Escherichia coli and Enterobacteriaceae Counts, Virulence Gene Profile, Antimicrobial Resistance, and Biofilm Formation Capacity during Pig Slaughter Stages
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Origin
2.2. Enterobacteriaceae and E. coli Count
2.3. E. coli Isolation
2.4. Isolate Antibiogram
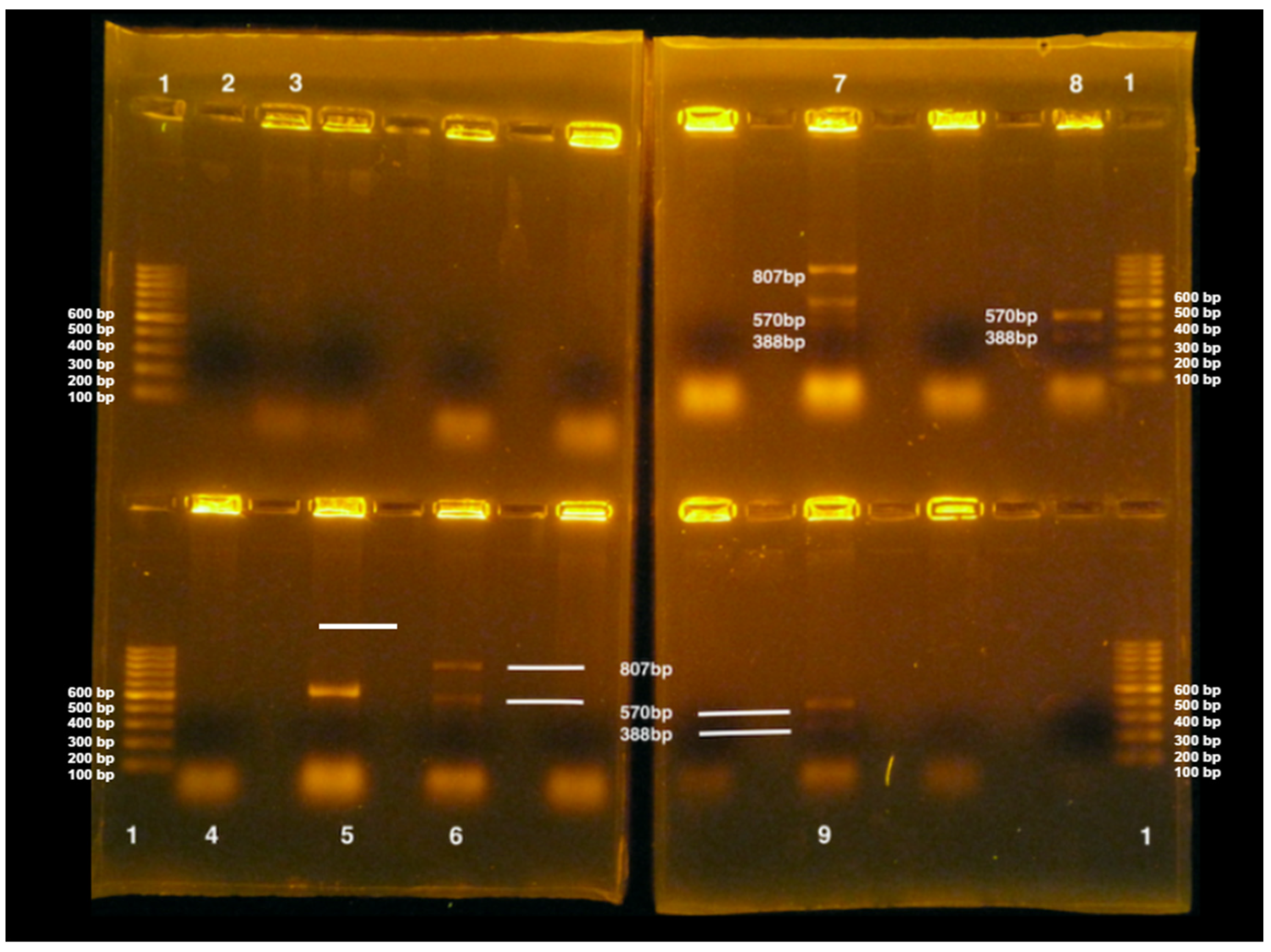
2.5. E. coli Virulence Gene Detection
2.6. In Vitro Biofilm Formation Capacity Evaluation
3. Results and Discussion
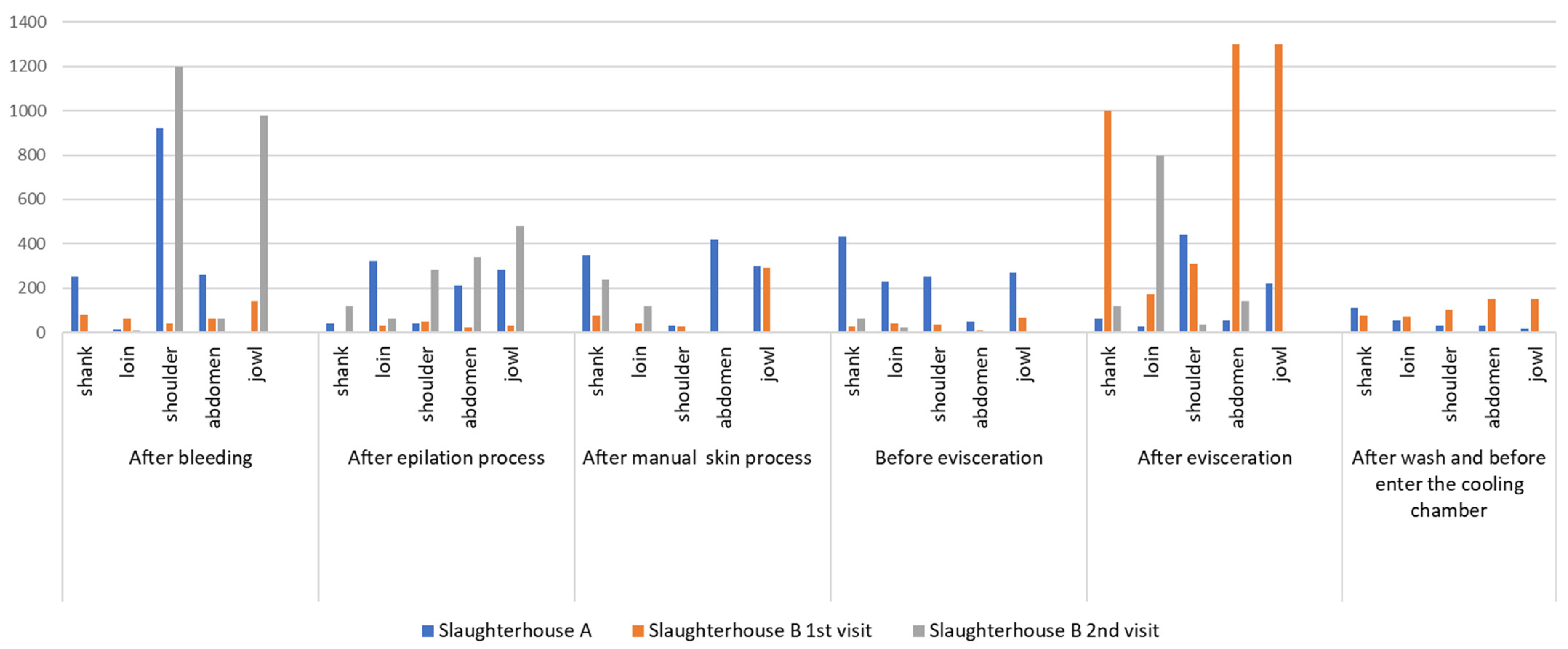
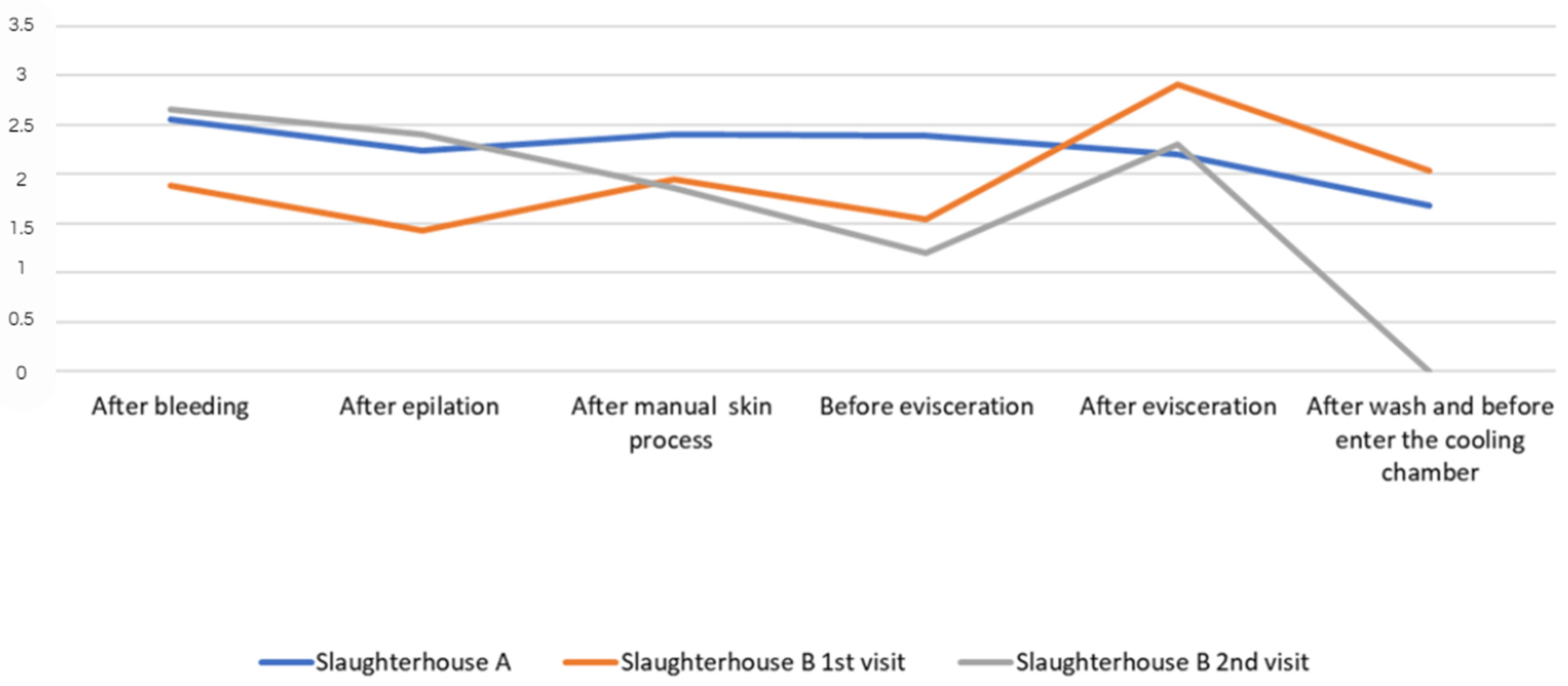
