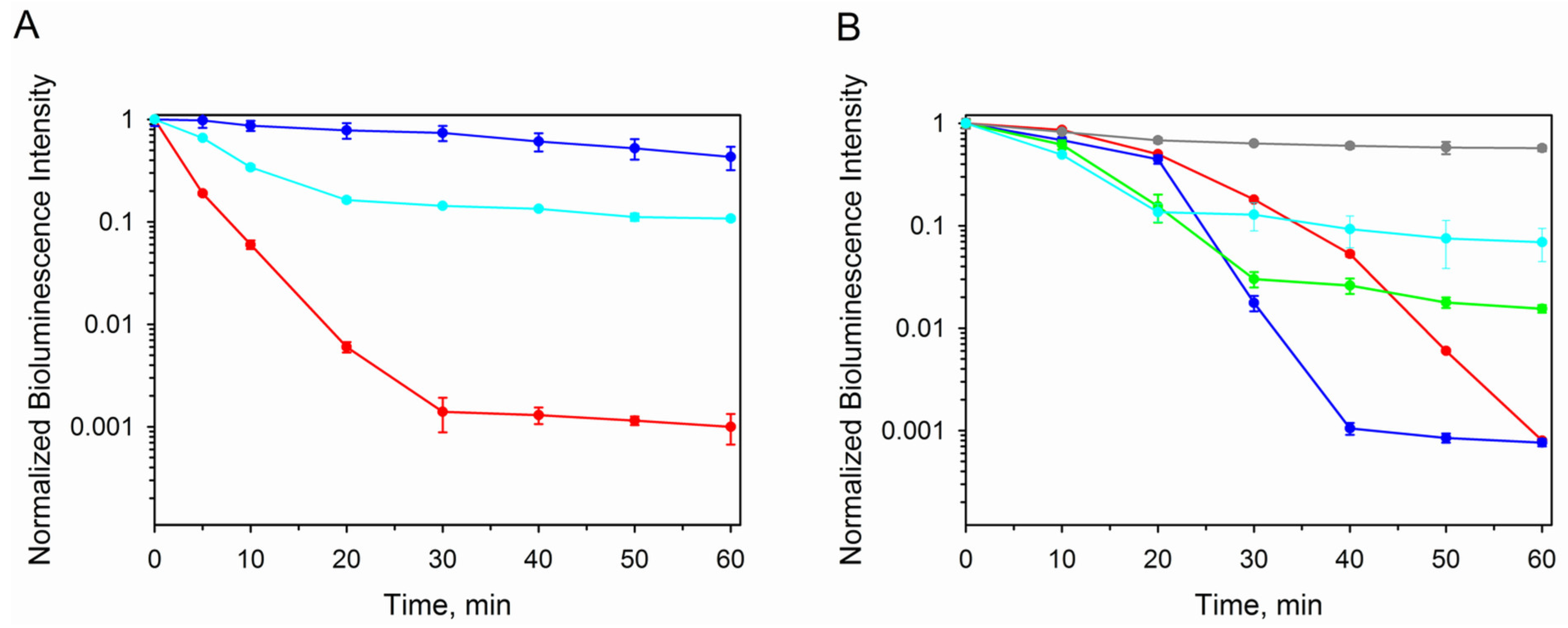
3.2.3. Photo- and Thermoinactivation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shimomura, O. Bioluminescence: Chemical Principles and Methods; World Scientific Publishing Co.: Singapore, 2006. [Google Scholar]
- Markova, S.V.; Vysotski, E.S. Coelenterazine-dependent luciferases. Biochemistry 2015, 80, 714–732. [Google Scholar] [CrossRef] [PubMed]
- Vysotski, E.S.; Lee, J. Ca2+-regulated photoproteins: Structural insight into the bioluminescence mechanism. Acc. Chem. Res. 2004, 37, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Burakova, L.P.; Vysotski, E.S. Recombinant Ca2+-regulated photoproteins of ctenophores: Current knowledge and application prospects. Appl. Microbiol. Biotechnol. 2019, 103, 5929–5946. [Google Scholar] [CrossRef] [PubMed]
- Haddock, S.H.D.; Moline, M.A.; Case, J.F. Bioluminescence in the sea. Ann. Rev. Mar. Sci. 2010, 2, 443–493. [Google Scholar] [CrossRef]
- Haddock, S.H.D.; Case, J.F. Bioluminescence spectra of shallow and deep-sea gelatinous zooplankton: Ctenophores, medusae and siphonophores. Mar. Biol. 1999, 133, 571–582. [Google Scholar] [CrossRef]
- Prasher, D.; McCann, R.O.; Cormier, M.J. Cloning and expression of the cDNA coding for aequorin, a bioluminescent calcium-binding protein. Biochem. Biophys. Res. Commun. 1985, 126, 1259–1268. [Google Scholar] [CrossRef]
- Inouye, S.; Noguchi, M.; Sakaki, Y.; Takagi, Y.; Miyata, T.; Iwanaga, S.; Miyata, T.; Tsuji, F.I. Cloning and sequence analysis of cDNA for the luminescent protein aequorin. Proc. Natl. Acad. Sci. USA 1985, 82, 3154–3158. [Google Scholar] [CrossRef]
- Prasher, D.C.; McCann, R.O.; Longiaru, M.; Cormier, M.J. Sequence comparisons of complementary DNAs encoding aequorin isotypes. Biochemistry 1987, 26, 1326–1332. [Google Scholar] [CrossRef]
- Gurskaya, N.G.; Fradkov, A.F.; Pounkova, N.I.; Staroverov, D.B.; Bulina, M.E.; Yanushevich, Y.G.; Labas, Y.A.; Lukyanov, S.; Lukyanov, K.A. A colourless green fluorescent protein homologue from the nonfluorescent hydromedusa Aequorea coerulescens and its fluorescent mutants. Biochem. J. 2003, 373, 403–408. [Google Scholar] [CrossRef]
- Xia, N.S.; Luo, W.X.; Zhang, J.; Xie, X.Y.; Yang, H.J.; Li, S.W.; Chen, M.; Ng, M.H. Bioluminescence of Aequorea macrodactyla, a common jellyfish species in the East China Sea. Mar. Biotechnol. 2002, 4, 155–162. [Google Scholar]
- Fagan, T.F.; Ohmiya, Y.; Blinks, J.R.; Inouye, S.; Tsuji, F.I. Cloning, expression and sequence analysis of cDNA for the Ca2+-binding photoprotein, mitrocomin. FEBS Lett. 1993, 333, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Burakova, L.P.; Natashin, P.V.; Markova, S.V.; Eremeeva, E.V.; Malikova, N.P.; Cheng, C.; Liu, Z.J.; Vysotski, E.S. Mitrocomin from the jellyfish Mitrocoma cellularia with deleted C-terminal tyrosine reveals a higher bioluminescence activity compared to wild type photoprotein. J. Photochem. Photobiol. B 2016, 162, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Inouye, S.; Tsuji, F.I. Cloning and sequence analysis of cDNA for the Ca2+-activated photoprotein, clytin. FEBS Lett. 1993, 315, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Inouye, S. Cloning, expression, purification and characterization of an isotype of clytin, a calcium-binding photoprotein from the luminous hydromedusa Clytia gregarium. J. Biochem. 