mRNA Abundance of Neurogenic Factors Correlates with Hearing Capacity in Auditory Brainstem Nuclei of the Rat
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal and Tissue Preparations
2.2. RNA Extraction from the Tissue of CN
2.3. cDNA Synthesis from RNA by Reverse Transcription
2.4. Rat Neurogenesis RT2 ProfilerTM PCR Array
2.5. Data Analysis and Statistical Evaluation
3. Results
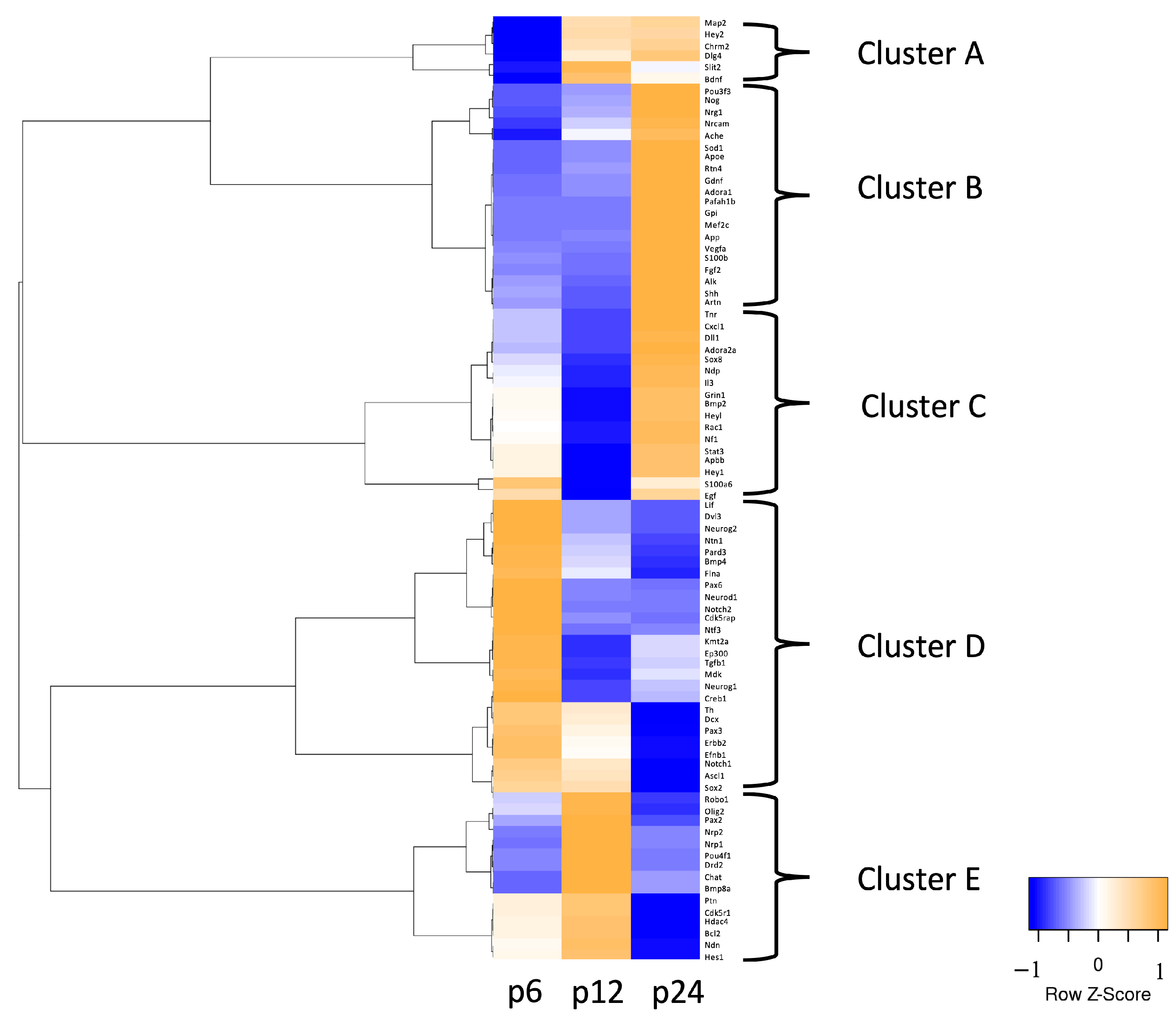
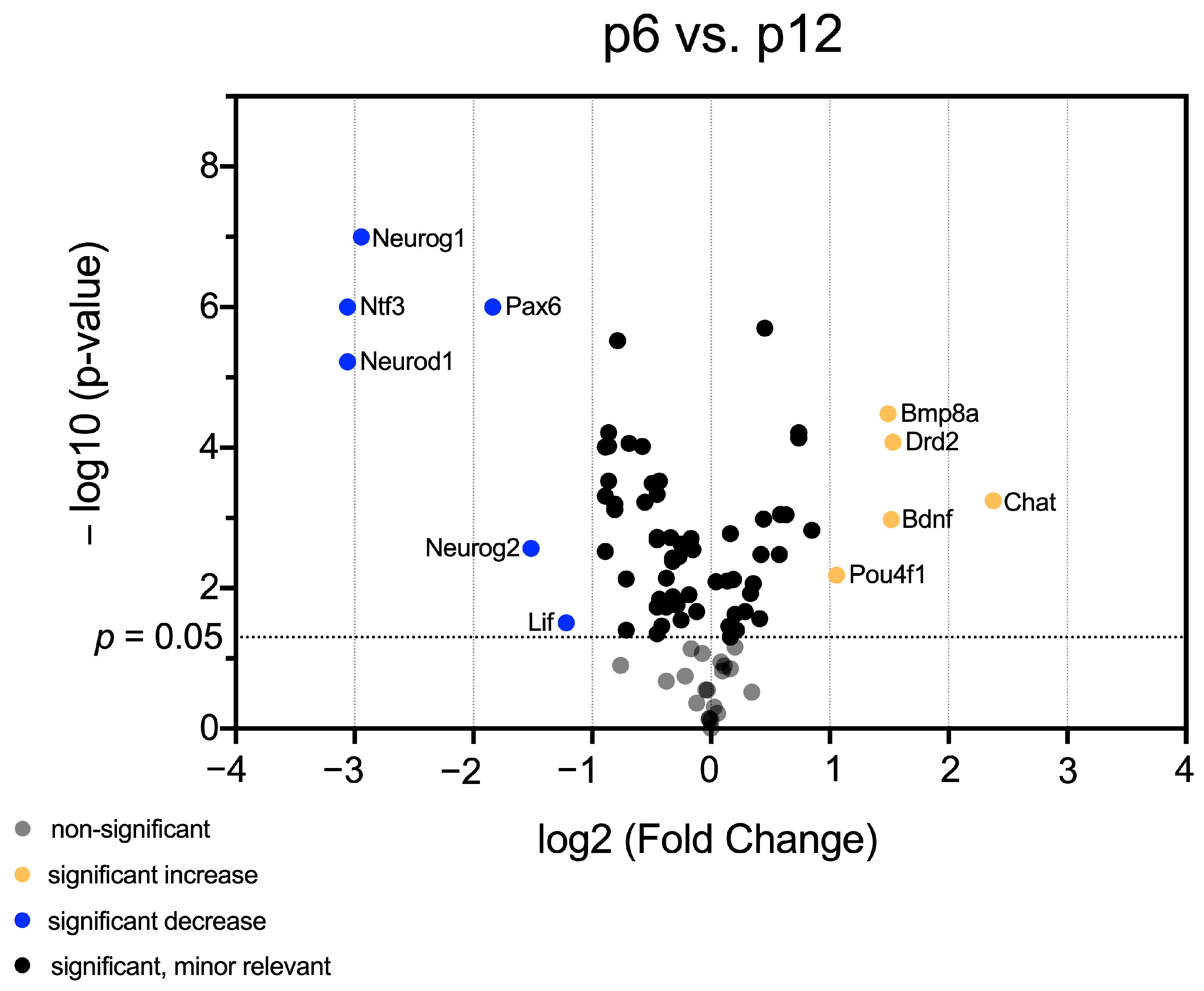
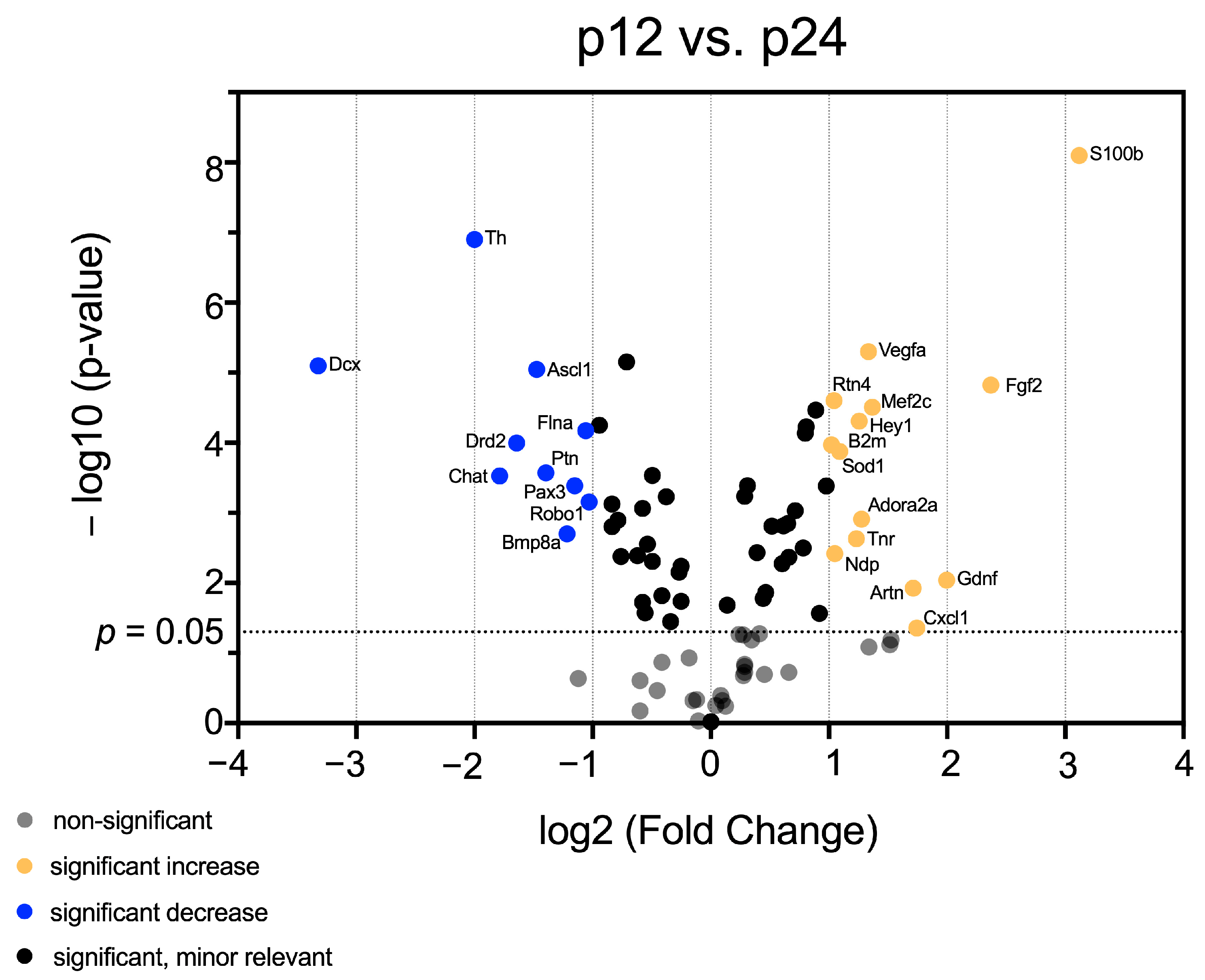
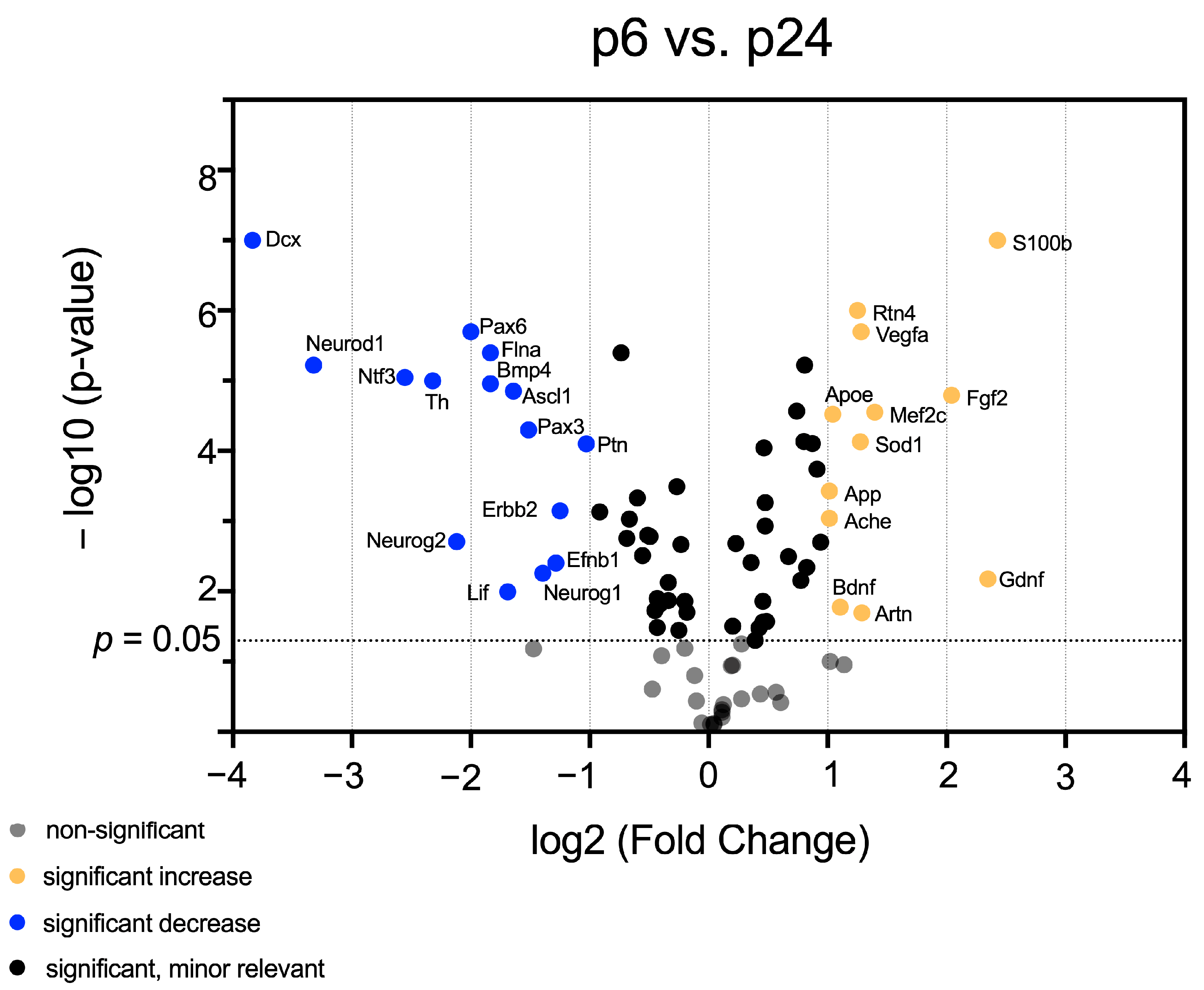
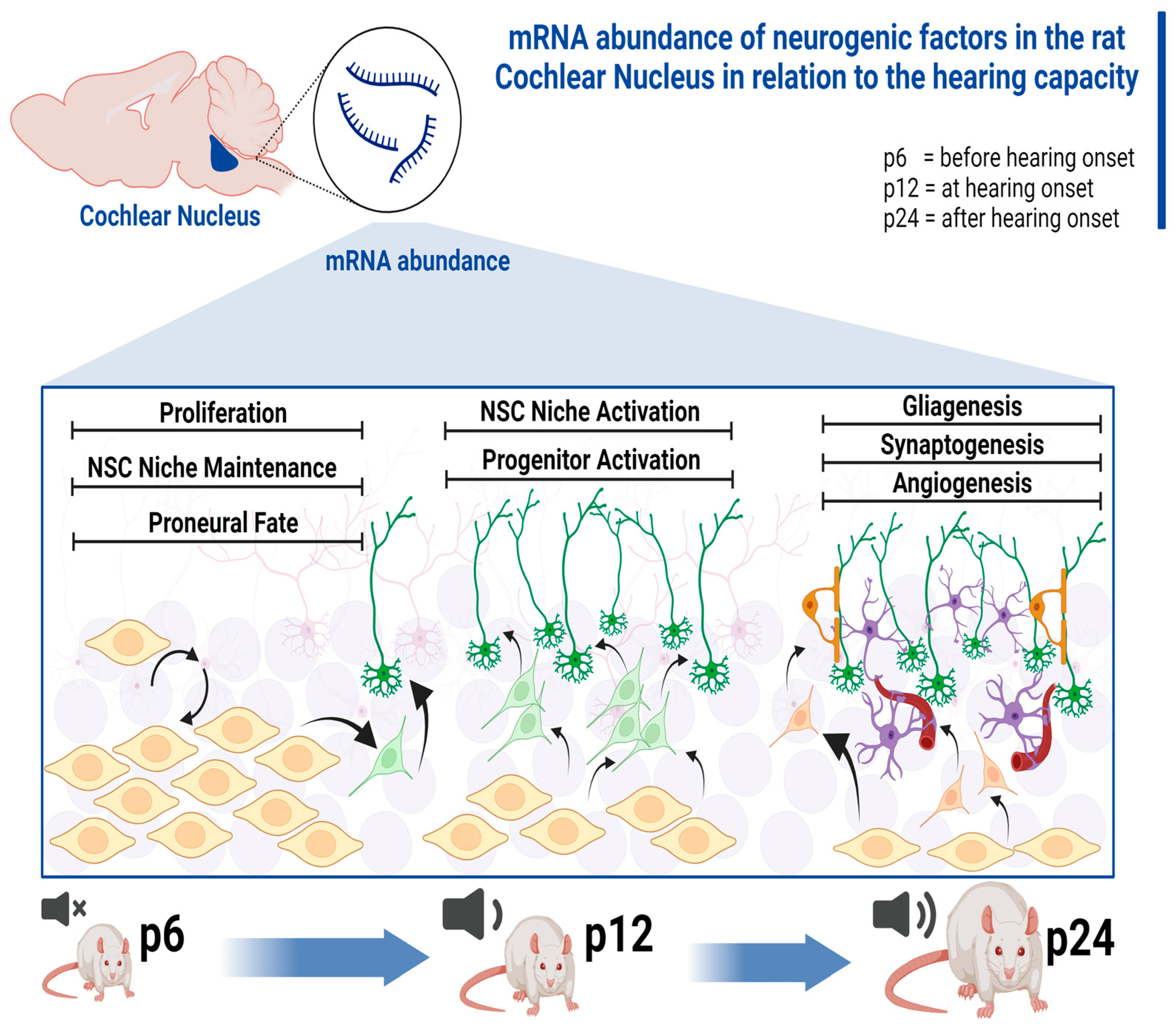
3.1. mRNA Abundance Patterns of Neurogenic Factors Correlate with Age in Maturing CN
3.2. Identification of Neurogenic Factors with High Importance for NSC Development and Neurogenesis in Maturing CN
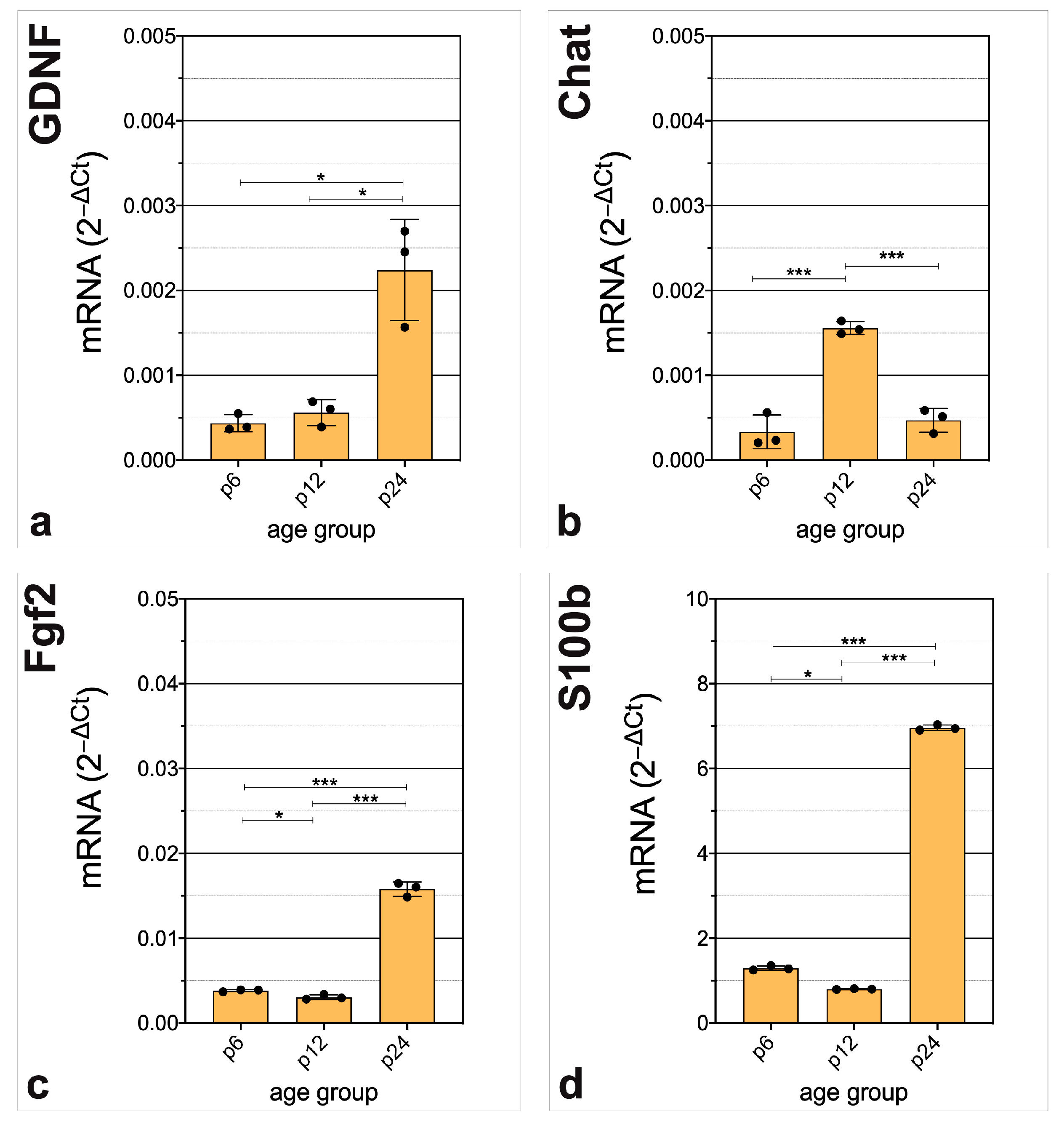
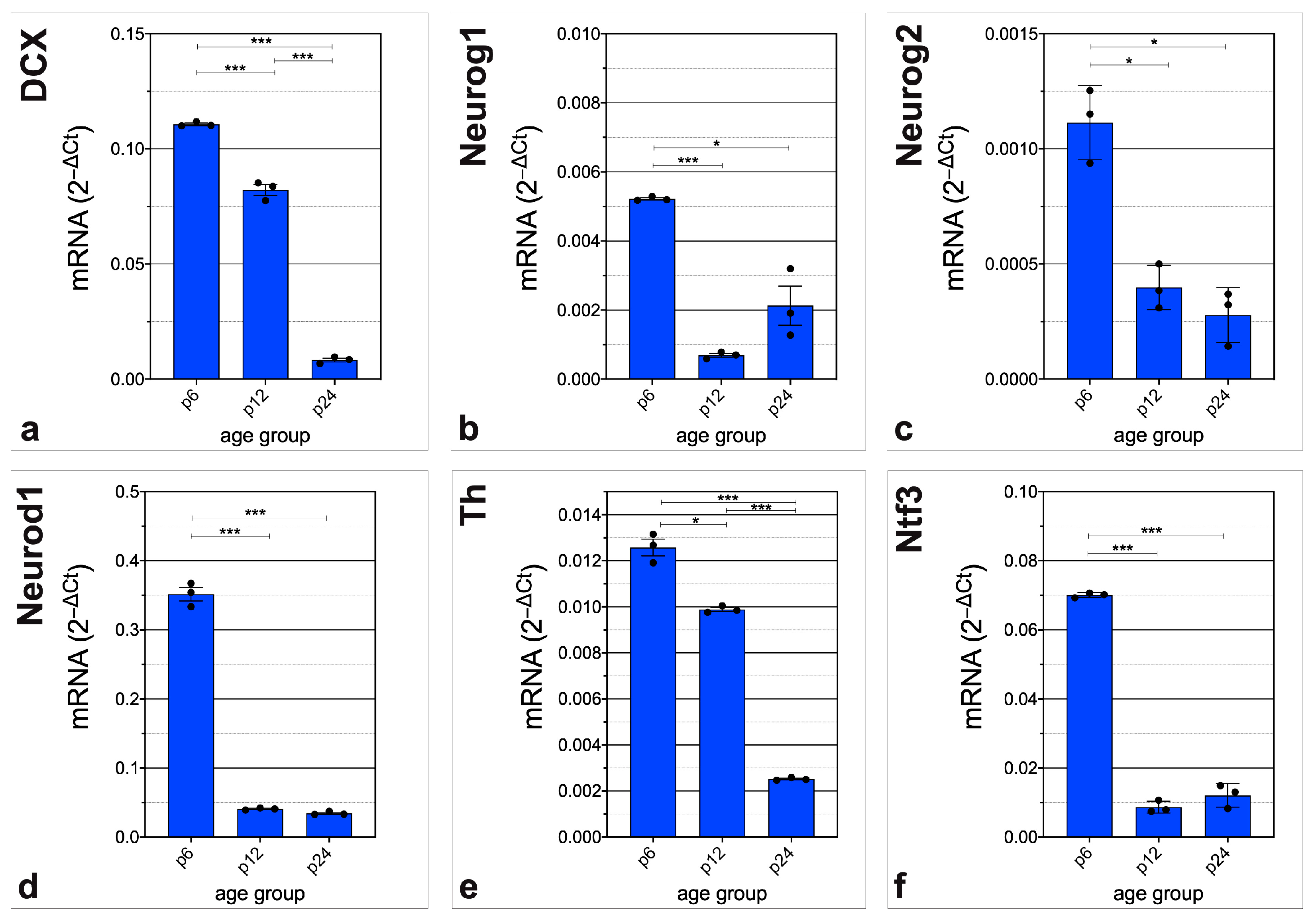
3.3. In-Depth Analysis of Selected Critical Neurogenic Factors in Maturing CN
4. Discussion
4.1. bHLH Family of Transcription Factors and Pax Gene Family Play Essential Roles in CN Neurogenesis and Regulation of NSC Pool on mRNA Level
4.2. NSC Niche-Activating Transcripts Are Elevated at the Hearing Onset
4.3. mRNA of Neurogenic Factors Stimulating Proliferation, Neuronal Migration, and Proneural Differentiation Is Abundant before Hearing Onset
4.4. Regulators of Gliogenesis, Neuritogenesis, Synaptogenesis, and Angiogenesis Are Prominent at the Transcriptional Level of CN after Hearing Onset
4.5. Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chadha, S.; Kamenov, K.; Cieza, A. The world report on hearing, 2021. Bull. World Health Organ. 2021, 99, 242. [Google Scholar] [CrossRef] [PubMed]
- Stojkovic, M.; Han, D.; Jeong, M.; Stojkovic, P.; Stankovic, K.M. Human induced pluripotent stem cells and CRISPR/Cas-mediated targeted genome editing: Platforms to tackle sensorineural hearing loss. Stem. Cells 2021, 39, 673–696. [Google Scholar] [CrossRef] [PubMed]
- Lahlou, G.; Calvet, C.; Giorgi, M.; Lecomte, M.J.; Safieddine, S. Towards the Clinical Application of Gene Therapy for Genetic Inner Ear Diseases. J. Clin. Med. 2023, 12, 1046. [Google Scholar] [CrossRef]
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods 2013, 10, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Tao, Y.; Lamas, V.; Huang, M.; Yeh, W.H.; Pan, B.; Hu, Y.J.; Hu, J.H.; Thompson, D.B.; Shu, Y.; et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature 2018, 553, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Martinez, J.; Patkaniowska, A.; Lendeckel, W.; Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001, 20, 6877–6888. [Google Scholar] [CrossRef]
- Yoshimura, H.; Shibata, S.B.; Ranum, P.T.; Moteki, H.; Smith, R.J.H. Targeted Allele Suppression Prevents Progressive Hearing Loss in the Mature Murine Model of Human TMC1 Deafness. Mol. Ther. 2019, 27, 681–690. [Google Scholar] [CrossRef]
- Gage, F.H. Mammalian neural stem cells. Science 2000, 287, 1433–1438. [Google Scholar] [CrossRef]
- Sohur, U.S.; Emsley, J.G.; Mitchell, B.D.; Macklis, J.D. Adult neurogenesis and cellular brain repair with neural progenitors, precursors and stem cells. Philos. Trans. R Soc. Lond B Biol. Sci. 2006, 361, 1477–1497. [Google Scholar] [CrossRef]
- Engert, J.; Spahn, B.; Bieniussa, L.; Hagen, R.; Rak, K.; Voelker, J. Neurogenic Stem Cell Niche in the Auditory Thalamus: In Vitro Evidence of Neural Stem Cells in the Rat Medial Geniculate Body. Life 2023, 13, 1188. [Google Scholar] [CrossRef]
- Hu, Z.; Tao, L.; Liu, Z.; Jiang, Y.; Deng, X. Identification of Neural Stem Cells from Postnatal Mouse Auditory Cortex In Vitro. Stem. Cells Dev. 2019, 28, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Oshima, K.; Grimm, C.M.; Corrales, C.E.; Senn, P.; Martinez Monedero, R.; Geleoc, G.S.; Edge, A.; Holt, J.R.; Heller, S. Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J. Assoc. Res. Otolaryngol. 2007, 8, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Rak, K.; Volker, J.; Frenz, S.; Scherzed, A.; Radeloff, A.; Hagen, R.; Mlynski, R. Dynamic changes of the neurogenic potential in the rat cochlear nucleus during post-natal development. Exp. Brain Res. 2013, 226, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Rask-Andersen, H.; Bostrom, M.; Gerdin, B.; Kinnefors, A.; Nyberg, G.; Engstrand, T.; Miller, J.M.; Lindholm, D. Regeneration of human auditory nerve. In vitro/in video demonstration of neural progenitor cells in adult human and guinea pig spiral ganglion. Hear. Res. 2005, 203, 180–191. [Google Scholar] [CrossRef]
