Early Monitoring of Donor-Derived Cell-Free DNA in Kidney Allograft Recipients Followed-Up for Two Years: Experience of One Center
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jimenez-Coll, V.; Llorente, S.; Boix, F.; Alfaro, R.; Galián, J.A.; Martinez-Banaclocha, H.; Botella, C.; Moya-Quiles, M.R.; Muro-Pérez, M.; Minguela, A.; et al. Monitoring of Serological, Cellular and Genomic Biomarkers in Transplantation, Computational Prediction Models and Role of Cell-Free DNA in Transplant Outcome. Int. J. Mol. Sci. 2023, 24, 3908. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, J.M.; Knight, S.R. Non-invasive biomarkers in monitoring kidney allograft health. Curr. Opin. Organ. Transplant. 2019, 24, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, A.; Gwinner, W.; Hiss, M.; Radermacher, J.; Mengel, M.; Haller, H. Safety and Adequacy of Renal Transplant Protocol Biopsies. Am. J. Transplant. 2005, 5, 1992–1996. [Google Scholar] [CrossRef] [PubMed]
- Rush, D.N.; Gibson, I.W. Subclinical Inflammation in Renal Transplantation. Transplantation 2019, 103, e139–e145. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Coll, V.; El kaaoui El band, J.; Llorente, S.; González-López, R.; Fernández-González, M.; Martínez-Banaclocha, H.; Galián, J.A.; Botella, C.; Moya-Quiles, M.R.; Minguela, A.; et al. All That Glitters in cfDNA Analysis Is Not Gold or Its Utility Is Completely Established due to Graft Damage: A Critical Review in the Field of Transplantation. Diagnostics 2023, 13, 1982. [Google Scholar] [CrossRef]
- Haas, M.; Loupy, A.; Lefaucheur, C.; Roufosse, C.; Glotz, D.; Seron, D.; Nankivell, B.J.; Halloran, P.F.; Colvin, R.B.; Akalin, E.; et al. The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell–mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am. J. Transplant. 2018, 18, 293–307. [Google Scholar] [CrossRef]
- Marsh, C.L.; Kurian, S.M.; Rice, J.C.; Whisenant, T.C.; David, J.; Rose, S.; Schieve, C.; Lee, D.; Case, J.; Barrick, B.; et al. Application of TruGraf v1: A Novel Molecular Biomarker for Managing Kidney Transplant Recipients with Stable Renal Function. Transplant. Proc. 2019, 51, 722–728. [Google Scholar] [CrossRef]