2008, 143, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Markova, S.V.; Burakova, L.P.; Frank, L.A.; Golz, S.; Korostileva, K.A.; Vysotski, E.S. Green-fluorescent protein from the bioluminescent jellyfish Clytia gregaria: cDNA cloning, expression, and characterization of novel recombinant protein. Photochem. Photobiol. Sci. 2010, 9, 757–765. [Google Scholar] [CrossRef]
- Fourrage, C.; Swann, K.; Gonzalez Garcia, J.R.; Campbell, A.K.; Houliston, E. An endogenous green fluorescent protein-photoprotein pair in Clytia hemisphaerica eggs shows co-targeting to mitochondria and efficient bioluminescence energy transfer. Open Biol. 2014, 4, 130206. [Google Scholar] [CrossRef]
- Illarionov, B.A.; Markova, S.V.; Bondar, V.S.; Vysotski, E.S.; Gitelson, J.I. Cloning and expression of cDNA for the Ca2+-activated photoprotein obelin from the hydroid polyp Obelia longissimi. Dokl. Akad. Nauk 1992, 326, 911–913. [Google Scholar]
- Illarionov, B.A.; Bondar, V.S.; Illarionova, V.A.; Vysotski, E.S. Sequence of the cDNA encoding the Ca2+-activated photoprotein obelin from the hydroid polyp Obelia longissima. Gene 1995, 153, 273–274. [Google Scholar] [CrossRef]
- Markova, S.V.; Vysotski, E.S.; Blinks, J.R.; Burakova, L.P.; Wang, B.C.; Lee, J. Obelin from the bioluminescent marine hydroid Obelia geniculata: Cloning, expression, and comparison of some properties with those of other Ca2+-regulated photoproteins. Biochemistry 2002, 41, 2227–2236. [Google Scholar] [CrossRef]
- Golz, S.; Markova, S.; Burakova, L.; Frank, L.; Vysotski, E. Isolated Berovin Photoprotein and Use Thereof. WO 2005021591-A1, 13 August 2004. [Google Scholar]
- Markova, S.V.; Burakova, L.P.; Golz, S.; Malikova, N.P.; Frank, L.A.; Vysotski, E.S. The light-sensitive photoprotein berovin from the bioluminescent ctenophore Beroe abyssicola: A novel type of Ca2+-regulated photoprotein. FEBS J. 2012, 279, 856–870. [Google Scholar] [CrossRef]
- Golz, S.; Markova, S.; Burakova, L.; Frank, L.; Vysotski, E. Isolated Photoprotein Bolinopsin, and the Use Thereof. WO/2005/000885, 18 June 2004. [Google Scholar]
- Jafarian, V.; Sariri, R.; Hosseinkhani, S.; Aghamaali, M.R.; Sajedi, R.H.; Taghdir, M.; Hassannia, S. A unique EF-hand motif in mnemiopsin photoprotein from Mnemiopsis leidyi: Implication for its low calcium sensitivity. Biochem. Biophys. Res. Commun. 