- Völker, J.; Engert, J.; Völker, C.; Bieniussa, L.; Schendzielorz, P.; Hagen, R.; Rak, K. Isolation and Characterization of Neural Stem Cells from the Rat Inferior Colliculus. Stem. Cells Int. 2019, 2019, 5831240. [Google Scholar] [CrossRef]
- Geal-Dor, M.; Freeman, S.; Li, G.; Sohmer, H. Development of hearing in neonatal rats: Air and bone conducted ABR thresholds. Hear. Res. 1993, 69, 236–242. [Google Scholar] [CrossRef]
- Schwartz, M.S.; Otto, S.R.; Shannon, R.V.; Hitselberger, W.E.; Brackmann, D.E. Auditory brainstem implants. Neurotherapeutics 2008, 5, 128–136. [Google Scholar] [CrossRef]
- Tanic, N.; Perovic, M.; Mladenovic, A.; Ruzdijic, S.; Kanazir, S. Effects of aging, dietary restriction and glucocorticoid treatment on housekeeping gene expression in rat cortex and hippocampus-evaluation by real time RT-PCR. J. Mol. Neurosci. 2007, 32, 38–46. [Google Scholar] [CrossRef]
- Yao, L.; Chen, X.; Tian, Y.; Lu, H.; Zhang, P.; Shi, Q.; Zhang, J.; Liu, Y. Selection of housekeeping genes for normalization of RT-PCR in hypoxic neural stem cells of rat in vitro. Mol. Biol. Rep. 2012, 39, 569–576. [Google Scholar] [CrossRef]
- Skidmore, A.F.; Beebee, T.J. Rat lactate dehydrogenase A and B subunit concentrations are not regulated by mRNA abundance in liver and heart. FEBS Lett. 1990, 270, 67–70. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome. Biol. 2002, 3, research0034. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Plante, T.B.; Cushman, M. Choosing color palettes for scientific figures. Res. Pract. Thromb. Haemost. 2020, 4, 176–180. [Google Scholar] [CrossRef] [PubMed]
- D’Haeseleer, P. How does gene expression clustering work? Nat. Biotechnol. 2005, 23, 1499–1501. [Google Scholar] [CrossRef]
- Blake, J.A.; Ziman, M.R. Pax genes: Regulators of lineage specification and progenitor cell maintenance. Development 2014, 141, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Maschhoff, K.L.; Baldwin, H.S. Molecular determinants of neural crest migration. Am. J. Med. Genet. 2000, 97, 280–288. [Google Scholar] [CrossRef]
- Sansom, S.N.; Griffiths, D.S.; Faedo, A.; Kleinjan, D.J.; Ruan, Y.; Smith, J.; van Heyningen, V.; Rubenstein, J.L.; Livesey, F.J. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009, 5, e1000511. [Google Scholar] [CrossRef]
- Li, H.; Liu, H.; Corrales, C.E.; Mutai, H.; Heller, S. Correlation of Pax-2 expression with cell proliferation in the developing chicken inner ear. J. Neurobiol. 2004, 60, 61–70. [Google Scholar] [CrossRef]
- Jahan, I.; Kersigo, J.; Pan, N.; Fritzsch, B. Neurod1 regulates survival and formation of connections in mouse ear and brain. Cell Tissue Res. 2010, 341, 95–110. [Google Scholar] [CrossRef]
- Hulme, A.J.; Maksour, S.; St-Clair Glover, M.; Miellet, S.; Dottori, M. Making neurons, made easy: The use of Neurogenin-2 in neuronal differentiation. Stem. Cell Rep. 2022, 17, 14–34. [Google Scholar] [CrossRef]
- Ma, Q.; Anderson, D.J.; Fritzsch, B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J. Assoc. Res. Otolaryngol. 2000, 1, 129–143. [Google Scholar] [CrossRef]
- Kageyama, R.; Ohtsuka, T.; Kobayashi, T. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development 2007, 134, 1243–1251. [Google Scholar] [CrossRef]
- Kim, W.Y.; Fritzsch, B.; Serls, A.; Bakel, L.A.; Huang, E.J.; Reichardt, L.F.; Barth, D.S.; Lee, J.E. NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development 2001, 128, 417–426. [Google Scholar] [CrossRef]
- Bertrand, N.; Castro, D.S.; Guillemot, F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002, 3, 517–530. [Google Scholar] [CrossRef]
- Sakamoto, M.; Hirata, H.; Ohtsuka, T.; Bessho, Y.; Kageyama, R. The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J. Biol. Chem. 2003, 278, 44808–44815. [Google Scholar] [CrossRef] [PubMed]
