Novel Compounds Derived from DFPM Induce Root Growth Arrest through the Specific VICTR Alleles of Arabidopsis Accessions
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Plant Materials and Root Growth Assay
2.3. Reporter Gene Expression Analysis
2.4. Quantitative Real-Time PCR
2.5. Comparative Sequence Analysis
2.6. Map-Based Cloning
3. Results
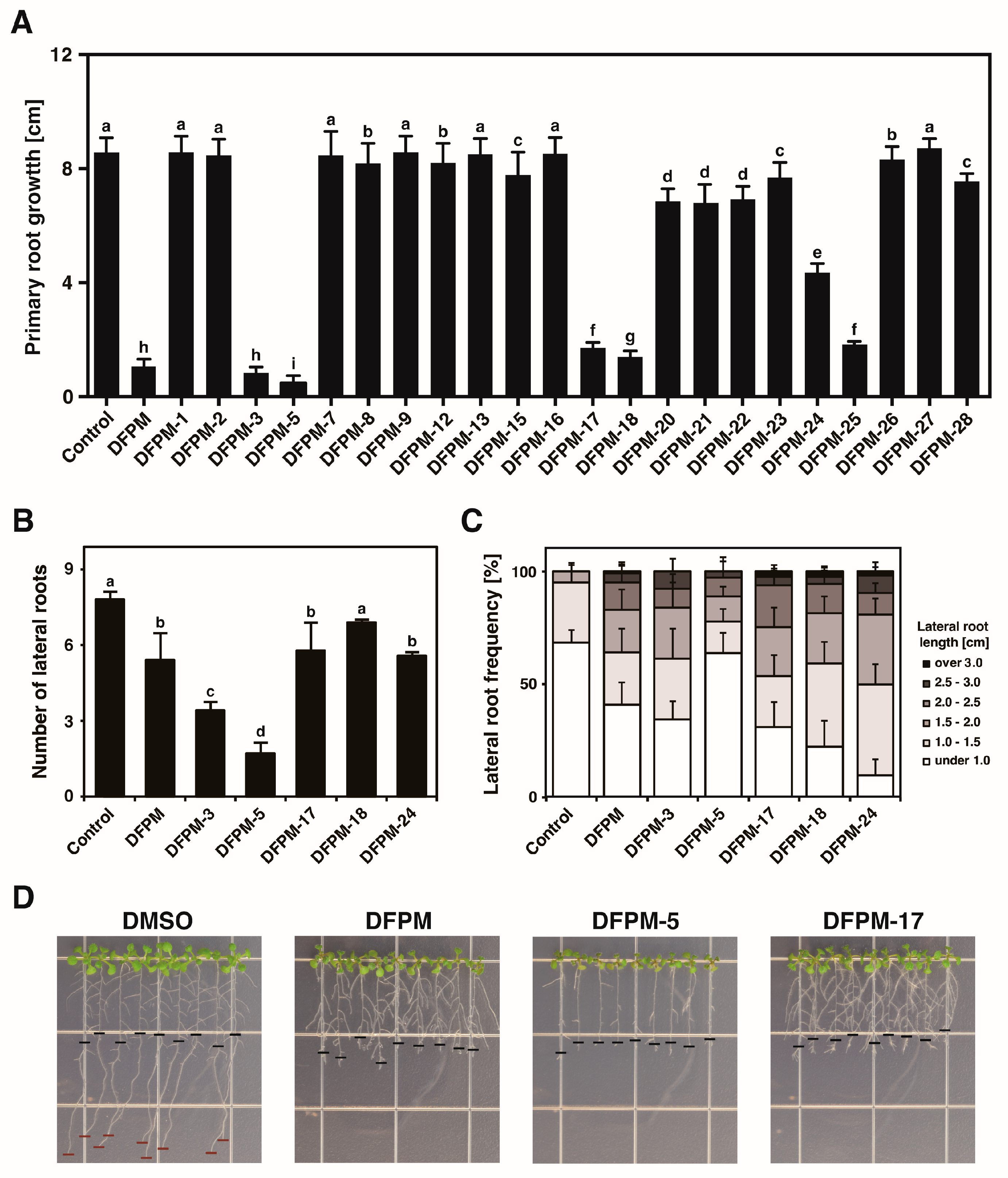
3.1. DFPM Derivatives Have Specific Effects on Root Growth