- Van Loon, E.; Giral, M.; Anglicheau, D.; Lerut, E.; Dubois, V.; Rabeyrin, M.; Brouard, S.; Roedder, S.; Spigarelli, M.G.; Rabant, M.; et al. Diagnostic performance of kSORT, a blood-based mRNA assay for non-invasive detection of rejection after kidney transplantation: A retrospective multicenter cohort study. Am. J. Transplant. 2021, 21, 740–750. [Google Scholar] [CrossRef]
- Bontha, S.V.; Maluf, D.G.; Mueller, T.F.; Mas, V.R. Systems Biology in Kidney Transplantation: The Application of Multi-Omics to a Complex Model. Am. J. Transplant. 2017, 17, 11–21. [Google Scholar] [CrossRef]
- Udomkarnjananun, S.; Kerr, S.J.; Townamchai, N.; van Besouw, N.M.; Hesselink, D.A.; Baan, C.C. Donor-specific ELISPOT assay for predicting acute rejection and allograft function after kidney transplantation: A systematic review and meta-analysis. Clin. Biochem. 2021, 94, 1–11. [Google Scholar] [CrossRef]
- Abdulhadi, T.; Alrata, L.; Dubrawka, C.; Amurao, G.; Kalipatnapu, S.M.; Isaac, C.; Rodrigues, S.; Flores, K.M.; Alsabbagh, D.Y.; Alomar, O.; et al. Donor-derived cell free DNA as a biomarker in kidney transplantation. Pharmacogenomics 2023, 24, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Kataria, A.; Kumar, D.; Gupta, G. Donor-derived Cell-free DNA in Solid-organ Transplant Diagnostics: Indications, Limitations, and Future Directions. Transplantation 2021, 105, 1203–1211. [Google Scholar] [CrossRef]
- Martuszewski, A.; Paluszkiewicz, P.; Król, M.; Banasik, M.; Kepinska, M. Donor-Derived Cell-Free DNA in Kidney Transplantation as a Potential Rejection Biomarker: A Systematic Literature Review. J. Clin. Med. 2021, 10, 193. [Google Scholar] [CrossRef] [PubMed]
- Kueht, M.L.; Dongur, L.P.; Cusick, M.; Stevenson, H.L.; Mujtaba, M. The Current State of Donor-Derived Cell-Free DNA Use in Allograft Monitoring in Kidney Transplantation. J. Pers. Med. 2022, 12, 1700. [Google Scholar] [CrossRef] [PubMed]