2011, 413, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Aghamaali, M.R.; Jafarian, V.; Sariri, R.; Molakarimi, M.; Rasti, B.; Taghdir, M.; Sajedi, R.H.; Hosseinkhani, S. Cloning, sequencing, expression and structural investigation of mnemiopsin from Mnemiopsis leidyi: An attempt toward understanding Ca2+-regulated photoproteins. Protein J. 2011, 30, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Powers, M.L.; McDermott, A.G.; Shaner, N.; Haddock, S.H. Expression and characterization of the calcium-activated photoprotein from the ctenophore Bathocyroe fosteri: Insights into light-sensitive photoproteins. Biochem. Biophys. Res. Commun. 2013, 431, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Nakayama, S.; Kretsinger, R.H. Classification and evolution of EF-hand proteins. Biometals 1998, 11, 277–295. [Google Scholar] [CrossRef] [PubMed]
- Strynadka, N.C.; James, M.N. Crystal structures of the helix-loop-helix calcium-binding proteins. Annu. Rev. Biochem. 1989, 58, 951–998. [Google Scholar] [CrossRef]
- Head, J.F.; Inouye, S.; Teranishi, K.; Shimomura, O. The crystal structure of the photoprotein aequorin at 2.3 Å resolution. Nature 2000, 405, 372–376. [Google Scholar] [CrossRef]
- Liu, Z.J.; Vysotski, E.S.; Deng, L.; Lee, J.; Rose, J.P.; Wang, B.C. Atomic resolution structure of obelin: Soaking with calcium enhances electron density of the second oxygen atom substituted at the C2-position of coelenterazine. Biochem. Biophys. Res. Commun. 2003, 311, 433–439. [Google Scholar] [CrossRef]
- Titushin, M.S.; Feng, Y.; Stepanyuk, G.A.; Li, Y.; Markova, S.V.; Golz, S.; Wang, B.C.; Lee, J.; Wang, J.; Vysotski, E.S.; et al. NMR-derived topology of a GFP-photoprotein energy transfer complex. J. Biol. Chem. 2010, 285, 40891–40900. [Google Scholar] [CrossRef]
- Liu, Z.J.; Stepanyuk, G.A.; Vysotski, E.S.; Lee, J.; Markova, S.V.; Malikova, N.P.; Wang, B.C. Crystal structure of obelin after Ca2+-triggered bioluminescence suggests neutral coelenteramide as the primary excited state. Proc. Natl. Acad. Sci. USA 2006, 103, 2570–2575. [Google Scholar] [CrossRef]
- Deng, L.; Markova, S.V.; Vysotski, E.S.; Liu, Z.J.; Lee, J.; Rose, J.; Wang, B.C. Crystal structure of a Ca2+-discharged photoprotein: Implications for mechanisms of the calcium trigger and bioluminescence. J. Biol. Chem. 2004, 279, 33647–33652. [Google Scholar] [CrossRef]
- Deng, L.; Vysotski, E.S.; Markova, S.V.; Liu, Z.J.; Lee, J.; Rose, J.; Wang, B.C. All three Ca2+-binding loops of photoproteins bind calcium ions: The crystal structures of calcium-loaded apo-aequorin and apo-obelin. Protein Sci. 2005, 14, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Stepanyuk, G.A.; Liu, Z.J.; Burakova, L.P.; Lee, J.; Rose, J.; Vysotski, E.S.; Wang, B.C. Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca2+-loaded apoprotein conformation state. Biochim. Biophys. Acta 2013, 1834, 2139–2146. [Google Scholar] [CrossRef]