- Jalali, A.; Bassuk, A.G.; Kan, L.; Israsena, N.; Mukhopadhyay, A.; McGuire, T.; Kessler, J.A. HeyL promotes neuronal differentiation of neural progenitor cells. J. Neurosci. Res. 2011, 89, 299–309. [Google Scholar] [CrossRef]
- Nakazaki, H.; Reddy, A.C.; Mania-Farnell, B.L.; Shen, Y.W.; Ichi, S.; McCabe, C.; George, D.; McLone, D.G.; Tomita, T.; Mayanil, C.S. Key basic helix-loop-helix transcription factor genes Hes1 and Ngn2 are regulated by Pax3 during mouse embryonic development. Dev. Biol. 2008, 316, 510–523. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, M.; Ang, S.L.; Shiota, K.; Nakanishi, S.; Kageyama, R.; Guillemot, F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995, 9, 3136–3148. [Google Scholar] [CrossRef]
- Lo, L.; Dormand, E.; Greenwood, A.; Anderson, D.J. Comparison of the generic neuronal differentiation and neuron subtype specification functions of mammalian achaete-scute and atonal homologs in cultured neural progenitor cells. Development 2002, 129, 1553–1567. [Google Scholar] [CrossRef] [PubMed]
- Snapyan, M.; Lemasson, M.; Brill, M.S.; Blais, M.; Massouh, M.; Ninkovic, J.; Gravel, C.; Berthod, F.; Gotz, M.; Barker, P.A.; et al. Vasculature guides migrating neuronal precursors in the adult mammalian forebrain via brain-derived neurotrophic factor signaling. J. Neurosci. 2009, 29, 4172–4188. [Google Scholar] [CrossRef] [PubMed]
- Ylikoski, J.; Pirvola, U.; Moshnyakov, M.; Palgi, J.; Arumae, U.; Saarma, M. Expression patterns of neurotrophin and their receptor mRNAs in the rat inner ear. Hear. Res. 1993, 65, 69–78. [Google Scholar] [CrossRef]
- Majdzadeh, N.; Wang, L.; Morrison, B.E.; Bassel-Duby, R.; Olson, E.N.; D’Mello, S.R. HDAC4 inhibits cell-cycle progression and protects neurons from cell death. Dev. Neurobiol. 2008, 68, 1076–1092. [Google Scholar] [CrossRef] [PubMed]
- Copray, S.; Balasubramaniyan, V.; Levenga, J.; de Bruijn, J.; Liem, R.; Boddeke, E. Olig2 overexpression induces the in vitro differentiation of neural stem cells into mature oligodendrocytes. Stem. Cells 2006, 24, 1001–1010. [Google Scholar] [CrossRef]
- Fujimoto, I.; Hasegawa, K.; Fujiwara, K.; Yamada, M.; Yoshikawa, K. Necdin controls EGFR signaling linked to astrocyte differentiation in primary cortical progenitor cells. Cell Signal 2016, 28, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Wang, M.; Wang, P.; Wang, L.; Wu, Q.; Guo, W. Neural Stem Cells Behave as a Functional Niche for the Maturation of Newborn Neurons through the Secretion of PTN. Neuron 2019, 101, 32–44.e6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xue, Y.Y.; Lu, S.D.; Wang, Y.; Zhang, L.M.; Huang, Y.L.; Signore, A.P.; Chen, J.; Sun, F.Y. Bcl-2 enhances neurogenesis and inhibits apoptosis of newborn neurons in adult rat brain following a transient middle cerebral artery occlusion. Neurobiol. Dis. 2006, 24, 345–356. [Google Scholar] [CrossRef]
- Lopez-Bendito, G.; Flames, N.; Ma, L.; Fouquet, C.; Di Meglio, T.; Chedotal, A.; Tessier-Lavigne, M.; Marin, O. Robo1 and Robo2 cooperate to control the guidance of major axonal tracts in the mammalian forebrain. J. Neurosci. 2007, 27, 3395–3407. [Google Scholar] [CrossRef]
- Yuan, W.; Zhou, L.; Chen, J.H.; Wu, J.Y.; Rao, Y.; Ornitz, D.M. The mouse SLIT family: Secreted ligands for ROBO expressed in patterns that suggest a role in morphogenesis and axon guidance. Dev. Biol. 1999, 212, 290–306. [Google Scholar] [CrossRef]
- Erkman, L.; McEvilly, R.J.; Luo, L.; Ryan, A.K.; Hooshmand, F.; O’Connell, S.M.; Keithley, E.M.; Rapaport, D.H.; Ryan, A.F.; Rosenfeld, M.G. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature 1996, 381, 603–606. [Google Scholar] [CrossRef]
- Ohtsuka, T.; Ishibashi, M.; Gradwohl, G.; Nakanishi, S.; Guillemot, F.; Kageyama, R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999, 18, 2196–2207. [Google Scholar] [CrossRef]
- Castella, P.; Sawai, S.; Nakao, K.; Wagner, J.A.; Caudy, M. HES-1 repression of differentiation and proliferation in PC12 cells: Role for the helix 3-helix 4 domain in transcription repression. Mol. Cell Biol. 2000, 20, 6170–6183. [Google Scholar] [CrossRef]
- Ng, T.; Ryu, J.R.; Sohn, J.H.; Tan, T.; Song, H.; Ming, G.L.; Goh, E.L. Class 3 semaphorin mediates dendrite growth in adult newborn neurons through Cdk5/FAK pathway. PLoS ONE 2013, 8, e65572. [Google Scholar] [CrossRef]