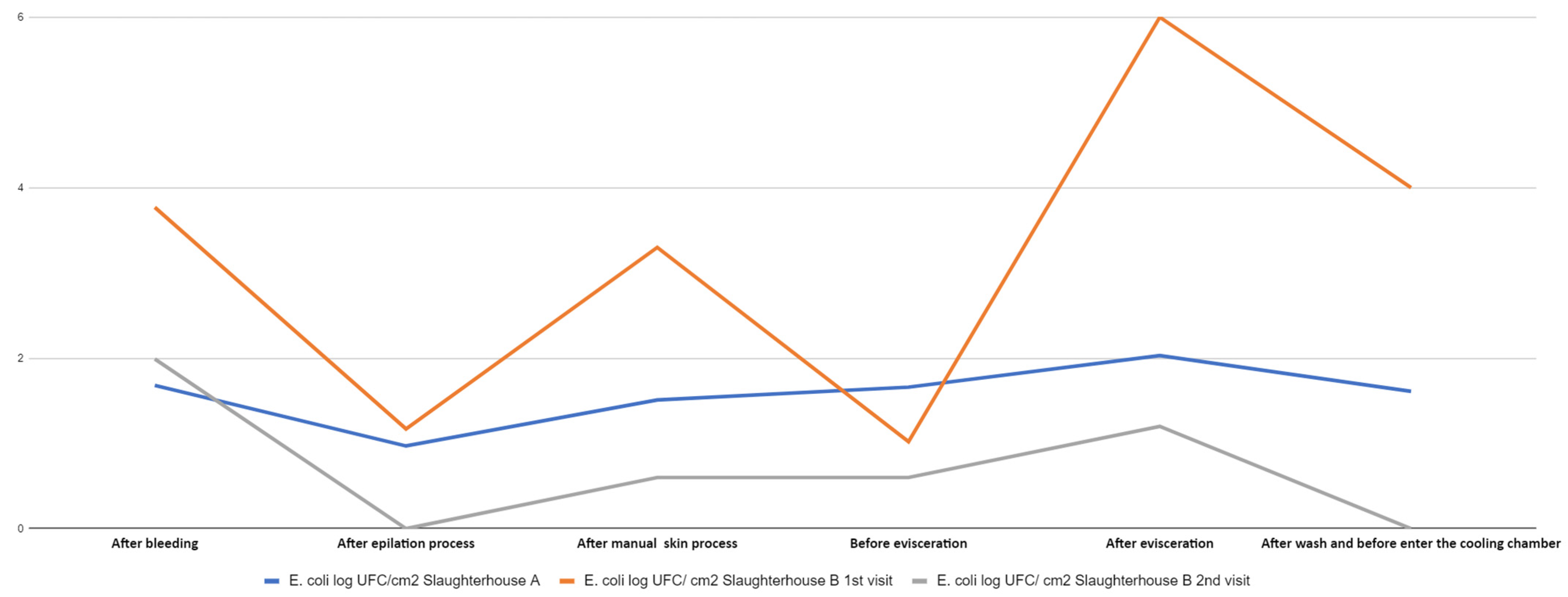
3.1. Enterobacteriaceae and E. coli Count
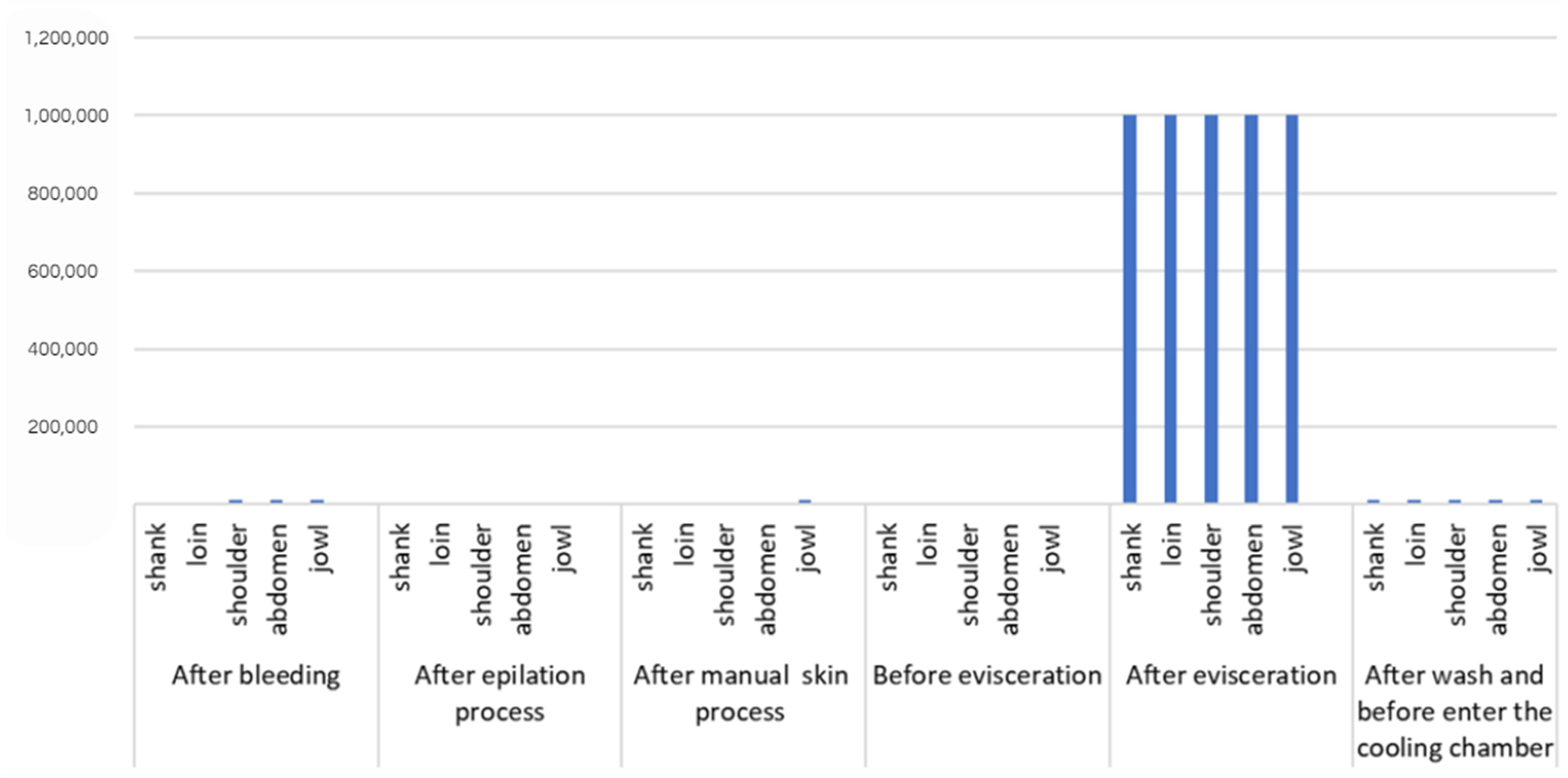
3.2. E. coli Counts
3.3. E. coli Isolates
3.4. Antibiogram of E. coli Isolates
3.5. Eae, stx1, and stx2 Virulence Gene Detection in E. coli
3.6. In Vitro Biofilm Formation Capacity Evaluation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Associação Brasileira de Proteína Animal. Relatório Anual. 2021. Available online: https://abpa-br.org/abpa-relatorio-anual/ (accessed on 3 February 2020).