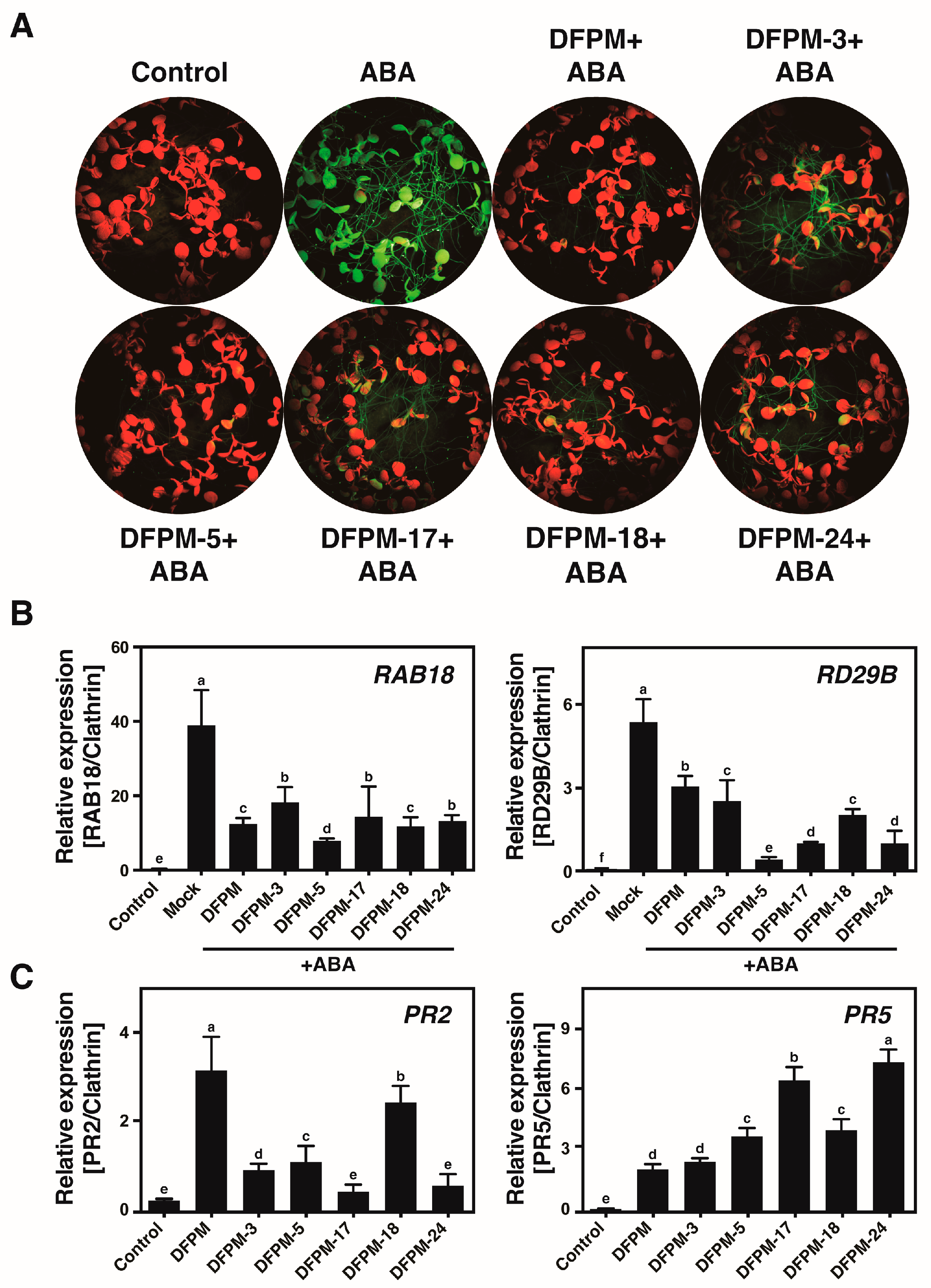
3.2. Selected Chemicals Affect Expression Patterns of the ABA-Responsive Genes and the Pathogen-Responsive Genes
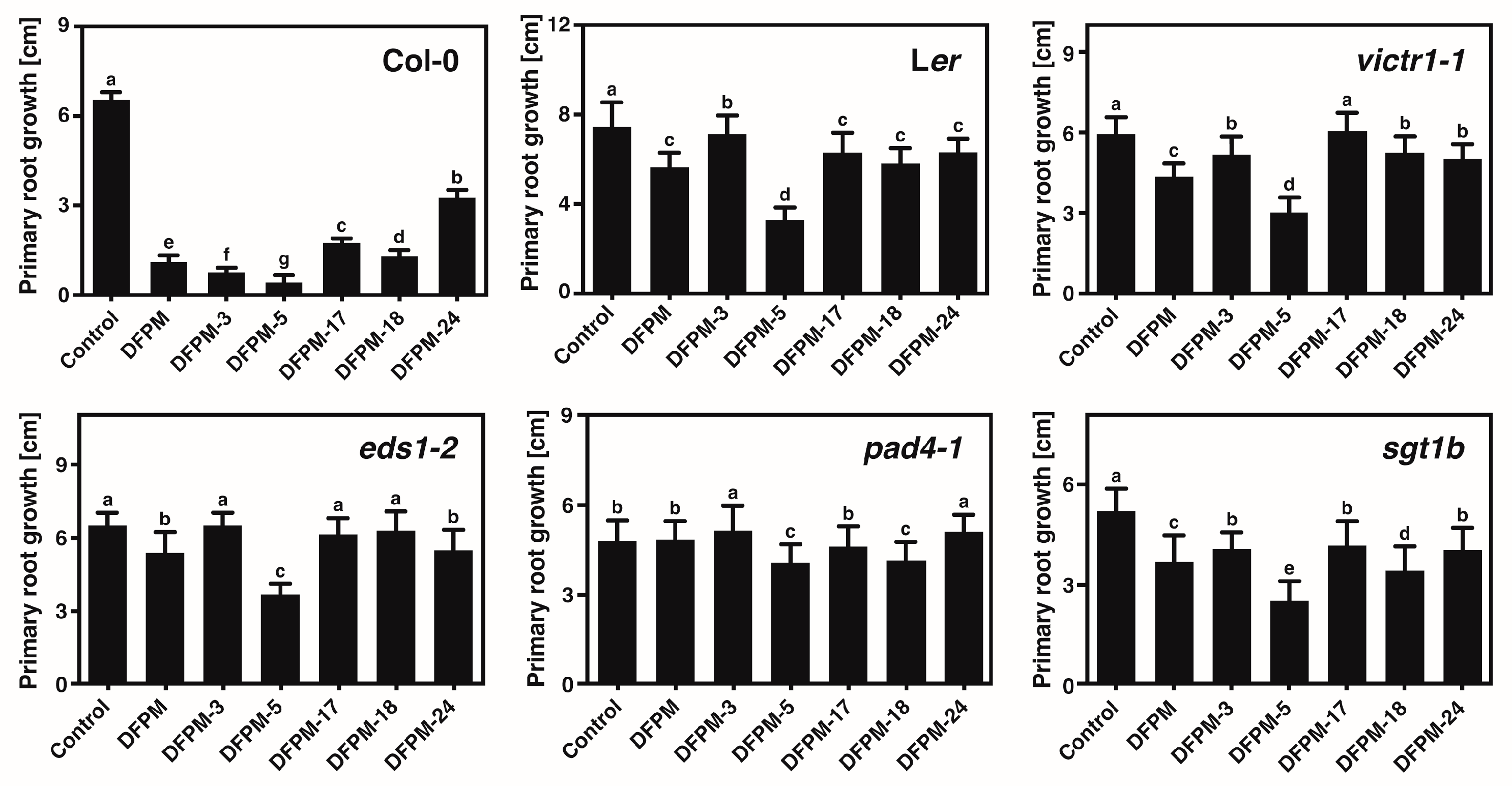
3.3. Selected DFPM Derivatives May