- Sahlgren, C.M.; Mikhailov, A.; Vaittinen, S.; Pallari, H.M.; Kalimo, H.; Pant, H.C.; Eriksson, J.E. Cdk5 regulates the organization of Nestin and its association with p35. Mol. Cell Biol. 2003, 23, 5090–5106. [Google Scholar] [CrossRef]
- Uchida, T.; Ishiguro, K.; Ohnuma, J.; Takamatsu, M.; Yonekura, S.; Imahori, K. Precursor of cdk5 activator, the 23 kDa subunit of tau protein kinase II: Its sequence and developmental change in brain. FEBS Lett. 1994, 355, 35–40. [Google Scholar] [CrossRef]
- Armenteros, T.; Andreu, Z.; Hortiguela, R.; Lie, D.C.; Mira, H. BMP and WNT signalling cooperate through LEF1 in the neuronal specification of adult hippocampal neural stem and progenitor cells. Sci. Rep. 2018, 8, 9241. [Google Scholar] [CrossRef] [PubMed]
- Hiramoto, T.; Kanda, Y.; Satoh, Y.; Takishima, K.; Watanabe, Y. Dopamine D2 receptor stimulation promotes the proliferation of neural progenitor cells in adult mouse hippocampus. Neuroreport 2007, 18, 659–664. [Google Scholar] [CrossRef]
- Paez-Gonzalez, P.; Asrican, B.; Rodriguez, E.; Kuo, C.T. Identification of distinct ChAT(+) neurons and activity-dependent control of postnatal SVZ neurogenesis. Nat. Neurosci. 2014, 17, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Wei, L.; Zha, D.J.; Qiao, L.; Lu, L.J.; Chen, F.Q.; Qiu, J.H. Exposure to acoustic stimuli promotes the development and differentiation of neural stem cells from the cochlear nuclei through the clusterin pathway. Int. J. Mol. Med. 2015, 35, 637–644. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Code, R.A.; Carr, C.E. Choline acetyltransferase-immunoreactive cochlear efferent neurons in the chick auditory brainstem. J. Comp. Neurol. 1994, 340, 161–173. [Google Scholar] [CrossRef]
- Howell, D.M.; Morgan, W.J.; Jarjour, A.A.; Spirou, G.A.; Berrebi, A.S.; Kennedy, T.E.; Mathers, P.H. Molecular guidance cues necessary for axon pathfinding from the ventral cochlear nucleus. J. Comp. Neurol. 2007, 504, 533–549. [Google Scholar] [CrossRef]
- Reynolds, B.A.; Rietze, R.L. Neural stem cells and neurospheres—Re-evaluating the relationship. Nat. Methods 2005, 2, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.S.; Hattiangady, B.; Shetty, A.K. The window and mechanisms of major age-related decline in the production of new neurons within the dentate gyrus of the hippocampus. Aging Cell 2006, 5, 545–558. [Google Scholar] [CrossRef]
- Kuhn, H.G.; Dickinson-Anson, H.; Gage, F.H. Neurogenesis in the dentate gyrus of the adult rat: Age-related decrease of neuronal progenitor proliferation. J. Neurosci. 1996, 16, 2027–2033. [Google Scholar] [CrossRef] [PubMed]
- Shetty, G.A.; Hattiangady, B.; Shetty, A.K. Neural stem cell- and neurogenesis-related gene expression profiles in the young and aged dentate gyrus. Age 2013, 35, 2165–2176. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.R.; Wen, G.; Lepier, A.; Schroeder, T.; Gotz, M. Par-complex proteins promote proliferative progenitor divisions in the developing mouse cerebral cortex. Development 2008, 135, 11–22. [Google Scholar] [CrossRef]
- Houlihan, S.L.; Lanctot, A.A.; Guo, Y.; Feng, Y. Upregulation of neurovascular communication through filamin abrogation promotes ectopic periventricular neurogenesis. eLife 2016, 5, e17823. [Google Scholar] [CrossRef]
- Lu, H.; Song, X.; Wang, F.; Wang, G.; Wu, Y.; Wang, Q.; Wang, Y.; Yang, G.Y.; Zhang, Z. Hyperexpressed Netrin-1 Promoted Neural Stem Cells Migration in Mice after Focal Cerebral Ischemia. Front. Cell Neurosci. 2016, 10, 223. [Google Scholar] [CrossRef]
- Hu, Z.; Wei, D.; Johansson, C.B.; Holmstrom, N.; Duan, M.; Frisen, J.; Ulfendahl, M. Survival and neural differentiation of adult neural stem cells transplanted into the mature inner ear. Exp. Cell Res. 2005, 302, 40–47. [Google Scholar] [CrossRef]
- Nakashima, K.; Yanagisawa, M.; Arakawa, H.; Taga, T. Astrocyte differentiation mediated by LIF in cooperation with BMP2. FEBS Lett. 1999, 457, 43–46. [Google Scholar] [CrossRef]
- Yanagisawa, M.; Takizawa, T.; Ochiai, W.; Uemura, A.; Nakashima, K.; Taga, T. Fate alteration of neuroepithelial cells from neurogenesis to astrocytogenesis by bone morphogenetic proteins. Neurosci. Res. 2001, 41, 391–396. [Google Scholar] [CrossRef]