- Instituto Brasileiro de Geografia e Estatística. Produção da Pecuária Municipal 2020. Rio de Janeiro. 2021. Available online: http://www.cidades.ibge.gov.br/brasil/df/brasilia/pesquisa/18/16459 (accessed on 13 June 2020).
- Morés, N.; Amaral, A.L.; Lima, G.J.M.M.; Costa, O.A.D.; Miele, M. Produção de Suínos em Família Sem Uso Coletivo de Antimicrobianos, 1st ed.; 2018. Available online: https://www.embrapa.br/busca-de-publicacoes/-/publicacao/1093916/producao-de-suinos-em-familia-sem-uso-coletivo-de-antimicrobianos (accessed on 5 July 2021).
- Wilbert, C.A.; Mores, N.; Klein, C.H.; Lima, G.J.M.M.; Bassi, N.S.S. Sistema de Produção de Suínos em Família Sem o Uso Coletivo de Antimicrobianos—Regulamento. Embrapa Suínos Aves 2019, 557, 6. Available online: https://www.embrapa.br/busca-de-publicacoes/-/publicacao/1102053/sistema-de-producao-de-suinos-em-familia-sem-o-uso-coletivo-de-antimicrobianos-regulamento (accessed on 9 February 2023).
- Lima, G.J.M.M. Estratégias no auxílio da redução do uso de antimicrobianos na produção de suínos. Rev. Científica Produção Anim. 2019, 20, 31–37. [Google Scholar]
- Neitzke, D.C.; Roza, C.R.; Weber, F.H. Segurança dos alimentos: Contaminação por Salmonella no abate de suínos. Campinas 2017, 20, e2015062. [Google Scholar] [CrossRef]
- Food and Agriculture Organization/World/World Health Organization (WHO) Guidelines for Risk Analysis of Foodborne Antimicrobial Resistance. 2011. Available online: https://www.fao.org/fao-who-codexalimentarius/sh-proxy/en/?lnk=1&url=https%253A%252F%252Fworkspace.fao.org%252Fsites%252Fcodex%252FStandards%252FCXG%2B77-2011%252FCXG_077e.pdf (accessed on 3 August 2021).
- Samulak, R.L.; Zanetti, G.F.; Rodrigues, S.A.; Bittencourt, J.V.M. Condição higiênico–sanitária de abatedouro frigorífico e fábrica de embutidos no estado do Paraná. Rev. Bras. Tecnol. Agroind. 2011, 5, 408–417. [Google Scholar] [CrossRef]
- Abebe, E.; Gugsa, G.; Ahmed, M. Review on major food-borne zoonotic bacterial pathogens. J. Trop Med. 2020, 2020, 4674235. [Google Scholar] [CrossRef]
- Newell, D.G.; Koopmans, M.; Verhoef, L.; Duizer, E.; Aidara-Kane, A.; Sprong, H.; Opsteegh, M.; Langelaar, M.; Threfall, J.; Scheutz, F.; et al. Foodborne diseases: The challenges of 20 years ago persist, while new ones continue to emerge. Int. J. Food Microbiol. 2012, 139, s3–s15. [Google Scholar] [CrossRef]
- Whitham, H.K.; Sundararaman, P.; Dewey-Matti, D.; Manikonda, K.; Marshall, K.; Griffin, P.M.; Gleason, B.L.; Subramhanya, S.; Crowe, S.J. Novel Outbreak-Associated Food Vehicles, United States. Emerg. Infect. Dis. 2021, 27, 2554–2559. [Google Scholar] [CrossRef]
- Brasil. Ministério da Agricultura, Pecuária e Abastecimento. Instrução Normativa n° 60, de 20 de Dezembro de 2018. Estabelece o Controle Microbiológico em Carcaça de Suínos e em Carcaça e Carne de Bovinos em Abatedouros Frigoríficos, Registrados No Departamento de Inspeção de Produtos de Origem Animal (DIPOA), Com Objetivo de Avaliar a Higiene do Processo e Reduzir a Prevalência de Agentes Patogênicos. 2018. Available online: https://www.gov.br/agricultura/pt-br/acesso-a-informacao/participacao-social/consultas-publicas/documentos/Portaria7518ConsultapblicaINmicrobiologiacarcaas.pdf (accessed on 17 October 2022).
- Brasil. Ministério da Saúde. Portaria GM/MS N° 888, de 4 de Maio de 2021. Altera o Anexo XX da Portaria de Consolidação n° 5/GM/MS, de 28 de Setembro de 2017, Para Dispor Sobre os Procedimentos de Controle e de Vigilância da Qualidade da Água Para Consumo Humano e Seu Padrão de Potabilidade. 2021. Available online: https://bvsms.saude.gov.br/bvs/saudelegis/gm/2021/prt0888_24_05_2021_rep.html (accessed on 30 November 2023).
- Corbellini, L.G.; Júnior, A.B.; Costa, E.F.; Duarte, A.S.; Albuquerque, E.R.; Kich, J.D.; Cardoso, M.; Nauta, M. Effect of slaughterhouse and day of sampling on the probability of a pig carcass being Salmonella-positive according to the Enterobacteriaceae count in the largest Brazilian pork production region. Int. J. Food Microbiol. 2016, 2, 58–66. [Google Scholar] [CrossRef]
- Barco, L.; Belluco, S.; Roccato, A.; Ricci, A. Escherichia coli and Enterobacteriaceae counts on pig and ruminant carcasses along the slaughterline, factors influencing the counts and relationship between visual fecal contamination of carcasses and counts: A review. EFSA Support. Publ. 2014, 11, 634E. [Google Scholar] [CrossRef]
- World (WHO). E. coli. Fact Sheets. 2018. Available online: https://www.who.int/news-room/fact-sheets/detail/e-coli (accessed on 3 February 2020).