- Bloom, R.D.; Bromberg, J.S.; Poggio, E.D.; Bunnapradist, S.; Langone, A.J.; Sood, P.; Matas, A.J.; Mehta, S.; Mannon, R.B.; Sharfuddin, A.; et al. Cell-Free DNA and Active Rejection in Kidney Allografts. J. Am. Soc. Nephrol. 2017, 28, 2221–2232. [Google Scholar] [CrossRef]
- Huang, E.; Jordan, S.C. Donor-derived cell-free DNA in kidney transplantation: Evolving concepts and potential limitations. Kidney Int. 2022, 101, 676–677. [Google Scholar] [CrossRef]
- Loupy, A.; Mengel, M.; Haas, M. Thirty Years of the International Banff Classification for Allograft Pathology: The Past, Present, and Future of Kidney Transplant Diagnostics; Kidney International Elsevier B.V.: Amsterdam, The Netherlands, 2022; Volume 101, pp. 678–691. [Google Scholar]
- Callemeyn, J.; Lamarthée, B.; Koenig, A.; Koshy, P.; Thaunat, O.; Naesens, M. Allorecognition and the Spectrum of Kidney Transplant Rejection; Kidney International Elsevier B.V.: Amsterdam, The Netherlands, 2022; Volume 101, pp. 692–710. [Google Scholar]
- Bu, L.; Gupta, G.; Pai, A.; Anand, S.; Stites, E.; Moinuddin, I.; Bowers, V.; Jain, P.; Axelrod, D.A.; Weir, M.R.; et al. Clinical outcomes from the Assessing Donor-derived cell-free DNA. Monitoring Insights of kidney Allografts with Longitudinal surveillance (ADMIRAL) study. Kidney Int. 2022, 101, 793–803. [Google Scholar] [CrossRef]
- Halloran, P.F.; Reeve, J.; Madill-Thomsen, K.S.; Demko, Z.; Prewett, A.; Billings, P.; Trifecta Investigators. The Trifecta Study: Comparing Plasma Levels of Donor-derived Cell-Free DNA with the Molecular Phenotype of Kidney Transplant Biopsies. J. Am. Soc. Nephrol. 2022, 33, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Aubert, O.; Ursule-Dufait, C.; Brousse, R.; Gueguen, J.; Racapé, M.; Raynaud, M.; Van Loon, E.; Pagliazzi, A.; Huang, E.; Jordan, S.C.; et al. Cell-free DNA for the detection of kidney allograft rejection. Nat. Med. 2024, 30, 2320–2327. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stites, E.; Kumar, D.; Olaitan, O.; John Swanson, S.; Leca, N.; Weir, M.; Bromberg, J.; Melancon, J.; Agha, I.; Fattah, H.; et al. High levels of dd-cfDNA identify patients with TCMR 1A and borderline allograft rejection at elevated risk of graft injury. Am. J. Transplant. 