- Etheridge, S.L.; Ray, S.; Li, S.; Hamblet, N.S.; Lijam, N.; Tsang, M.; Greer, J.; Kardos, N.; Wang, J.; Sussman, D.J.; et al. Murine dishevelled 3 functions in redundant pathways with dishevelled 1 and 2 in normal cardiac outflow tract, cochlea, and neural tube development. PLoS Genet. 2008, 4, e1000259. [Google Scholar] [CrossRef]
- Buchman, J.J.; Tseng, H.C.; Zhou, Y.; Frank, C.L.; Xie, Z.; Tsai, L.H. Cdk5rap2 interacts with pericentrin to maintain the neural progenitor pool in the developing neocortex. Neuron 2010, 66, 386–402. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.D.; Kumari, U.; Xiao, Z.C.; Tan, E.K. Notch as a molecular switch in neural stem cells. IUBMB Life 2010, 62, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, S.; Srivatsa, S.; Nityanandam, A.; Tarabykin, V. Ntf3 acts downstream of Sip1 in cortical postmitotic neurons to control progenitor cell fate through feedback signaling. Development 2014, 141, 3324–3330. [Google Scholar] [CrossRef] [PubMed]
- Fritzsch, B.; Farinas, I.; Reichardt, L.F. Lack of neurotrophin 3 causes losses of both classes of spiral ganglion neurons in the cochlea in a region-specific fashion. J. Neurosci. 1997, 17, 6213–6225. [Google Scholar] [CrossRef]
- Herold, S.; Jagasia, R.; Merz, K.; Wassmer, K.; Lie, D.C. CREB signalling regulates early survival, neuronal gene expression and morphological development in adult subventricular zone neurogenesis. Mol. Cell Neurosci. 2011, 46, 79–88. [Google Scholar] [CrossRef]
- Huang, Y.C.; Shih, H.Y.; Lin, S.J.; Chiu, C.C.; Ma, T.L.; Yeh, T.H.; Cheng, Y.C. The epigenetic factor Kmt2a/Mll1 regulates neural progenitor proliferation and neuronal and glial differentiation. Dev. Neurobiol. 2015, 75, 452–462. [Google Scholar] [CrossRef]
- Kandasamy, M.; Lehner, B.; Kraus, S.; Sander, P.R.; Marschallinger, J.; Rivera, F.J.; Trumbach, D.; Ueberham, U.; Reitsamer, H.A.; Strauss, O.; et al. TGF-beta signalling in the adult neurogenic niche promotes stem cell quiescence as well as generation of new neurons. J. Cell Mol. Med. 2014, 18, 1444–1459. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Kawasaki, T. Histone acetyltransferase EP300 regulates the proliferation and differentiation of neural stem cells during adult neurogenesis and regenerative neurogenesis in the zebrafish optic tectum. Neurosci. Lett. 2021, 756, 135978. [Google Scholar] [CrossRef]
- Ishikawa, E.; Ooboshi, H.; Kumai, Y.; Takada, J.; Nakamura, K.; Ago, T.; Sugimori, H.; Kamouchi, M.; Kitazono, T.; Ibayashi, S.; et al. Midkine gene transfer protects against focal brain ischemia and augments neurogenesis. J. Neurol. Sci. 2009, 285, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Conover, J.C.; Doetsch, F.; Garcia-Verdugo, J.M.; Gale, N.W.; Yancopoulos, G.D.; Alvarez-Buylla, A. Disruption of Eph/ephrin signaling affects migration and proliferation in the adult subventricular zone. Nat. Neurosci. 2000, 3, 1091–1097. [Google Scholar] [CrossRef] [PubMed]
- Couillard-Despres, S.; Winner, B.; Schaubeck, S.; Aigner, R.; Vroemen, M.; Weidner, N.; Bogdahn, U.; Winkler, J.; Kuhn, H.G.; Aigner, L. Doublecortin expression levels in adult brain reflect neurogenesis. Eur. J. Neurosci. 2005, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ferri, A.L.; Cavallaro, M.; Braida, D.; Di Cristofano, A.; Canta, A.; Vezzani, A.; Ottolenghi, S.; Pandolfi, P.P.; Sala, M.; DeBiasi, S.; et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 2004, 131, 3805–3819. [Google Scholar] [CrossRef]
- Nishimura, K.; Weichert, R.M.; Liu, W.; Davis, R.L.; Dabdoub, A. Generation of induced neurons by direct reprogramming in the mammalian cochlea. Neuroscience 2014, 275, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Sakurada, K.; Ohshima-Sakurada, M.; Palmer, T.D.; Gage, F.H. Nurr1, an orphan nuclear receptor, is a transcriptional activator of endogenous tyrosine hydroxylase in neural progenitor cells derived from the adult brain. Development 1999, 126, 4017–4026. [Google Scholar] [CrossRef]
- Schmid, R.S.; McGrath, B.; Berechid, B.E.; Boyles, B.; Marchionni, M.; Sestan, N.; Anton, E.S. Neuregulin 1-erbB2 signaling is required for the establishment of radial glia and their transformation into astrocytes in cerebral cortex. Proc. Natl. Acad. Sci. USA 2003, 100, 4251–4256. [Google Scholar] [CrossRef]