- Viltrop, A.; Niine, T.; Tobias, T.; Sassu, E.L.; Bartolo, I.D.; Pavoni, E.; Alborali, G.L.; Burow, E.; Smith, R.P. Slaughter practices and their effectiveness in controlling microbial species Salmonella Contamination of Pig Carcasses. J. Food Prot. 2023, 86, 100171. [Google Scholar] [CrossRef] [PubMed]
- Pissetti, C.; Werlang, G.O.; Biesus, L.L.; Kich, J.D.; Cardoso, M.R.d.I. Detecção de Salmonella entericae Listeria monocytogenes em carcaças suínas na etapa de pré-resfriamento. Acta Sci. Vet. 2002, 40, 1–8. [Google Scholar]
- Zero, R.C.; Rodrigues, J.O. Salmonella: Riscos, transmissão e controle na cadeia de produção suína–revisão da literatura. Nucl. Anim. 2017, 9, 129–141. [Google Scholar] [CrossRef][Green Version]
- Paim, D.S.; Pissetti, C.; Vieira, T.R.; Werlang, G.O.; Costa, E.F.; Kich, J.D.; Cardoso, M. Enumeration, Antimicrobial Resistance and Typing of Salmonella enterica: Profile of Strains Carried in the Intestinal Contents of Pigs at Slaughter in Southern Brazil. Acta Sci. Vet. 2019, 47, 1636. [Google Scholar] [CrossRef]
- Viana, C.; Grossi, J.L.; Sereno, M.J.; Yamatogi, R.S.; Bersot, L.S.; Call, D.R.; Nero, L.A. Phenotypic and genotypic characterization of non-typhoidal Salmonella isolated from a Brazilian pork production chain. Food Res. Int. 2020, 137, 109406. [Google Scholar] [CrossRef]
- Rodrigues, G.R.; Carvalho, I.R.; Gandra, E.E.A.; Carbonera, N.; Demari, G.H.; Demari, A.C.; Lautenchleger, F.; Moura, N.B.; Martins, T.S.; Scheer, E.H.; et al. Reflexes and microbiological interrelationships during pork processing and bases. Braz. J. Dev. 2021, 7, 25395–25430. [Google Scholar] [CrossRef]
- Rizzotto, D.; Montes, J.H.; Kich, J.D.; Pepipolli, V.; Bianchi, I.; Oliveira Júnior, J.M.; Duval, E.H.; Schewegler, E.; Moreira, F. Salmonella enterica and enterobacteria in pig carcasses processed on different days after slaughter Pesq. Agropec. Bras. 2022, 57, e02813. [Google Scholar] [CrossRef]
- Pinto, A.P.S. Swine carcass microbiological evaluation, hazard analysis, and critical control points (HACCP) in a slaughterhouse in Minas Gerais, Brazil. Rev. Soc. Ven. Microbiol. 2004, 24, 83–88. [Google Scholar]
- Costa, E.F.; Cardoso, M.; Kich, J.D.; Corbellini, L.G. Qualitative risk assessment approach for foodborne microbial hazards. Microb. Risk Anal. 2020, 15, 1000105. [Google Scholar] [CrossRef]
- Food and Agriculture Organization/World Health Organization (FAO/WHO). Risk Assessment of Microbiological Hazards in Food. 1999. Available online: https://apps.who.int/iris/handle/10665/65973 (accessed on 6 March 2023).
- Davanzo, E.F.A.; Santos, R.L.; Castro, V.H.L.; Palma, J.M.; Pribul, B.R.; Dallago, B.S.L.; Fuga, B.; Medeiros, M.; Almeida, S.S.T.; Costa, H.M.B.; et al. Molecular characterization of Salmonella spp. and Listeria monocytogenes strains from biofilms in cattle and poultry slaughterhouses located in the federal District and State of Goiás, Brazil. PLoS ONE 2021, 16, e0259687. [Google Scholar] [CrossRef]
- Matsubara, E.N. Condição Higiênico-Sanitária de Meias-Carcaças de Suínos Após o Abate e Depois do Resfriamento e Análise da Utilização de Lista de Verificação Para Avaliar Boas Práticas no Abate de Suínos. Master’s Thesis, University of São Paulo, São Paulo, Brazil, 2005. Available online: https://www.teses.usp.br/teses/disponiveis/10/10134/tde-30102006-113224/publico/EstherNaomiMatsubara.pdf (accessed on 3 February 2023).
- Brasil. Ministério da Agricultura, Pecuária e Abastecimento. Manual de Coleta de Amostras de Produtos de Origem Animal. 2017. Available online: https://www.gov.br/agricultura/pt-br/assuntos/inspecao/produtos-animal/anuario-dos-programas-de-controle-de-alimentos-de-origem-animal-do-dipoa/manual-de-coleta-de-amostras-de-produtos-de-origem-animal.pdf (accessed on 2 October 2022).
- Cê, E.R. Influências das Etapas do Processo de Abate de Suínos na Prevalência de Patógenos e Níveis de Microrganismos Indicadores de Qualidade e Higiene. Master’s Thesis, Universidade Tecnológica do Paraná, Curitiba, Brazil, 2016. Available online: https://repositorio.utfpr.edu.br/jspui/handle/1/1665 (accessed on 5 December 2023).
- Silva, N.; Junqueira, V.C.A.; Silveira, N.F.A.; Tanowaki, M.H.; Gomes, R.A.R.; Okazaki, M.M. Manual de Métodos de Análise Microbiológica de Alimentos e Água, 5th ed.; Blucher: São Paulo, Brazil, 2017. [Google Scholar]
- ABNT NBR ISO 21528-2:2020; Microbiologia da Cadeia Produtiva de Alimentos—Método Horizontal Para Detecção e Enumeração de Enterobacteriaceae Parte 2: Método de Contagem de Colônias. 1st ed. Associação Brasileira de Normas Técnicas (ABNT): Rio de Janeiro, Brazil, 2020.
- M02-A11; Performance Standards for Antimicrobial Disc Susceptibility Tests: CLSI Document M02-A11. 11th ed. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2012. Available online: https://www.researchgate.net/file.PostFileLoader.html?id=58139aa4615e27240754da03&assetKey=AS%3A422233756704774%401477679780485 (accessed on 9 February 2023).
- CLSI M100-ED31:2021; Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. CLSI Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. Available online: http://em100.edaptivedocs.net/GetDoc.aspx?doc=CLSI%20M100%20ED32:2022&sbssok=CLSI%20M100%20ED32:2022%20TABLE%202A&format=HTML#CLSI%20M100%20ED32:2022%20TABLE%202A (accessed on 3 August 2023).
- China, B.; Pirson, V.; Mainil, J. Typing of Bovine Attaching and Effacing Escherichia coli by Multiplex In Vitro Amplification of Virulence-Associated Genes. Appl. Environ. Microbiol. 1996, 62, 3462–3465. [Google Scholar] [CrossRef]
- Djordjevic, D.; Wiedmann, M.; McLandsborough, L.A. Microtiter plate assay for Assessment of Listeria monocytogenes Biofilm Formation. Appl. Environ. Microbiol. 2002, 68, 2950–2958. [Google Scholar] [CrossRef] [PubMed]
- Borges, K.A.; Furian, T.Q.; Sousa, S.S.; Menezes, R.; Tondo, E.C.; Salle, C.T.P.; Moraes, H.L.S.; Nascimento, V.P. Biofilm-forming capacity of Salmonella serotypes at different temperatures. Pesq. Vet. Bras. 2018, 38, 71–76. [Google Scholar] [CrossRef]
- Stepanovic, S.; Vukovic, D.; Dakic, I.; Savic, B.; Svabic-Vlahovic, M. A modified microtiter plate test was used to quantify Staphylococcal biofilm formation. J. Microbio. Methods. 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Wheatley, P.; Giotis, E.S.; McKevitt, A.I. Effects of slaughter on carcass contamination at an Irish pork production plant. Ir. Vet. J. 2014, 67, 1. [Google Scholar] [CrossRef]
- Bolton, D.J.; Pearce, R.A.; Sheridan, J.J.; Blair, I.S.; McDowell, D.A.; Harrigton, D. Washing and chilling as critical control points in pork slaughter hazard analysis and critical control point (HACCP) systems. J. Appl. Microbiol. 2002, 92, 893–902. [Google Scholar] [CrossRef]
- Brasil. Ministério da Agricultura, Pecuária e Abastecimento. Portaria n° 711, de 1° de Novembro de 1995. Aprova as Normas Técnicas de Instalações e Equipamentos Para Abate e Industrialização de Suínos. 1995. Available online: https://www.gov.br/agricultura/pt-br/assuntos/inspecao/produtos-animal/empresario/arquivos/Portaria7111995alteradaportarian13042018.pdf/view (accessed on 5 January 2023).
- Montes, J.H.; Rozzoto, D.W.; Bianchi, I.; Oliveira, J.M.; Peripolli, V.; Moreira, F. Contaminação por Enterobactérias em Carcaças Suínas ao Longo da Linha de Abate. In Mostra Nacional de Iniciação Científica e Tecnologia Interdisciplinar—XII MICTI—IFC Campus Brusque. 2019. Available online: https://www.google.com/search?client=safari&rls=en&q=.+Contamina%C3%A7%C3%A3o+por+enterobact%C3%A9rias+em+carca%C3%A7as+su%C3%ADnas+ao+longo+da+linha+de+abate.&ie=UTF-8&oe=UTF-8 (accessed on 22 March 2022).