2020, 20, 2491–2498. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, E.; Haas, M.; Gillespie, M.; Sethi, S.; Peng, A.; Najjar, R.; Vo, A.; Jordan, S.C. An Assessment of the Value of Donor-derived Cell-free DNA Surveillance in Patients with Preserved Kidney Allograft Function. Transplantation 2023, 107, 274–282, Erratum in Transplantation 2022, 106, e514. [Google Scholar] [CrossRef] [PubMed]
- Furness, P.N.; Taub, N.; Convergence of European Renal Transplant Pathology Assessment Procedures (CERTPAP) Project. International variation in the interpretation of renal transplant biopsies: Report of the CERTPAP Project. Kidney Int. 2001, 60, 1998–2012. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.; Cornell, L.D.; Smith, M.; Cortese, C.; Geiger, X.; Alexander, M.P.; Ryan, M.; Park, W.; Alvarez, M.C.M.; Schinstock, C.; et al. A method tos reduce variability in scoring antibody-mediated rejection in renal allografts: Implications for clinical trials—A retrospective study. Transplant. Int. 2019, 32, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Obrișcă, B.; Butiu, M.; Sibulesky, L.; Bakthavatsalam, R.; Smith, K.D.; Gimferrer, I.; Warner, P.; Ismail, G.; Leca, N. Combining donor-derived cell-free DNA and donor specific antibody testing as non-invasive biomarkers for rejection in kidney transplantation. Sci. Rep. 2022, 12, 15061. [Google Scholar] [CrossRef]
- Butiu, M.; Obrisca, B.; Sibulesky, L.; Bakthavatsalam, R.; Smith, K.D.; Gimferrer, I.; Warner, P.; Ismail, G.; Leca, N. Donor-derived Cell-free DNA Complements De Novo Class II DSA in Detecting Late Alloimmune Injury Post Kidney Transplantation. Transplant. Direct. 2022, 8, e1285. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Halloran, P.F.; Reeve, J.; Madill-Thomsen, K.S.; Kaur, N.; Ahmed, E.; Cantos, C.; Al Haj Baddar, N.; Demko, Z.; Liang, N.; Swenerton, R.K.; et al. Combining Donor-derived Cell-free DNA Fraction and Quantity to Detect Kidney Transplant Rejection Using Molecular Diagnoses and Histology as Confirmation. Transplantation 2022, 106, 2435–2442. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| NR (Banff 1) | BR (Banff 3) | ABMR (Banff 2) | TCMR (Banff 4) | Total | |