- Trigueiros-Cunha, N.; Renard, N.; Humbert, G.; Tavares, M.A.; Eybalin, M. Catecholamine-independent transient expression of tyrosine hydroxylase in primary auditory neurons is coincident with the onset of hearing in the rat cochlea. Eur. J. Neurosci. 2003, 18, 2653–2662. [Google Scholar] [CrossRef]
- Brown, J.P.; Couillard-Despres, S.; Cooper-Kuhn, C.M.; Winkler, J.; Aigner, L.; Kuhn, H.G. Transient expression of doublecortin during adult neurogenesis. J. Comp. Neurol. 2003, 467, 1–10. [Google Scholar] [CrossRef]
- Manohar, S.; Paolone, N.A.; Bleichfeld, M.; Hayes, S.H.; Salvi, R.J.; Baizer, J.S. Expression of doublecortin, a neuronal migration protein, in unipolar brush cells of the vestibulocerebellum and dorsal cochlear nucleus of the adult rat. Neuroscience 2012, 202, 169–183. [Google Scholar] [CrossRef]
- Bunton-Stasyshyn, R.K.; Saccon, R.A.; Fratta, P.; Fisher, E.M. SOD1 Function and Its Implications for Amyotrophic Lateral Sclerosis Pathology: New and Renascent Themes. Neuroscientist 2015, 21, 519–529. [Google Scholar] [CrossRef]
- Guizzetti, M.; Costa, P.; Peters, J.; Costa, L.G. Acetylcholine as a mitogen: Muscarinic receptor-mediated proliferation of rat astrocytes and human astrocytoma cells. Eur. J. Pharmacol. 1996, 297, 265–273. [Google Scholar] [CrossRef]
- Tanie, Y.; Tanabe, N.; Kuboyama, T.; Tohda, C. Extracellular Neuroleukin Enhances Neuroleukin Secretion From Astrocytes and Promotes Axonal Growth in vitro and in vivo. Front. Pharmacol. 2018, 9, 1228. [Google Scholar] [CrossRef]
- Gross, R.E.; Mehler, M.F.; Mabie, P.C.; Zang, Z.; Santschi, L.; Kessler, J.A. Bone morphogenetic proteins promote astroglial lineage commitment by mammalian subventricular zone progenitor cells. Neuron 1996, 17, 595–606. [Google Scholar] [CrossRef]
- Dasgupta, B.; Gutmann, D.H. Neurofibromin regulates neural stem cell proliferation, survival, and astroglial differentiation in vitro and in vivo. J. Neurosci. 2005, 25, 5584–5594. [Google Scholar] [CrossRef]
- Takouda, J.; Katada, S.; Imamura, T.; Sanosaka, T.; Nakashima, K. SoxE group transcription factor Sox8 promotes astrocytic differentiation of neural stem/precursor cells downstream of Nfia. Pharmacol. Res. Perspect. 2021, 9, e00749. [Google Scholar] [CrossRef]
- Rickmann, M.; Wolff, J.R. S100 protein expression in subpopulations of neurons of rat brain. Neuroscience 1995, 67, 977–991. [Google Scholar] [CrossRef] [PubMed]
- Tramontina, F.; Conte, S.; Goncalves, D.; Gottfried, C.; Portela, L.V.; Vinade, L.; Salbego, C.; Goncalves, C.A. Developmental changes in S100B content in brain tissue, cerebrospinal fluid, and astrocyte cultures of rats. Cell Mol. Neurobiol. 2002, 22, 373–378. [Google Scholar] [CrossRef] [PubMed]
- El-Husseini, A.E.; Schnell, E.; Chetkovich, D.M.; Nicoll, R.A.; Bredt, D.S. PSD-95 involvement in maturation of excitatory synapses. Science 2000, 290, 1364–1368. [Google Scholar] [CrossRef]
- Rohrbough, J.; Kent, K.S.; Broadie, K.; Weiss, J.B. Jelly Belly trans-synaptic signaling to anaplastic lymphoma kinase regulates neurotransmission strength and synapse architecture. Dev. Neurobiol. 2013, 73, 189–208. [Google Scholar] [CrossRef] [PubMed]
- Adachi, M.; Lin, P.Y.; Pranav, H.; Monteggia, L.M. Postnatal Loss of Mef2c Results in Dissociation of Effects on Synapse Number and Learning and Memory. Biol. Psychiatry 2016, 80, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Euteneuer, S.; Yang, K.H.; Chavez, E.; Leichtle, A.; Loers, G.; Olshansky, A.; Pak, K.; Schachner, M.; Ryan, A.F. Glial cell line-derived neurotrophic factor (GDNF) induces neuritogenesis in the cochlear spiral ganglion via neural cell adhesion molecule (NCAM). Mol. Cell Neurosci. 2013, 54, 30–43. [Google Scholar] [CrossRef]
- Park, S.; Hong, Y.W. Transcriptional regulation of artemin is related to neurite outgrowth and actin polymerization in mature DRG neurons. Neurosci. Lett. 2006, 404, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.Y.; Tang, B.L. Cell autonomous function of Nogo and reticulons: The emerging story at the endoplasmic reticulum. J. Cell Physiol. 2008, 216, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Hoe, H.S.; Magill, L.A.; Guenette, S.; Fu, Z.; Vicini, S.; Rebeck, G.W. FE65 interaction with the ApoE receptor ApoEr2. J. Biol. Chem. 2006, 281, 24521–24530. [Google Scholar] [CrossRef] [PubMed]