- Spescha, C.; Stephan, R.; Zweifel, C. Microbiological contamination of pig carcasses at different stages of slaughter in two European Union-proven abattoirs. J. Food Prot. 2006, 69, 2568–2575. [Google Scholar] [CrossRef]
- Warriner, K.; Aldsworth, T.G.; Kaur, S.; Dodd, C.E. Cross-contamination of carcasses and equipment during pork processing. J. Appl. Microbiol. 2002, 93, 169–177. [Google Scholar] [CrossRef]
- Nastasijevic, I.; Lakicevic, B.; Raseta, M.; Djordjevic, V.Z.; Jankovic, V.; Mrdovic, B.; Brankovic-Lazic, I. Evaluation of pig welfare in a lairage and process hygiene in a single abattoir. Sci. J. Meat Technol. Quot. 2018, 59, 8–22. [Google Scholar] [CrossRef]
- Zwirzitz, B.; Wetzels, S.U.; Dixon, E.D.; Stessl, B.; Zaiser, A.; Rabanser, I.; Thalguter, S.; Pinior, B.; Roch, F.F.; Strachan, C.; et al. Sources and transmission routes of microbial populations throughout meat processing facilities. J. Biofilm Microbiome. 2020, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Buncic, S.; Sofos, J. Interventions to control Salmonella contamination during poultry, cattle, and pig slaughter. Food Res. Int. 2012, 45, 641–655. [Google Scholar] [CrossRef]
- Velebit, B.; Lakicevic, B.; Semenova, A.A.; Revutskaya, N.M.; Yushina, Y.K.; Nasonova, V.V. Factors Influencing Microbial Transmission in Meat Processing Plants. Theor. Pract. Meat Process. 2021, 6, 183–190. [Google Scholar] [CrossRef]
- Ghafir, Y.; China, B.; Dierick, K.; Zutter, L.; Daube, G. Hygiene indicator microorganisms for selected pathogens on beef, pork, and poultry meat in Belgium. J. Food Prot. 2008, 71, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Machado, L.A.P.; Lucca, F.; Alves, J.; Pozzobon, A.; Bustamante-filho, I.C. Prevalência e genotipagem de Escherichia coli patogênica em carcaças de suínos abatidos em frigoríficos comerciais na Região Sul do Brasil. Rev. Bras. Hig. Sanidade Anim. 2014, 8, 128. [Google Scholar]
- Mangal, P.; Rao, R.; Joshi, R. Isolation, identification, and antibiotic sensitivity pattern of Escherichia coli in raw pork: A cross-sectional study. J. Entomol. Zool. Stud. 2018, 6, 395–398. [Google Scholar]
- Drummond, V.O.; Perecmanis, S. Genes de enterotoxinas e perfil antimicrobiano de Escherichia coli isoladas de suínos hígidos no Distrito Federal. Arq. Bras. Med. Veter-E Zootec. 2013, 54, 1005–1009. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant, and pandrug-resistant bacteria: An international expert proposal for interim standard definitions of acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Pissetti, C.; Werlang, G.O.; Kich, J.D.; Cardoso, M. Genotyping and antimicrobial resistance in Escherichia coli from pig carcasses. Pesqui. Veter-Bras. 2017, 37, 1253–1260. [Google Scholar] [CrossRef]
- Kim, H.; Baek, H.; Lee, S.; Jang, Y.; Jung, S.; Kim, A.; Choe, N.N. Prevalence and antimicrobial resistance of Salmonella spp. and Escherichia coli isolated from pigs at slaughterhouses in Korea. Afr. J. Microbiol. Res. 2011, 15, 823–830. [Google Scholar] [CrossRef][Green Version]
- Bischoff, K.M.; White, D.G.; Hume, M.E.; Poole, T.L.; Nisbet, D.J. The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple drug resistance in swine Escherichia coli. FEMS Microbiol. Lett. 2005, 243, 285–291. [Google Scholar] [CrossRef]
- Moennighoff, C.; Thomas, N.; Niehaus, F.; Hartmann, M.; Menrath, A.; Merkel, J.; Detlefsen, H.; Kreienbrock, L.; Hennig-Pauka, I. Phenotypic antimicrobial resistance in Escherichia coli strains isolated from swine husbandry in Northwestern Germany: Temporal patterns in samples from laboratory practice from 2006 to 2017. BMC Vet. Res. 2020, 16, 37. [Google Scholar] [CrossRef]
- World Health Organisation (WHO). Antimicrobial Resistance in the Food Chain. Food Safety. 2017. Available online: https://www.who.int/news-room/questions-and-answers/item/antimicrobial-resistance-in-the-food-chain (accessed on 9 October 2020).
- Bouvet, J.; Montet, M.P.; Rossel, R.; Le Roux, A.; Bavai, C.; Ray-Gueniot, S.; Mazuy, C.; Atrache, V.; Vernozy-Rozand, C. Effects of slaughter on pig carcass contamination with verotoxin-producing Escherichia coli and E. coli O157:H7. Int. J. Food Microbiol. 2002, 77, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Paton, A.W.; Paton, J.C. Detection and characterization of Shiga toxigenic Escherichia coli using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 1998, 36, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Brasil. Ministério da Agricultura, Pecuária e Abastecimento. Instrução Normativa n° 09, de 27 de Junho de 2003. Proíbe a Fabricação, a Manipulação, o Fracionamento, a Comercialização, a Importação e o Uso Dos Princípios Ativos Cloranfenicol Nitrofuranos e os Produtos Que Contenham Estes Princípios Ativos, Para Uso Veterinário e Suscetível de Emprego Na Alimentação de Todos os Animais e Insetos. 2003. Available online: https://www.gov.br/agricultura/pt-br/assuntos/insumos-agropecuarios/insumos-pecuarios/alimentacao-animal/arquivos-alimentacao-animal/legislacao/instrucao-normativa-no-9-de-27-de-junho-de-2003.pdf (accessed on 26 February 2023).
- Tseng, M.; Fratamico, P.M.; Manning, S.D. Shiga toxin-producing Escherichia coli in swine: A public health perspective. Anim. Health Res. Rev. 2014, 15, 63–75. [Google Scholar] [CrossRef]
- Chassagne, L.; Pradel, N.; Robin, F.; Livrelli, V.; Bonnet, R.; Delmas, J. Detection of stx1, stx2, and eae genes of enterohemorrhagic Escherichia coli using SYBR Green in a real-time polymerase chain reaction. Diag. Microb. Infec. Dis. 2009, 64, 98–101. [Google Scholar] [CrossRef]
- Zhang, P.; Essendoubi, S.; Keenliside, J.; Reuter, T.; Stanford, K.; King, R.; Lu, P.; Yang, X. Genomic analysis of Shiga toxin-producing Escherichia coli O157:H7 from cattle and pork production-related environments. NPJ Sci. Food. 2021, 5, 15. [Google Scholar] [CrossRef]