- Burakova, L.P.; Natashin, P.V.; Malikova, N.P.; Niu, F.; Pu, M.; Vysotski, E.S.; Liu, Z.J. All Ca2+-binding loops of light-sensitive ctenophore photoprotein berovin bind magnesium ions: The spatial structure of Mg2+-loaded apo-berovin. J. Photochem. Photobiol. B 2016, 154, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Molakarimi, M.; Gorman, M.A.; Mohseni, A.; Pashandi, Z.; Taghdir, M.; Naderi-Manesh, H.; Sajedi, R.H.; Parker, M.W. Reaction mechanism of the bioluminescent protein mnemiopsin1 revealed by X-ray crystallography and QM/MM simulations. J. Biol. Chem. 2019, 294, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Burakova, L.P.; Lyakhovich, M.S.; Mineev, K.S.; Petushkov, V.N.; Zagitova, R.I.; Tsarkova, A.S.; Kovalchuk, S.I.; Yampolsky, I.V.; Vysotski, E.S.; Kaskova, Z.M. Unexpected coelenterazine degradation products of Beroe abyssicola photoprotein photoinactivation. Org. Lett. 2021, 23, 6846–6849. [Google Scholar] [CrossRef] [PubMed]
- Burakova, L.P.; Stepanyuk, G.A.; Eremeeva, E.V.; Vysotski, E.S. Role of certain amino acid residues of the coelenterazine-binding cavity in bioluminescence of light-sensitive Ca2+-regulated photoprotein berovin. Photochem. Photobiol. Sci. 2016, 15, 691–704. [Google Scholar] [CrossRef]
- Malikova, N.P.; Eremeeva, E.V.; Gulnov, D.V.; Natashin, P.V.; Nemtseva, E.V.; Vysotski, E.S. Specific activities of hydromedusan Ca2+-regulated photoproteins. Photochem. Photobiol. 2022, 98, 275–283. [Google Scholar] [CrossRef]
- Burakova, L.P.; Kolmakova, A.A.; Vysotski, E.S. Recombinant light-sensitive photoprotein berovin from ctenophore Beroe abyssicola: Bioluminescence and absorbance characteristics. Biochem. Biophys. Res. Commun. 2022, 624, 23–27. [Google Scholar] [CrossRef]
- Tomilin, F.N.; Rogova, A.V.; Burakova, L.P.; Tchaikovskaya, O.N.; Avramov, P.V.; Fedorov, D.G.; Vysotski, E.S. Unusual shift in the visible absorption spectrum of an active ctenophore photoprotein elucidated by time-dependent density functional theory. Photochem. Photobiol. Sci. 2021, 20, 559–570. [Google Scholar] [CrossRef]
- Eremeeva, E.V.; Natashin, P.V.; Song, L.; Zhou, Y.; van Berkel, W.J.; Liu, Z.J.; Vysotski, E.S. Oxygen activation of apo-obelin–coelenterazine complex. Chembiochem 2013, 14, 739–745. [Google Scholar] [CrossRef]
- Ahdritz, G.; Bouatta, N.; Kadyan, S.; Xia, Q.; Gerecke, W.; O’Donnell, T.J.; Berenberg, D.; Fisk, I.; Zanichelli, N.; Zhang, B.; et al. OpenFold: Retraining AlphaFold2 yields new insights into its learning mechanisms and capacity for generalization. Nat. Methods 2024, 21, 1514–1524. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Eremeeva, E.V.; Markova, S.V.; Westphal, A.H.; Visser, A.J.; van Berkel, W.J.; Vysotski, E.S. The intrinsic fluorescence of apo-obelin and apo-aequorin and use of its quenching to characterize coelenterazine binding. FEBS Lett. 2009, 583, 1939–1944. [Google Scholar] [CrossRef]
- Bollen, Y.J.M.; Nabuurs, S.M.; van Berkel, W.J.H.; van Mierlo, C.P.M. Last in, first out: The role of cofactor binding in flavodoxin folding. J. Biol. Chem. 2005, 280, 7836–7844. [Google Scholar] [CrossRef]
- Edelhoch, H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 1967, 6, 1948–1954. [Google Scholar] [CrossRef]
- Allen, D.G.; Blinks, J.R.; Prendergast, F.G. Aequorin luminescence: Relation of light emission to calcium concentration—A calcium-independent component. Science 1977, 195, 996–998. [Google Scholar] [CrossRef]