- Soldano, A.; Hassan, B.A. Beyond pathology: APP, brain development and Alzheimer’s disease. Curr. Opin. Neurobiol. 2014, 27, 61–67. [Google Scholar] [CrossRef]
- Xie, W.; Stribley, J.A.; Chatonnet, A.; Wilder, P.J.; Rizzino, A.; McComb, R.D.; Taylor, P.; Hinrichs, S.H.; Lockridge, O. Postnatal developmental delay and supersensitivity to organophosphate in gene-targeted mice lacking acetylcholinesterase. J. Pharmacol. Exp. Ther. 2000, 293, 896–902. [Google Scholar]
- Masuho, I.; Mototani, Y.; Sahara, Y.; Asami, J.; Nakamura, S.; Kozasa, T.; Inoue, T. Dynamic expression patterns of G protein-regulated inducer of neurite outgrowth 1 (GRIN1) and its colocalization with Galphao implicate significant roles of Galphao-GRIN1 signaling in nervous system. Dev. Dyn. 2008, 237, 2415–2429. [Google Scholar] [CrossRef]
- Kamegai, M.; Niijima, K.; Kunishita, T.; Nishizawa, M.; Ogawa, M.; Araki, M.; Ueki, A.; Konishi, Y.; Tabira, T. Interleukin 3 as a trophic factor for central cholinergic neurons in vitro and in vivo. Neuron 1990, 4, 429–436. [Google Scholar] [CrossRef]
- Safaei, R.; Fischer, I. Regulation of microtubule-associated protein 2 (MAP2) mRNA expression during rat brain development. J. Mol. Neurosci. 1989, 1, 189–198. [Google Scholar] [CrossRef]
- Hines, T.J.; Gao, X.; Sahu, S.; Lange, M.M.; Turner, J.R.; Twiss, J.L.; Smith, D.S. An Essential Postdevelopmental Role for Lis1 in Mice. eNeuro 2018, 5. [Google Scholar] [CrossRef]
- Sakurai, T. The role of NrCAM in neural development and disorders--beyond a simple glue in the brain. Mol. Cell Neurosci. 2012, 49, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Markus, E.J.; Petit, T.L. Neocortical synaptogenesis, aging, and behavior: Lifespan development in the motor-sensory system of the rat. Exp. Neurol. 1987, 96, 262–278. [Google Scholar] [CrossRef] [PubMed]
- Stover, T.; Gong, T.L.; Cho, Y.; Altschuler, R.A.; Lomax, M.I. Expression of the GDNF family members and their receptors in the mature rat cochlea. Brain Res. Mol. Brain Res. 2000, 76, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Choi-Lundberg, D.L.; Bohn, M.C. Ontogeny and distribution of glial cell line-derived neurotrophic factor (GDNF) mRNA in rat. Brain Res. Dev. Brain Res. 1995, 85, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Raab, S.; Beck, H.; Gaumann, A.; Yuce, A.; Gerber, H.P.; Plate, K.; Hammes, H.P.; Ferrara, N.; Breier, G. Impaired brain angiogenesis and neuronal apoptosis induced by conditional homozygous inactivation of vascular endothelial growth factor. Thromb. Haemost. 2004, 91, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Guo, D.H.; Hernandez, C.M.; Stranahan, A.M. Endothelial Adora2a Activation Promotes Blood-Brain Barrier Breakdown and Cognitive Impairment in Mice with Diet-Induced Insulin Resistance. J. Neurosci. 2019, 39, 4179–4192. [Google Scholar] [CrossRef]
- Ye, X.; Smallwood, P.; Nathans, J. Expression of the Norrie disease gene (Ndp) in developing and adult mouse eye, ear, and brain. Gene Expr. Patterns 2011, 11, 151–155. [Google Scholar] [CrossRef]
- Ogunshola, O.O.; Stewart, W.B.; Mihalcik, V.; Solli, T.; Madri, J.A.; Ment, L.R. Neuronal VEGF expression correlates with angiogenesis in postnatal developing rat brain. Brain Res. Dev. Brain Res. 2000, 119, 139–153. [Google Scholar] [CrossRef]
- Lu, F.; Wei, L.; Yang, C.; Qiao, Y.; Liu, Y.S.; Chen, X.D.; Wang, J.; Shi, Z.H.; Chen, F.Q.; Zha, D.J.; et al. Nrg1/ErbB2 regulates differentiation and apoptosis of neural stem cells in the cochlear nucleus through PI3K/Akt pathway. Neurosci. Lett. 2021, 751, 135803. [Google Scholar] [CrossRef]
- Gu, F.; Hata, R.; Ma, Y.J.; Tanaka, J.; Mitsuda, N.; Kumon, Y.; Hanakawa, Y.; Hashimoto, K.; Nakajima, K.; Sakanaka, M. Suppression of Stat3 promotes neurogenesis in cultured neural stem cells. J. Neurosci. Res. 2005, 81, 163–171. [Google Scholar] [CrossRef]
- Chen, L.; Melendez, J.; Campbell, K.; Kuan, C.Y.; Zheng, Y. Rac1 deficiency in the forebrain results in neural progenitor reduction and microcephaly. Dev. Biol. 2009, 325, 162–170. [Google Scholar] [CrossRef]