|---|---|---|---|---|---|
| Patients (N, %) | 18 (60%) | 3 (10%) | 5 (16.66%) | 4 (13.33%) | 30 (100%) |
| Recipient Age (average mean ± SD) | 50.56 ± 11.75 | 46.67 ± 6.02 | 48.60 ± 5.85 | 46.50 ± 8.50 | 49.30 ± 9.93 |
| Recipient Sex (% men/% women) | 72.2/27.8 | 66.7/33.3 | 60/40 | 75/25 | 70/30 |
| Indication for Transplant (N, %) | |||||
| DN | 5 (16.7%) | 0 (0%) | 1 (3.3%) | 2 (6.7%) | 8 (26.7%) |
| IgAN | 4 (13.3%) | 0 (0%) | 1 (3.3%) | 1 (3.3%) | 6 (20%) |
| LN | 1 (3.3%) | 0 (0%) | 2 (6.7%) | 0 (0%) | 3 (10%) |
| GN | 2 (6.7%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (6.7%) |
| PKD | 2 (6.7%) | 1(3.3%) | 1(3.3%) | 0 (0%) | 4 (13.3%) |
| CKDu | 4 (13.3%) | 2 (6.7%) | 0 (0%) | 1 (3.3%) | 7 (23.3%) |
| Previous Infections (N, %) | |||||
| Negative Serology | 4 (13.3%) | 0 (0%) | 0 (0%) | 0 (0%) | 4 (13.3%) |
| CMV | 7 (23.3%) | 2(6.7%) | 3 (10%) | 1 (3.3%) | 13 (%) |
| EBV | 2 (6.7%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (6.7%) |
| BKV | 1 (3.3%) | 0 (0%) | 1 (3.3%) | 1 (3.3%) | 3 (10%) |
| HIV | 1 (3.3%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (3.3%) |
| CMV + EBV | 3 (10%) | 0 (0%) | 0 (0%) | 2 (6.7%) | 5 (16.7%) |
| CMV + HIV | 0 (0%) | 0 (0%) | 1 (3.3%) | 0 (0%) | 1 (3.3%) |
| CMV + BKV | 0 (0%) | 0 (0%) | 0 (0%) | 1 (3.3%) | 1 (3.3%) |
| CMV + HSV | 0 (0%) | 1 (3.3%) | 0 (0%) | 0 (0%) | 1 (3.3%) |
| EBV + HBV | 1 (3.3%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (3.3%) |
| Other Pathological History (N, %) | |||||
| HBP | 17 (56.6%) | 2 (6.7%) | 4 (13.3%) | 4 (13.3%) | 27 (90%) |
| Obesity | 4 (13.3%) | 0 (0%) | 0 (0%) | 2 (6.7%) | 6 (20%) |
| DM | 7 (23.3%) | 1(3.3%) | 2 (6.7%) | 2 (6.7%) | 12 (40%) |
| AI | 4 (13.3%) | 0 (0%) | 2 (6.7%) | 3 (10%) | 9 (30%) |
| Hyperuricemia | 5 (%) | 1 (3.3%) | 0 (0%) | 1 (3.3%) | 7 (23.3%) |