- Rosengren, L.B.; Waldner, C.L.; Reid-Smith, R.J. Associations between antimicrobial resistance phenotypes, antimicrobial resistance genes, and virulence genes in fecal Escherichia coli isolates from healthy grow-finishing pigs. Appl. Environ. Microbiol. 2009, 75, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.X.; Lu, D.H.; Chen, Z.L.; Wang, X.M.; Chen, J.R.; Liu, Y.H.; Liao, X.P.; Liu, J.H.; Zeng, Z.L. High prevalence and widespread distribution of multi-resistant Escherichia coli isolates in pigs and poultry in China. Vet. J. 2011, 187, 99–103. [Google Scholar] [CrossRef]
- Gemeda, B.A.; Assefa, A.; Jaleta, M.B.; Amenu, K.; Wieland, B. Antimicrobial resistance in Ethiopia: A systematic review and meta-analysis of prevalence in foods, food handlers, animals, and the environment. One Health 2021, 13, 100286. [Google Scholar] [CrossRef]
- Tadesse, D.A.; Zhao, S.; Tong, E.; Ayers, S.; Singh, A.; Bartholomew, M.J.; McDermott, P.F. Antimicrobial Drug Resistance in Escherichia coli from Humans and Food Animals, United States, 1950–2002. Emerg. Inf. Dis. 2012, 18, 741–749. [Google Scholar] [CrossRef]
- Milan, C.; Timm, C.D. Avaliação da capacidade de formar biofilme de Escherichia coli produtoras de shiga toxina. Arq. Ciências Veterinárias Zool. UNIPAR 2013, 16, 31–33. [Google Scholar]
- Sanchez, C.J.; Mende, K.; Beckius, K.M.; Akers, K.S.; Romano, D.R.; Wenke, J.C.; Murray, C.K. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infec. Dis. 2013, 3, 47. [Google Scholar] [CrossRef] [PubMed]

| Gene | Sequence of Nucleotides (5′-3′) | Size (pb) | Annealing Temperature (°C) | Reference |
|---|---|---|---|---|
| Stx-1 | F-AGA GCG ATG TTA CGG TTT G R-TTG CCC CCA GAG TGG ATG | 388 | 50 | [35] |
| Stx-2 | F-TGG GTT TTT CTT CGG TAT C R-GAC ATT CTG GTT GAC TCT CTT | 807 | 50 | [35] |
| eae | F-AGG CTT CGT CAC AGT TG R-CCA TCG TCA CCA GAG GA | 570 | 50 | [35] |
| Slaughter Technological Processing Stage | Visits (Slaughterhouse A and B) | Microorganism | Carcass Part/Point (Count in CFU/cm2) | Average Carcass Count (log CFU/cm2) | ||||
|---|---|---|---|---|---|---|---|---|
| 1 Shank | 2 Loin | 3 Shoulder | 4 Abdomen | 5 Jowl | ||||
| After bleeding | 1st visit (slaughterhouse A) | Enterobacteriaceae | 2.5 × 102 2.3log | 1.3 × 101 1.1log | 9.2 × 102 2.9log | 2.6 × 102 2.4log | US* 0 | 2.55 |
| E. coli | 1.0 × 102 2log | 0.2 × 101 0.3log | 7.0 × 101 1.84log | 2.1 × 101 1.3log | US* 0 | 1.68 | ||
| 1st visit (slaughterhouse B) | Enterobacteriaceae | 8.0 × 101 1.9log | 6.1 × 101 1.7log | 4.0 × 101 1.6log | 6.0 × 101 1.7log | 1.4 × 102 2.1log | 1.88 | |
| E. coli | 0.4 × 101 0.6log | 0.3 × 101 0.4log | 104 4log | 104 4log | 104 4log | 3.77 | ||
| 2nd visit (slaughterhouse B) | Enterobacteriaceae | NG** 0 | 0.8 × 101 0.9log | 1.2 × 103 3.07log | 0.6 × 102 1.7log | 9.8 × 102 2.9log | 2.65 | |
| E. coli | NG** 0 | NG** 0 | 2.8 × 102 2.4log | NG** 0 | 2.1 × 102 2.3log | 1.99 | ||
| Slaughter Technological Processing Stage | Visits (Slaughterhouse A and B) | Microorganism | Carcass Part/Point (Count in CFU/cm2) | Average Carcass Count (log CFU/cm2) | ||||
|---|---|---|---|---|---|---|---|---|
| 1 Shank | 2 Loin | 3 Shoulder | 4 Abdomen | 5 Jowl | ||||
| After epilation process | 1st visit (slaughterhouse A) | Enterobacteriaceae | 3.9 × 101 1.5log | 3.2 × 102 2.5log | 3.8 × 101 1.5log | 2.1 × 102 2.3log | 2.8 × 102 2.4log | 2.24 |
| E. coli | 2.1 × 101 1.3log | 1.3 × 101 1.1log | 0.7 × 101 0.8log | 0.2 × 101 0.3log | 0.4 × 101 0.6log | 0.97 | ||
| 1st visit (slaughterhouse B) | Enterobacteriaceae | 0.4 × 101 0.6log | 2.9 × 101 1.4log | 4.9 × 101 1.6log | 2.0 × 101 1.3log | 3.3 × 101 1.5log | 1.43 | |
| E. coli | 1.3 × 101 1.1log | 1.2 × 101 1log | 1.9 × 101 1.2log | 0.9 × 101 0.9log | 2.1 × 101 | 1.17 | ||
| 2nd visit (slaughterhouse B) | Enterobacteriaceae | 1.2 × 102 2log | 6.0 × 101 1.7log | 2.8 × 102 2.4log | 3.4 × 102 2.5log | 4.8 × 102 2.6log | 2.40 | |
| E. coli | NG** | NG** | NG** | NG** | NG** | 0 | ||
| Slaughter Technological Processing Stage | Visits (Slaughterhouse A and B) | Microorganism | Carcass Part/Point (Count in CFU/cm2) | Average Carcass Count (log CFU/cm2) | ||||
|---|---|---|---|---|---|---|---|---|
| 1 Shank | 2 Loin | 3 Shoulder | 4 Abdomen | 5 Jowl | ||||
| After manual skin process | 1st visit (slaughterhouse A) | Enterobacteriaceae | 3.5 × 102 2.5log | US* 0 | 3.2 × 101 1.5log | 4.2 × 102 2.6log | 3.0 × 102 2.4log | 2.4 |
| E. coli | 9.8 × 101 1.9log | 1.0 × 101 1log | 0.9 × 101 0.9log | 1.3 × 101 1.1log | US* 0 | 1.51 | ||
| 1st visit (slaughterhouse B) | Enterobacteriaceae | 7.7 × 101 1.8log | 4.0 × 101 1.6log | 2.8 × 101 1.4log | 0.4 × 101 0.6log | 2.9 × 102 2.4log | 1.94 | |
| E. coli | 2.9 × 101 1.4log | 3.0 × 101 1.4log | 1.5 × 101 1.1log | 1.1 × 101 1log | 104 4log | 3.30 | ||
| 2nd visit (slaughterhouse B) | Enterobacteriaceae | 2.4 × 102 2.3log | 1.2 × 102 2log | NG** | 0.5 × 101 0.6log | NG** | 1.86 | |
| E. coli | NG** | NG** | 2.0 × 101 1.3log | NG** | NG** | 0.6 | ||
| Slaughter Technological Processing Stage | Visits (Slaughterhouse A and B) | Microorganism | Carcass Part/Point (Count in CFU/cm2) | Average Carcass Count (log CFU/cm2) | ||||
|---|---|---|---|---|---|---|---|---|
| 1 Shank | 2 Loin | 3 Shoulder | 4 Abdomen | 5 Jowl | ||||
| Before evisceration | 1st visit (slaughterhouse A) | Enterobacteriaceae | 4.3 × 103 2.6log | 2.3 × 102 2.3log | 2.5 × 102 2.3log | 5.0 × 101 1.6log | 2.7 × 102 2.4log | 2.39 |
| E. coli | 1.7 × 102 2.2log | 2.4 × 101 1.3log | NG** | 2.7 × 101 1.4log | 0.9 × 101 0.9log | 1.66 | ||
| 1st visit (slaughterhouse B) | Enterobacteriaceae | 2.7 × 101 31.4log | 3.9 × 101 1.5log | 3.6 × 101 1.5log | 0.7 × 101 0.8log | 6.6 × 101 1.8log | 1.54 | |
| E. coli | 1.1 × 101 1log | 1.3 × 101 1.1log | 1.4 × 101 1.1log | 0.7 × 101 0.8log | 0.8 × 101 0.9log | 1.02 | ||