- Eremeeva, E.V.; Vysotski, E.S. Exploring bioluminescence function of the Ca2+-regulated photoproteins with site-directed mutagenesis. Photochem. Photobiol. 2019, 95, 8–23. [Google Scholar] [CrossRef]
- Shimomura, O.; Teranishi, K. Light-emitters involved in the luminescence of coelenterazine. Luminescence 2000, 15, 51–58. [Google Scholar] [CrossRef]
- Ding, B.W.; Eremeeva, E.V.; Vysotski, E.S.; Liu, Y.J. Luminescence activity decreases when v-coelenterazine replaces coelenterazine in calcium-regulated photoprotein-A theoretical and experimental study. Photochem. Photobiol. 2020, 96, 1047–1060. [Google Scholar] [CrossRef]
- Moroz, L.L. Brief history of Ctenophora. Methods Mol. Biol. 2024, 2757, 1–26. [Google Scholar]
| Photoprotein | Yield of Active Protein, % | Lspecific, RLU, ×109 | LCa-free, RLU, ×10−8 | BL λmax/ Shoulder λmax, nm | FL λmax/ Shoulder λmax, nm |
|---|---|---|---|---|---|
| Berovin (BAwt) | 65 ± 5 | 300 ± 0.22 (100%) | 0.4 | 490 * | 492/398 (pH 8.5) |
| 416 (pH 7.0) | |||||
| K90A | 10 ± 1 | 23.0 ± 0.23 (7.7%) | 17 | 441 | 418 |
| K90M | 14 ± 1 | 0.09 ± 0.003 (0.03%) | 300 | 445 | 412 |
| N107S | 13 ± 1 | 1.4 ± 0.06 (0.47%) | 71 | 490 | 428 |
| K90A/N107S | 8 ± 1 | 1.3 ± 0.2 (0.44%) | 480 | 402/450/530 | 420 |
| K90M/N107S | 10 ± 1 | 6.0 ± 0.13 (2.0%) | 58 | 415/500 | 420 |
| K90A/W103F/N107S | 8 ± 1 | 0.54 ± 0.027 (0.18%) | 37 | ND ** | 416 |
| K90M/W103F/N107S | 47 ± 2 | 7.0 ± 0.98 (2.3%) | 8.1 | 402/450 | 415 |
| K90A/W103F/N107V | 7 ± 1 | 1.1 ± 0.027 (0.37%) | 77 | 490 | 425 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burakova, L.P.; Ivanisenko, N.V.; Rukosueva, N.V.; Ivanisenko, V.A.; Vysotski, E.S. Design of Ctenophore Ca2+-Regulated Photoprotein Berovin Capable of Being Converted into Active Protein Under Physiological Conditions: Computational and Experimental Approaches. Life 2024, 14, 1508. https://doi.org/10.3390/life14111508
Burakova LP, Ivanisenko NV, Rukosueva NV, Ivanisenko VA, Vysotski ES. Design of Ctenophore Ca2+-Regulated Photoprotein Berovin Capable of Being Converted into Active Protein Under Physiological Conditions: Computational and Experimental Approaches. Life. 2024; 14(11):1508. https://doi.org/10.3390/life14111508
Chicago/Turabian StyleBurakova, Ludmila P., Nikita V. Ivanisenko, Natalia V. Rukosueva, Vladimir A. Ivanisenko, and Eugene S. Vysotski. 2024. "Design of Ctenophore Ca2+-Regulated Photoprotein Berovin Capable of Being Converted into Active Protein Under Physiological Conditions: Computational and Experimental Approaches" Life 14, no. 11: 1508. https://doi.org/10.3390/life14111508
APA StyleBurakova, L. P., Ivanisenko, N. V., Rukosueva, N. V., Ivanisenko, V. A., & Vysotski, E. S. (2024). Design of Ctenophore Ca2+-Regulated Photoprotein Berovin Capable of Being Converted into Active Protein Under Physiological Conditions: Computational and Experimental Approaches. Life, 14(11), 1508. https://doi.org/10.3390/life14111508