| Preformed anti-HLA Antibodies (N,%) | 1 (3.3%) | 1 (3.3%) | 1 (3.3%) | 0 (0%) | 3 (10%) |
| Previous Transplants (N,%) | 2 (6.7%) | 1 (3.3%) | 1 (3.3%) | 0 (0%) | 4 (13.3%) |
| Previous Transfusions (N,%) | 1 (3.3%) | 1 (3.3%) | 1 (3.3%) | 0 (0%) | 3 (10%) |
| Pregnancies (N,%) | 2 (6.7%) | 1 (3.3%) | 0 (0%) | 0 (0%) | 3 (10%) |
| Type of Transplant (N, %) | |||||
| Asystole | 0 (0%) | 1 (3.3%) | 0(0%) | 0 (0%) | 1 (3.3%) |
| Brain Death | 18 (60%) | 2 (6.7%) | 5 (16.6%) | 4 (13.3%) | 29 (96.6%) |
| Donor–Recipient Incompatibilities (ABCDRDQ) (Average ± SD) | 6.8 ± 2.2 | 8.2 ± 1.4 | 6.6 ± 2.8 | 6.5 ± 0.6 | 7.0 ± 1.9 |
| Post-Transplant Therapy (N, %) | |||||
| Tacrolimus | 18 (60%) | 3 (10%) | 5 (16.7%) | 4 (13.3%) | 30 (100%) |
| Cyclosporine | 13 (43.3%) | 2 (6.7%) | 5 (16.6%) | 2 (6.7%) | 22 (73.3%) |
| Everolimus | 4 (13.3%) | 1 (3.3%) | 2 (6.7%) | 1 (3.3%) | 8 (26.6%) |
| MMF | 3(10%) | 0 (0%) | 1 (3.3%) | 0 (0%) | 4 (13.3%) |
| NR (Banff 1) | BR (Banff 3) | ABMR (Banff 2) | TCMR (Banff 4) | Total | |
|---|---|---|---|---|---|
| Patients (N, %) | 18 (60%) | 3 (10%) | 5 (16.66%) | 4 (13.33%) | 30 (100%) |
| Post-transplant complications (N, %) | |||||
| No adverse events | 9 (30%) | 0 (0%) | 0 (0%) | 0 (0%) | 9 (30%) |
| Delayed graft function | 4 (13.3%) | 3 (10%) | 0 (0%) | 0 (0%) | 7 (23.3%) |
| Acute humoral rejection | 0 (0%) | 0 (0%) | 2 (6.7%) | 0 (0%) | 2 (6.7%) |
| Acute cellular rejection | 0 (0%) | 0 (0%) | 2 (6.7%) | 4 (13.3%) | 6 (20%) |
| Borderline acute rejection | 0 (0%) | 3 (10%) | 0 (0%) | 0 (0%) | 3 (10%) |
| Chronic humoral rejection | 0 (0%) | 0 (0%) | 3 (10%) | 0 (0%) | 3 (10%) |
| Urinary tract infection | 2 (6.7%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (6.7%) |
| Cyclosporine nephrotoxicity | 0 (0%) | 0 (0%) | 0 (0%) | 1 (3.3%) | 1 (3.3%) |
| Tacrolimus neurotoxicity | 0 (0%) | 0 (0%) | 1 (3.3%) | 0 (0%) | 1 (3.3%) |
| Interstitial nephritis | 1 (3.3%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (3.3%) |
| Pyelonephrosis | 1 (3.3%) | 1 (3.3%) | 0 (0%) | 0 (0%) | 2 (6.7%) |