| 2nd visit (slaughterhouse B) | Enterobacteriaceae | 0.6 × 101 1.7log | 2.0 × 101 1.3log | 0.1 × 101 0 | NG** | NG** | 1.2 | |
| E. coli | 2.0 × 101 1.3log | NG** | NG** | NG** | NG** | 0.6 | ||
| Slaughter Technological Processing Stage | Visits (Slaughterhouse A and B) | Microorganism | Carcass Part/Point (Count in CFU/cm2) | Average Carcass Count (log CFU/cm2) | ||||
|---|---|---|---|---|---|---|---|---|
| 1 Shank | 2 Loin | 3 Shoulder | 4 Abdomen | 5 Jowl | ||||
| After evisceration | 1st visit (slaughterhouse A) | Enterobacteriaceae | 6.2 × 101 1.7log | 2.8 × 101 1.4log | 4.4 × 102 2.6log | 5.4 × 101 1.7log | 2.2 × 102 2.3log | 2.20 |
| E. coli | 3.4 × 102 2.5log | 3.8 × 101 1.5log | 2.9 × 101 1.4log | US* | 2.9 × 101 1.4log | 2.03 | ||
| 1st visit (slaughterhouse B) | Enterobacteriaceae | 1.0 × 103 3log | 1.7 × 102 2.2log | 3.1 × 102 2.4log | 1.3 × 103 3.1log | 1.3 × 103 3.1log | 2.91 | |
| E. coli | 106 6log | 106 6log | 106 6log | 106 6log | 106 6log | 6.0 | ||
| 2nd visit (slaughterhouse B) | Enterobacteriaceae | 1.2 × 102 2log | 8.0 × 102 2.9log | 3.6 × 101 1.5log | 1.4 × 102 2.1log | 0.4 × 101 0.6log | 2.3 | |
| E. coli | 2.0 × 101 1.3log | 6.0 × 101 1.7log | NG** | NG** | NG** | 1.20 | ||
| Slaughter Technological Processing Stage | Visits (Slaughterhouse A and B) | Microorganism | Carcass Part/Point (Count in CFU/cm2) | Average Carcass Count (log CFU/cm2) | ||||
|---|---|---|---|---|---|---|---|---|
| 1 Shank | 2 Loin | 3 Shoulder | 4 Abdomen | 5 Jowl | ||||
| After wash and before enter the cooling chamber | 1st visit (slaughterhouse A) | Enterobacteriaceae | 1.1 × 102 2log | 5.1 × 101 1.6log | 3.3 × 101 1.5log | 3.2 × 101 1.5log | 1.8 × 101 1.2log | 1.68 |
| E. coli | 3.3 × 101 1.5log | 8.7 × 101 1.9log | 4.4 × 101 1.6log | 0.2 × 101 0.3log | 4.0 × 101 1.6log | 1.61 | ||
| 1st visit (slaughterhouse B) | Enterobacteriaceae | 7.5 × 101 1.8log | 7.2 × 101 1.8log | 1.0 × 102 2log | 1.5 × 102 2.1log | 1.5 × 102 2.1log | 2.03 | |
| E. coli | 104 4log | 104 4log | 104 4log | 104 4log | 104 4log | 4.0 | ||
| 2nd visit (slaughterhouse B) | Enterobacteriaceae | NG** 0 | NG** 0 | NG** 0 | NG** 0 | NG** 0 | 0 | |
| E. coli | NG** 0 | NG** 0 | NG** 0 | NG** 0 | NG** 0 | 0 | ||
| Slaughtering Stage | Slaughterhouse/Visit | Shank | Loin | Shoulder | Abdomen | Jowl |
|---|---|---|---|---|---|---|
| After bleeding | A (1st visit) | 1A | 1B | 1C | 1D | 1E |
| B (2nd visit) | 7A | 7B | 7C | 7D | 7E | |
| After epilation | A (1st visit) | 2A | 2B | 2C | ND | ND |
| B (2nd visit) | 8A | 8B | 8C | 8D | 8E | |
| After manual skin | A (1st visit) | 3A | 3B | ND | 3D | 3E |
| B (2nd visit) | 9A | 9B | 9C | 9D | 9E | |
| Before evisceration | A (1st visit) | 4A | ND | ND | 4D | 4E |
| B (2nd visit) | 10A | 10B | 10C | 10D | 10E | |
| After evisceration | A (1st visit) | 5A | 5B | 5C | 5D | ND |
| B (2nd visit) | 11A | 11B | 11C | 11D | 11E | |
| After final wash | A (1st visit) | 6A | 6B | ND | 6D | 6E |
| B (2nd visit) | 12A | 12B | ND | 12D | 12E |
| Antimicrobial | Resistant | Intermedite | Sensitive |
|---|---|---|---|
| Nalidixic acid | 1A-1B-1C-1D; 2A-2B-2C; 3A-3D; 4A-4E; 5A-5B-5C-5D; 6A-6B-6D-6E; 7B-7C-7D; 8B-8C-8D; 9A-9B-9D-9E; 10A-10C-10D; 11A-11B-11C-11D-11E; 12E | 3B-3E 7A-7E 8A | 1E; 4D 8E; 9C ‘10B-10E 12A-1-B-12D |
| Amoxicillin | 1A-1B-1C-1D-1E; 2A-2B-2C 3A-3B-3D-3E; 4A-4D-4E; 5A-5B-5C-5D; 6A-6B-6E; 7A-7B-7D-7E; 8A-8B-8C-8D-8E; 9A-9B-9D 10A, 10B, 10D 11A-11B-11C-11D-11E 12A-12B-12D-12E | - | 6D 7C 9-C-9E 1-C-10E |
| Ampicillin | 1A-1B-1C-1D-1E; 2A-2B-2C 3A-3B-3D-3E; 4A-4D-4E - 5A-5B-5C-5D; 6A-6B-6E 7A-7B-7D-7E 8A-8B-8C-8D-8E; 9A-9B-9D; 10A, 10B, 10D 11A-11B-11C-11D-11E 12A-12B-12D-12E | - | 6D 7C 9-C-9E 1-C-10E |
| Cefazolin | 1D 8A-8D 10A-10C | 1A-1B-1C-1E; 2A-2B-2C; 3A-3B-3D-3E; 4A-4D-4E; 5A-5B-5C-5D; 6A-6B-6D-6E 8B-8C-8E; 9A; 11E | 7A-7B-7C-7D-7E; 9B-9C-9D-9E; 10B-10D-10E; 11A-11B-11C-11D 12A-12B-12D-12E |
| Ceftazidime | 2A-2B 8A 9C | 1C | 1A-1B-1D-1E; 2C; 3A-3B-3D-3E; 4A-4D-4E; 5A-5B-5C-5D; 6A-6B-6D-6E 7A-7B-7C-7D-7E 8B-8C-8D-8E; 9A-9B-9D-9E; 10A-10B-10C-10D-10E 11A-11B-11C-11D-11E; 12A-12B-12D-12E |
| Ciprofloxacin | 1C-1D; 2C; 3B-3D-3E; 4E 5D; 6D 7C; 9A 10A-10D | 1A; 4A; 5B; 6A 7A-7E; 8A 9D; 10C | 1B-1E; 2A-2B; 3A 4D 5A-5C; 6B-6E 7B-7D; 8B-8C-8D-8E 9B-9C-9E; 10B-10E 11A-11B-11C-11D-11E 12A-12B-12D-12E |
| Chloramphenicol | 1A-1C-1D; 2A-2B-2C 3A-3B-3D-3E; 4A-4E 5A-5B-5D; 6A-6B-6D-6E 7A-7B-7C-7D-7E; 8A-8B-8C-8E; 9A-9B-9D-9E 10A-10B-10C-10D-10E 11A-11B-11C-11D-11E; 12E | 8D | 1B-1E 4D; 5C 9C 12A-12B-12D |
| Doxycycline | 1A-1B; 6D 7A-7B-7C-7D-7E 8A-8B-8C-8D-8E 9A-9B-9D-9E 10A-10B-10C-10D-10E 11A-11B-11C-11D-11E; 12E | 1D-1E | 1C; 2A-2B-2C 3A-3B-3D-3E; 4A-4D-4E 5A-5B-5C-5D 6A-6B-6E 9C 12A-12B-12D |
| Streptomycin | 1A-1C-1D-1E; 2A-2B-2C; 3A-3B-3D-3E; 4A-4D-4E; 5A-5C-5D; 6A-6B-6D-6E 7A-7B-7C-7D-7E 8A-8B-8C-8E; 9A-9B-9D; 10A-10B-10E 11A-11B-11C-11D-11E; 12E | 8D 9E 10C-10D | 1B 5B 9C 12A-12B-12D |
| Erythromycin | 1A-1B-1C-1D-1E; 2A-2B-2C 3A-3B-3D-3E; 4A-4D-4E 5A-5B-5C-5D; 6A-6B-6D-6E 7A; 8A-8B-8C-8D 10C-10D-10E 11A-11B-11C-11D-11E; 12E | - | 7B-7C-7D-7E; 8E 9A-9B-9C-9D-9E 10A-10B 12A-12B-12D |
| Gentamicin | 1C-1D 2C 4E; 5B-5D 7C | - | 1A-1B-1E; 2A-2B; 3A-3B-3D-3E; 4A-4D; 5A-5C; 6A-6B-6D-6E 7A-7B-7D-7E; 8A-8B-8C-8D-8E; 9A-9B-9C-9D-9E 10A-10B-10C-10D-10E 11A-11B-11C-11D-11E 12A-12B-12D-12E |
| Sulfonamide | 1A-1C-1D-1E; 2A-2B-2C 3A-3B-3D-3E; 4A-4D-4E 5A-5B-5C-5D; 6A-6B-6D-6E 7A-7B-7C-7D-7E; 8C-8E; 9A-9B-9E; 10A-10B-10C-10D-10E 11A-11B-11C-11D-11E 12D-12E | - | 1B 8A-8B-8D 9C-9D 12A-12B |
| Tetracycline | 1A-1B; 6D 7A-7B-7C-7D-7E 8A-8B-8C-8D-8E 9A-9B-9D-9E 10A-10B-10C-10D-10E 11A-11B-11C-11D-11E; 12E | - | 1C-1D-1E; 2A-2B-2C 3A-3B-3D-3E; 4A-4D-4E 5A-5B-5C-5D; 6A-6B-6E 9C 12A-12B-12D |