- Sumiyama, K.; Washio-Watanabe, K.; Saitou, N.; Hayakawa, T.; Ueda, S. Class III POU genes: Generation of homopolymeric amino acid repeats under GC pressure in mammals. J. Mol. Evol. 1996, 43, 170–178. [Google Scholar] [CrossRef]
- Lim, D.A.; Tramontin, A.D.; Trevejo, J.M.; Herrera, D.G.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Noggin antagonizes BMP signaling to create a niche for adult neurogenesis. Neuron 2000, 28, 713–726. [Google Scholar] [CrossRef]
- Balordi, F.; Fishell, G. Hedgehog signaling in the subventricular zone is required for both the maintenance of stem cells and the migration of newborn neurons. J. Neurosci. 2007, 27, 5936–5947. [Google Scholar] [CrossRef]
- Kawaguchi, D.; Furutachi, S.; Kawai, H.; Hozumi, K.; Gotoh, Y. Dll1 maintains quiescence of adult neural stem cells and segregates asymmetrically during mitosis. Nat. Commun. 2013, 4, 1880. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.K.; He, Y.; Park, J.S.; Bieri, G.; Snethlage, C.E.; Lin, K.; Gontier, G.; Wabl, R.; Plambeck, K.E.; Udeochu, J.; et al. beta2-microglobulin is a systemic pro-aging factor that impairs cognitive function and neurogenesis. Nat. Med. 2015, 21, 932–937. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.C.; Xiao, M.F.; Jakovcevski, I.; Sivukhina, E.; Hargus, G.; Cui, Y.F.; Irintchev, A.; Schachner, M.; Bernreuther, C. The extracellular matrix glycoprotein tenascin-R regulates neurogenesis during development and in the adult dentate gyrus of mice. J. Cell Sci. 2014, 127, 641–652. [Google Scholar] [CrossRef]
- Lin, Y.; He, M.; Zeng, X.; Sheng, X.; Wang, S.; Cui, F. 4-hydroxybenzophenone exposure inhibits mouse hippocampal neural stem cell proliferation in vitro by upregulating Cxcl1. Toxicol. In Vitro 2022, 82, 105367. [Google Scholar] [CrossRef]
- Silva, V.A.; Gomide, V.C.; Chadi, G. Fibroblast growth factor-2 immunoreactivity is present in the central and peripheral auditory pathways of adult rats. J. Morphol. 2005, 265, 141–151. [Google Scholar] [CrossRef]
- Ray, J.; Gage, F.H. Differential properties of adult rat and mouse brain-derived neural stem/progenitor cells. Mol. Cell Neurosci. 2006, 31, 560–573. [Google Scholar] [CrossRef] [PubMed]
- Tureyen, K.; Vemuganti, R.; Bowen, K.K.; Sailor, K.A.; Dempsey, R.J. EGF and FGF-2 infusion increases post-ischemic neural progenitor cell proliferation in the adult rat brain. Neurosurgery 2005, 57, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hu, J.; Zhou, L.; Pollard, S.M.; Smith, A. Interplay between FGF2 and BMP controls the self-renewal, dormancy and differentiation of rat neural stem cells. J. Cell Sci. 2011, 124, 1867–1877. [Google Scholar] [CrossRef] [PubMed]
- Kiyota, T.; Ingraham, K.L.; Jacobsen, M.T.; Xiong, H.; Ikezu, T. FGF2 gene transfer restores hippocampal functions in mouse models of Alzheimer’s disease and has therapeutic implications for neurocognitive disorders. Proc. Natl. Acad. Sci. USA 2011, 108, E1339–E1348. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Engert, J.; Doll, J.; Vona, B.; Ehret Kasemo, T.; Spahn, B.; Hagen, R.; Rak, K.; Voelker, J. mRNA Abundance of Neurogenic Factors Correlates with Hearing Capacity in Auditory Brainstem Nuclei of the Rat. Life 2023, 13, 1858. https://doi.org/10.3390/life13091858
Engert J, Doll J, Vona B, Ehret Kasemo T, Spahn B, Hagen R, Rak K, Voelker J. mRNA Abundance of Neurogenic Factors Correlates with Hearing Capacity in Auditory Brainstem Nuclei of the Rat. Life. 2023; 13(9):1858. https://doi.org/10.3390/life13091858
Chicago/Turabian StyleEngert, Jonas, Julia Doll, Barbara Vona, Totta Ehret Kasemo, Bjoern Spahn, Rudolf Hagen, Kristen Rak, and Johannes Voelker. 2023. "mRNA Abundance of Neurogenic Factors Correlates with Hearing Capacity in Auditory Brainstem Nuclei of the Rat" Life 13, no. 9: 1858. https://doi.org/10.3390/life13091858
APA StyleEngert, J., Doll, J., Vona, B., Ehret Kasemo, T., Spahn, B., Hagen, R., Rak, K., & Voelker, J. (2023). mRNA Abundance of Neurogenic Factors Correlates with Hearing Capacity in Auditory Brainstem Nuclei of the Rat. Life, 13(9), 1858. https://doi.org/10.3390/life13091858