| Biopsies (N, %) | 7 (23.3%) | 3 (10%) | 7 (23.3%) | 3 (10%) | 20 (6.6%) |
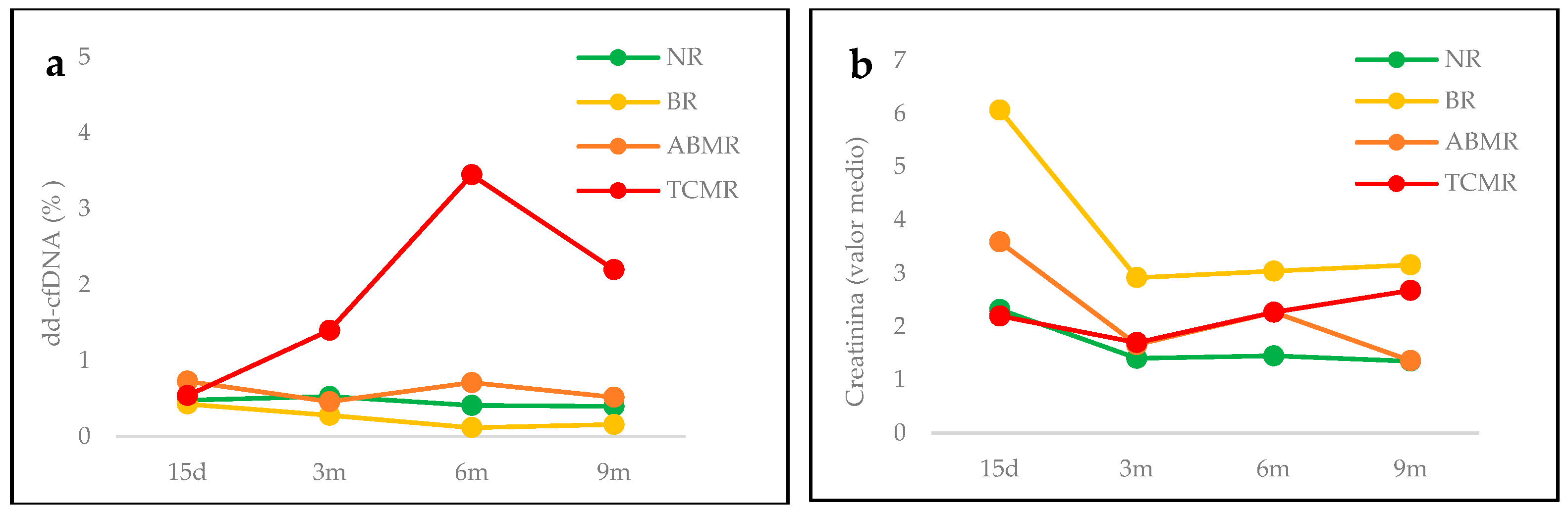
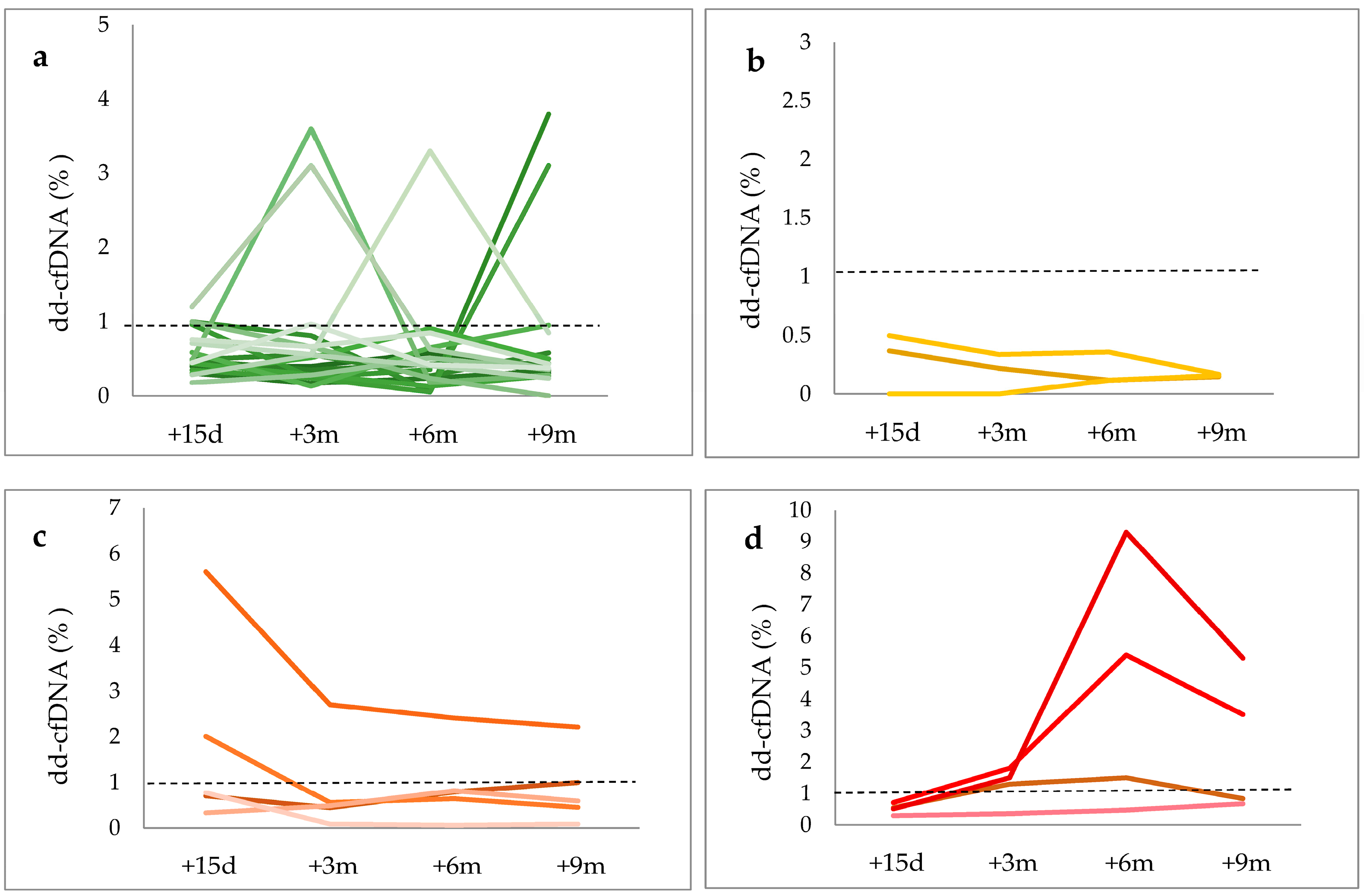
| dd-cfDNA (Average mean± SD, %) | 0.68 ± 0.79 | 0.35 ± 0.13 | 1.13 ± 1.31 | 2.12 ± 2.5 | 0.91 ± 1.32 |
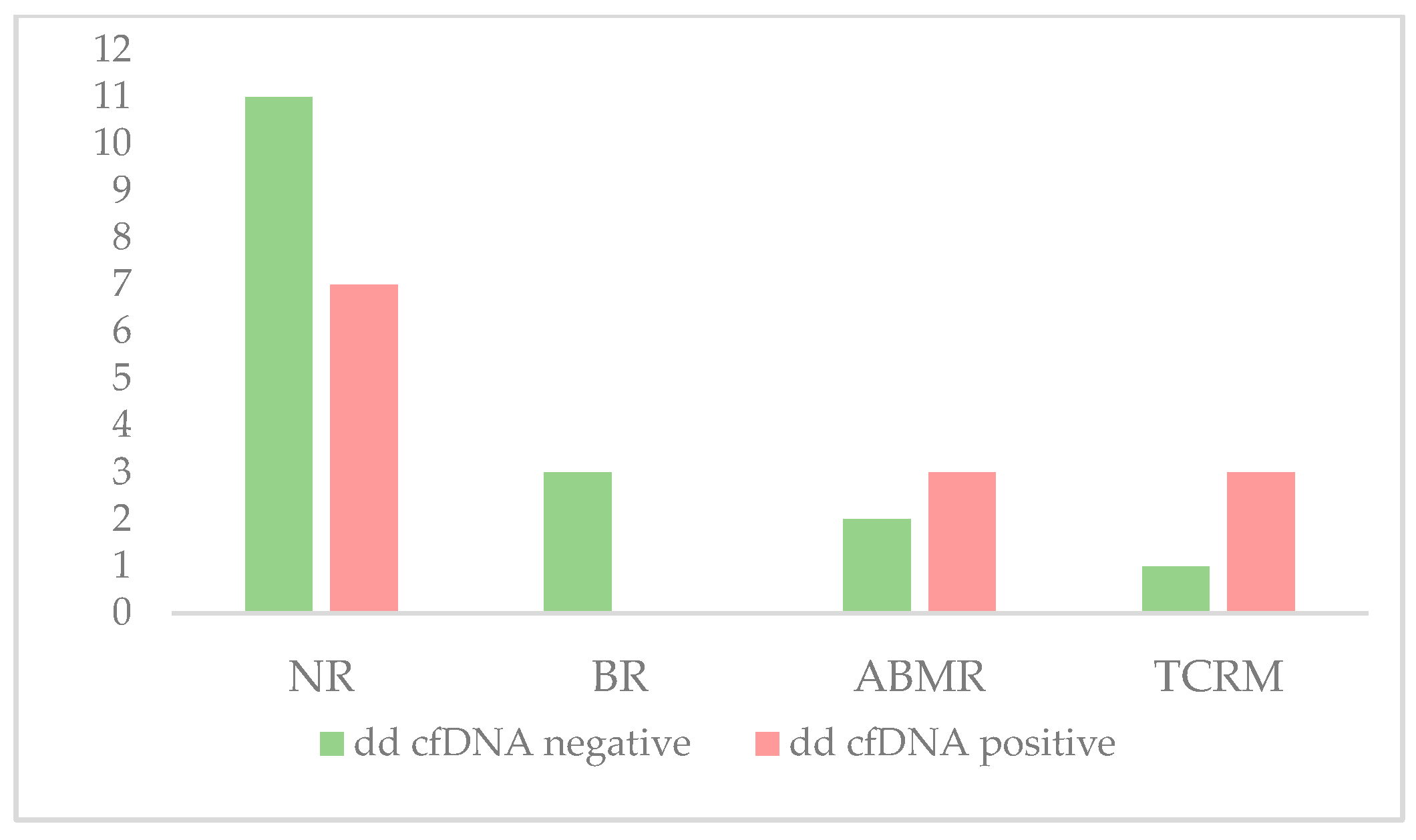
| dd-cfDNA < 1% (N, %) | 11 (36.6%) | 3 (10%) | 2 (6.7%) | 1 (3.3%) | 17 (56.6%) |
| dd-cfDNA ≥ 1% (N, %) | 7 (23.3%) | 0 (0%) | 3 (10%) | 3 (10%) | 13 (43.3%) |
| Creatinine (Average± SD, mg/mL) | 1.63 ± 1.05 | 3.8 ± 1.7 | 2.16 ± 1.47 | 2.3 ± 1.24 | 2.03 ± 1.38 |
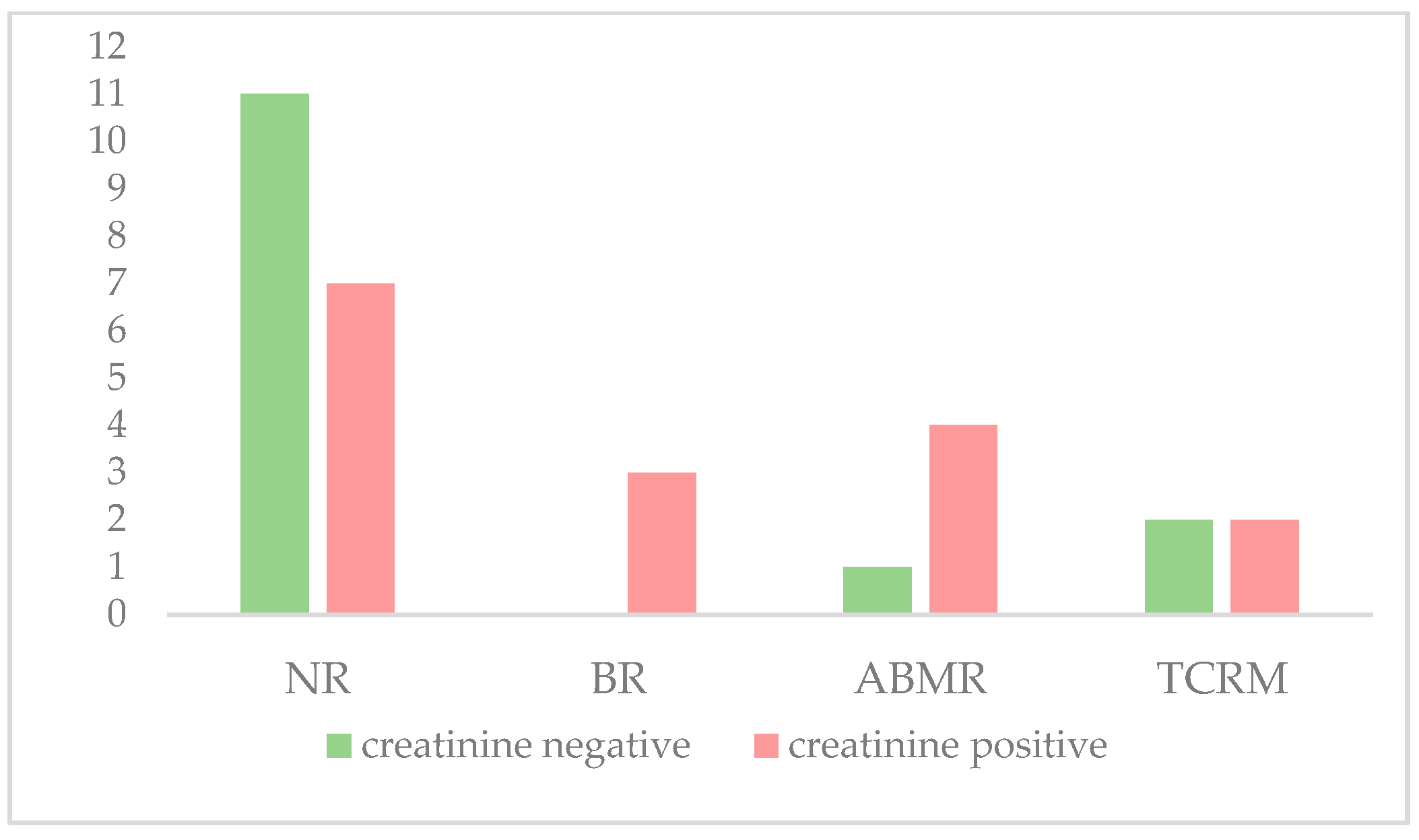
| Creatinine < 2 mg/mL | 11 (36.6%) | 0 (0%) | 1 (3.3%) | 2 (6.7%) | 14 (46.6%) |
| Creatinine ≥ 2 mg/ml | 7 (23.3%) | 3 (10%) | 4 (13.3%) | 2 (6.7%) | 16 (53.3%) |
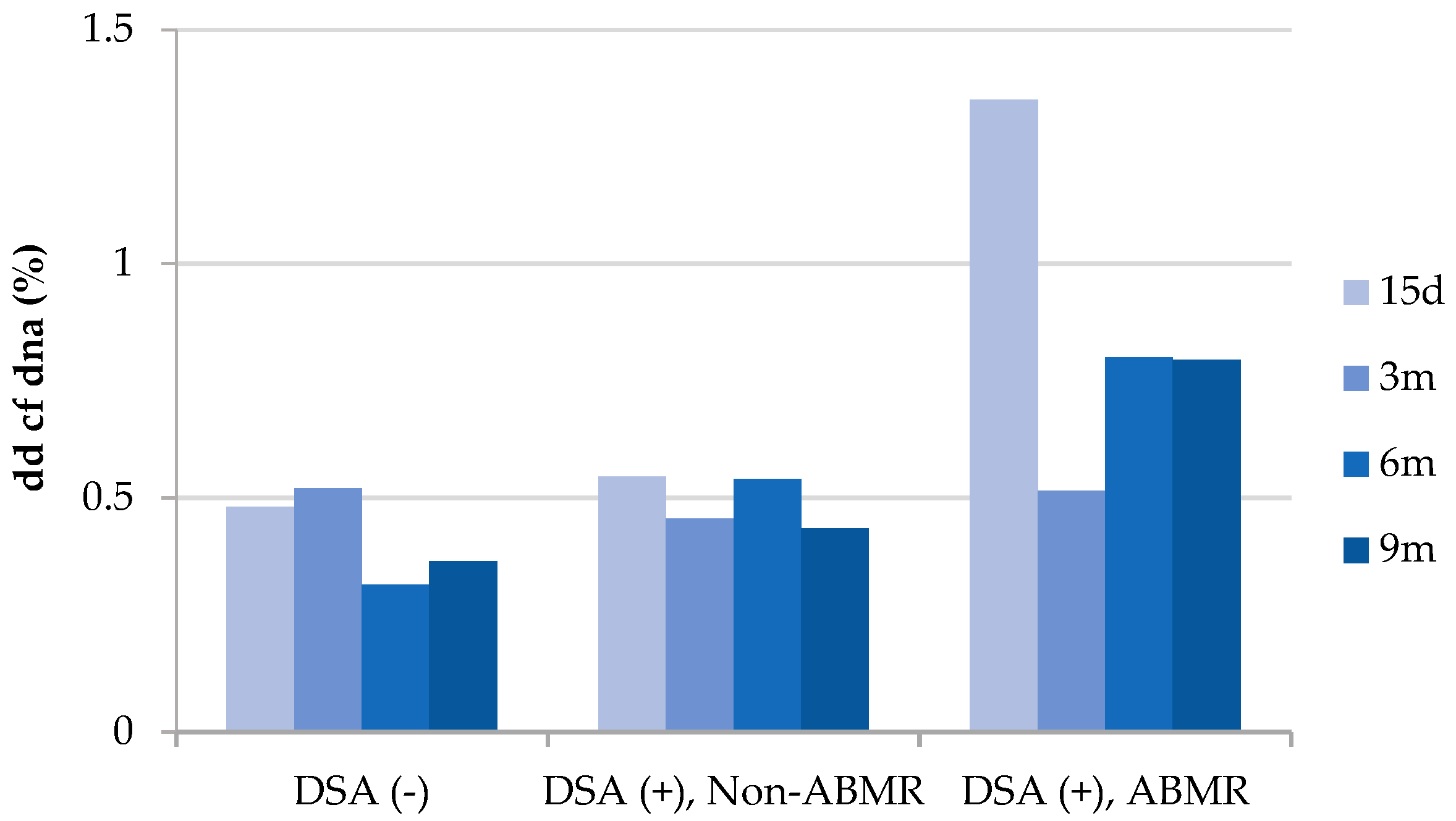
| Post-transplant anti-HLA DSAs (N, %) | |||||
| HLA-I | 1 (3.3%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (3.3%) |
| HLA-II | 4(13.3%) | 0 (0%) | 4 (13.3%) | 0 (0%) | 8 (26.7%) |
| HLA-I + HLA-II | 0 (0%) | 1 (3.3%) | 0 (0%) | 0 (0%) | 1 (3.3%) |
| Non-HLA antibodies | 0 (0%) | 0 (0%) | 1 (3.3%) | 0 (0%) | 1 (3.3%) |
| Banff | ID | cfDNA +15 d | cfDNA +3 m | cfDNA +6 m | cfDNA +9 m | Cr + 15 d | Cr + 3 m | Cr + 6 m | Cr + 9 m | Rejection Date |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | TX011 | 0.7 | 0.44 | 0.79 | 1 | 2.67 | 0.99 | 1.04 | 1.18 | 9 m |
| 2 | TX012 | 5.6 | 2.7 | 2.4 | 2.2 | 3.58 | 1.41 | 1.25 | 1.55 | 1 m |
| 2 | TX027 | 0.77 | 0.08 | 0.06 | 0.08 | 6.53 | 3.06 | 4.66 | 1.87 | 10 d |
| 2 | TX096 | 2 | 0.55 | 0.64 | 0.45 | 1.34 | 1.41 | 1.62 | 1.46 | 2 y |
| 2 | TX110 | 0.33 | 0.48 | 0.81 | 0.59 | 3.83 | 1.19 | 1.79 | 0.94 | 1 y 9 m |
| 4 | TX004 | 0.54 | 1.3 | 1.5 | 0.83 | 1.74 | 1.68 | 3.58 | 3.22 | 4 m |
| 4 | TX014 | 0.28 | 0.35 | 0.46 | 0.66 | 2.29 | 3.12 | 4.08 | 5.42 | 6 m |
| 4 | TX022 | 0.7 | 1.8 | 5.40 | 3.50 | 2.09 | 1.32 | 1.39 | 1.46 | 1 y 9 m |
| 4 | TX023 | 0.5 | 1.5 | 9.30 | 5.30 | 1.36 | 1.23 | 1.05 | 1.79 | 8 m |
| Rejection | BR | ABMR | TCRM | |
|---|---|---|---|---|