| Antimicrobial | Number of Resistant Isolates (%) | Number of Intermediate Resistance Isolates (%) | Number of Sensitive Isolates (%) | Total of Resistant and Intermediate Isolates (%) |
|---|---|---|---|---|
| Nalidixic acid (NAL) | 73% (38/52) | 9.6% (5/52) | 17.4% (9/52) | 82.6% (43/52) |
| Amoxicillin (AMO) | 88.4% (46/52) | 0% (0/0) | 11.6% (6/52) | 88.4% (46/52) |
| Ampicillin (AMP) | 88.4% (46/52) | 0% (0/0) | 11.6% (6/52) | 88.4% (46/52) |
| Cefazolin (CFZ) | 9.6% (5/52) | 51.9% (27/52) | 38.5% (20/52) | 61.5% (32/52) |
| Ceftazidime (CAZ) | 7.6% (4/52) | 2% (1/52) | 90.4% (47/52) | 9.6% (5/52) |
| Ciprofloxacin (CIP) | 25% (13/52) | 17.3% (9/52) | 57.7% (30/52) | 42.3% (22/52) |
| Chloramphenicol (CLO) | 82.6% (43/52) | 2% (1/52) | 15.4% (8/52) | 84.6% (44/52) |
| Doxycycline (DOX) | 53.7% (28/52) | 4% (2/52) | 42.3% (22/52) | 57.7% (30/52) |
| Streptomycin (EST) | 80.7% (42/52) | 7.7% (4/52) | 11.6% (6/52) | 88.4% (46/52) |
| Erythromycin (ERI) | 71.1% (37/52) | 0% (0/0) | 28.9% (15/52) | 71.1% (37/52) |
| Gentamicin (GEN) | 13.5% (7/52) | 0% (0/0) | 86.5% (45/52) | 13.5% (7/52) |
| Tetracycline (TET) | 53.7% (28/52) | 0% (0/0) | 46.3% (24/52) | 53.7% (28/52) |
| Sulfonamide (SUL) | 84.6% (44/52) | 0% (0/0) | 15.4% (8/52) | 84.6% (44/52) |
| Isolates | Slaughterhouse | Antimicrobial Resistance | Virulence Gene |
|---|---|---|---|
| 1B | A | AMO AMP ERI DOX TET NAL | eae |
| 1C | A | AMO AMP ERI EST GEN SUL CLO NAL CIP | eae |
| 1E | A | AMO AMP ERI EST SUL | eae, stx1 |
| 2A | A | AMO AMP CAZ ERI EST SUL CLO NAL | eae |
| 3A | A | AMO AMP EST ERI SUL CLO NAL | eae |
| 3B | A | AMO AMP ERI EST SUL CLO CIP | eae |
| 3E | A | AMO AMP ERI EST SUL CLO CIP | eae, stx2 |
| 4A | A | AMO AMP ERI EST SUL CLO NAL | eae, stx1 |
| 4D | A | AMO AMP ERI EST SUL | eae, stx1 |
| 5A | A | AMO AMP ERI EST SUL CLO NAL | eae |
| 5D | A | AMO AMP ERI EST GEN SUL CLO NAL CIP | eae, stx1, stx2 |
| 6B | A | AMO AMP ERI EST SUL CLO NAL | eae, stx1 |
| 6D | A | ERI EST SUL DOX TET CLO NAL CIP | eae |
| 7A | B | AMO AMP ERI EST SUL DOX TET CLO | stx1, stx2 |
| 7B | B | AMO AMP EST SUL DOX TET CLO NAL | stx1 |
| 7C | B | EST GEN SUL DOX TET CLO NAL CIP | stx1 |
| 7D | B | AMO AMP EST SUL DOX TET CLO NAL | stx1 |
| 7E | B | AMO AMP EST SUL DOX TET CLO | stx1 |
| 8A | B | AMO AMP CFZ CAZ ERI EST DOX TET CLO | eae, stx1, stx2 |
| 8B | B | AMO AMP ERI EST DOX TET CLO NAL | stx2 |
| 8E | B | AMO AMP EST SUL DOX TET CLO | eae |
| 9A | B | AMO AMP EST SUL DOX TET CLO NAL CIP | eae, stx2 |
| 9B | B | AMO AMP EST SUL DOX TET CLO NAL | eae |
| 9C | B | CAZ | stx1, stx2 |
| 10B | B | AMO AMP EST SUL DOX TET CLO | eae |
| 10C | B | CFZ ERI SUL DOX TET CLO NAL | stx1 |
| 10E | B | EST SUL DOX TET CLO | eae |
| 11A | B | AMO AMP EST SUL DOX TET CLO NAL | eae, stx2 |
| 11D | B | AMO AMP EST SUL DOX TET CLO NAL | eae, stx1, stx2 |
| 12A | B | AMO AMP ERI | eae, stx1 |
| 12B | B | AMO AMP | eae |
| 12D | B | AMO AMP ERI SUL | eae, stx1 |
| Isolates | Virulence Genes | Biofilm Formation Capacity | Antimicrobial Resistance | ||
|---|---|---|---|---|---|
| 37 °C | 24 °C | 10 °C | |||
| 1E | stx1 | NF | NF | NF | AMO-AMP-ERI-EST-SUL |
| 3E | stx2 | WF | MF | WF | AMO-AMP-ERI-EST-SUL-CLO-CIP |
| 4A | stx1 | SF | SF | SF | AMO-AMP-ERI-EST-SUL-CLO-NAL |
| 4D | stx1 | WF | WF | NF | AMO-AMP-ERI-EST-SUL |
| 5D | stx1, stx2 | MF | FFO | WF | AMO-AMP-ERI-EST-GEN-SUL-CLO-NAL-CIP |
| 6B | stx1 | NF | NF | NF | AMO-AMP-ERI-EST-SUL-CLO-NAL |
| 7A | stx1, stx2 | SF | SF | SF | AMO-AMP-ERI-EST-SUL-DOX-TET-CLO |
| 7B | stx1 | MF | SF | SF | AMO-AMP-EST-SUL-DOX-TET-CLO-NAL |
| 7C | stx1 | MF | WF | WF | EST-GEN-SUL-DOX-TET-CLO-NAL-CIP |
| 7D | stx1 | MF | MF | WF | AMO-AMP-EST-SUL-DOX-TET-CLO-NAL |
| 7E | stx1 | WF | NF | NF | AMO-AMP-EST-SUL-DOX-TET-CLO |
| 8A | stx1, stx2 | WF | WF | WF | AMO-AMP-CFZ-CAZ-ERI-EST-DOX-TET-CLO |
| 8B | stx2 | FM | WF | WF | AMO-AMP-ERI-EST-DOX-TET-CLO-NAL |
| 9A | stx2 | WF | WF | WF | AMO-AMP-EST-SUL-DOX-TET-CLO-NAL-CIP |
| 9C | stx1, stx2 | WF | MF | SF | CAZ |
| 10C | stx1 | WF | WF | WF | CFZ-ERI-SUL-DOX-TET-CLO-NAL |
| 11A | stx2 | MF | MF | SF | AMO-AMP-EST-SUL-DOX-TET-CLO-NAL |
| 11D | stx1, stx2 | MF | MF | WF | AMO-AMP-EST-SUL-DOX-TET-CLO-NAL |
| 12A | stx1 | WF | MF | SF | AMO-AMP-ERI |
| 12D | stx1 | WF | WF | WF | AMO-AMP-ERI-SUL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coelho, M.M.S.; Davanzo, E.F.A.; dos Santos, R.L.; Castro, V.H.d.L.; da Costa, H.M.B.; Dallago, B.S.L.; Perecmanis, S.; Santana, A.P. Escherichia coli and Enterobacteriaceae Counts, Virulence Gene Profile, Antimicrobial Resistance, and Biofilm Formation Capacity during Pig Slaughter Stages. Life 2024, 14, 1261. https://doi.org/10.3390/life14101261
Coelho MMS, Davanzo EFA, dos Santos RL, Castro VHdL, da Costa HMB, Dallago BSL, Perecmanis S, Santana AP. Escherichia coli and Enterobacteriaceae Counts, Virulence Gene Profile, Antimicrobial Resistance, and Biofilm Formation Capacity during Pig Slaughter Stages. Life. 2024; 14(10):1261. https://doi.org/10.3390/life14101261
Chicago/Turabian StyleCoelho, Madalena Maria Saldanha, Emilia Fernanda Agostinho Davanzo, Rebecca Lavarini dos Santos, Virgílio Hipólito de Lemos Castro, Hayanna Maria Boaventura da Costa, Bruno Stéfano Lima Dallago, Simone Perecmanis, and Angela Patrícia Santana. 2024. "Escherichia coli and Enterobacteriaceae Counts, Virulence Gene Profile, Antimicrobial Resistance, and Biofilm Formation Capacity during Pig Slaughter Stages" Life 14, no. 10: 1261. https://doi.org/10.3390/life14101261
APA StyleCoelho, M. M. S., Davanzo, E. F. A., dos Santos, R. L., Castro, V. H. d. L., da Costa, H. M. B., Dallago, B. S. L., Perecmanis, S., & Santana, A. P. (2024). Escherichia coli and Enterobacteriaceae Counts, Virulence Gene Profile, Antimicrobial Resistance, and Biofilm Formation Capacity during Pig Slaughter Stages. Life, 14(10), 1261. https://doi.org/10.3390/life14101261