| Sensitivity (%) | 50 | 0 | 60 | 75 |
| Specificity (%) | 61.11 | 61.11 | 61.11 | 61.11 |
| PPV (%) | 46.15 | 0 | 30 | 30 |
| NPV (%) | 64.71 | 78.57 | 84.62 | 91.67 |
| Rejection | BR | ABMR | TCRM | |
|---|---|---|---|---|
| Sensitivity (%) | 75 | 100 | 80 | 50 |
| Specificity (%) | 61.11 | 61.11 | 61.11 | 61.11 |
| PPV (%) | 56.25 | 30 | 36.36 | 22.22 |
| NPV (%) | 78.57 | 100 | 91.67 | 8462 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Botella, C.; Galián, J.A.; Jiménez-Coll, V.; Fernández-González, M.; Morales, F.; Martínez-Gómez, G.; González-López, R.; Alegría, M.J.; Moya, M.R.; Martinez-Banaclocha, H.; et al. Early Monitoring of Donor-Derived Cell-Free DNA in Kidney Allograft Recipients Followed-Up for Two Years: Experience of One Center. Life 2024, 14, 1491. https://doi.org/10.3390/life14111491
Botella C, Galián JA, Jiménez-Coll V, Fernández-González M, Morales F, Martínez-Gómez G, González-López R, Alegría MJ, Moya MR, Martinez-Banaclocha H, et al. Early Monitoring of Donor-Derived Cell-Free DNA in Kidney Allograft Recipients Followed-Up for Two Years: Experience of One Center. Life. 2024; 14(11):1491. https://doi.org/10.3390/life14111491
Chicago/Turabian StyleBotella, Carmen, José Antonio Galián, Víctor Jiménez-Coll, Marina Fernández-González, Francisco Morales, Gloria Martínez-Gómez, Rosana González-López, María José Alegría, María Rosa Moya, Helios Martinez-Banaclocha, and et al. 2024. "Early Monitoring of Donor-Derived Cell-Free DNA in Kidney Allograft Recipients Followed-Up for Two Years: Experience of One Center" Life 14, no. 11: 1491. https://doi.org/10.3390/life14111491
APA StyleBotella, C., Galián, J. A., Jiménez-Coll, V., Fernández-González, M., Morales, F., Martínez-Gómez, G., González-López, R., Alegría, M. J., Moya, M. R., Martinez-Banaclocha, H., Minguela, A., Legaz, I., Llorente, S., & Muro, M. (2024). Early Monitoring of Donor-Derived Cell-Free DNA in Kidney Allograft Recipients Followed-Up for Two Years: Experience of One Center. Life, 14(11), 1491. https://doi.org/10.3390/life14111491