Interfere with Root Growth through the Same Signal Transduction Pathway Controlled by DFPM
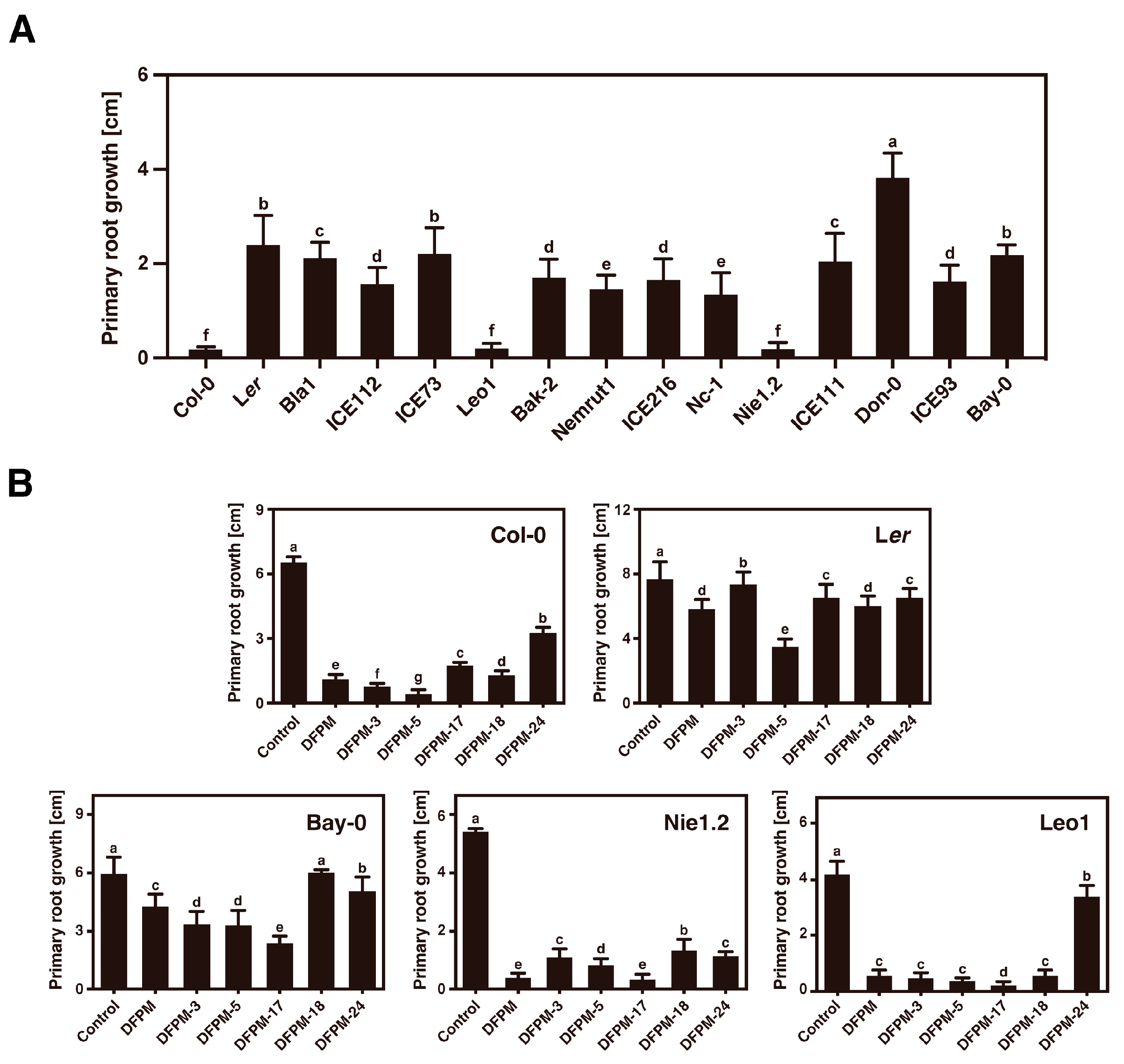
3.4. Selected DFPM Derivatives Cause Accession-Specific Root Growth Arrest in Col-0, Nie1.2, and Leo1
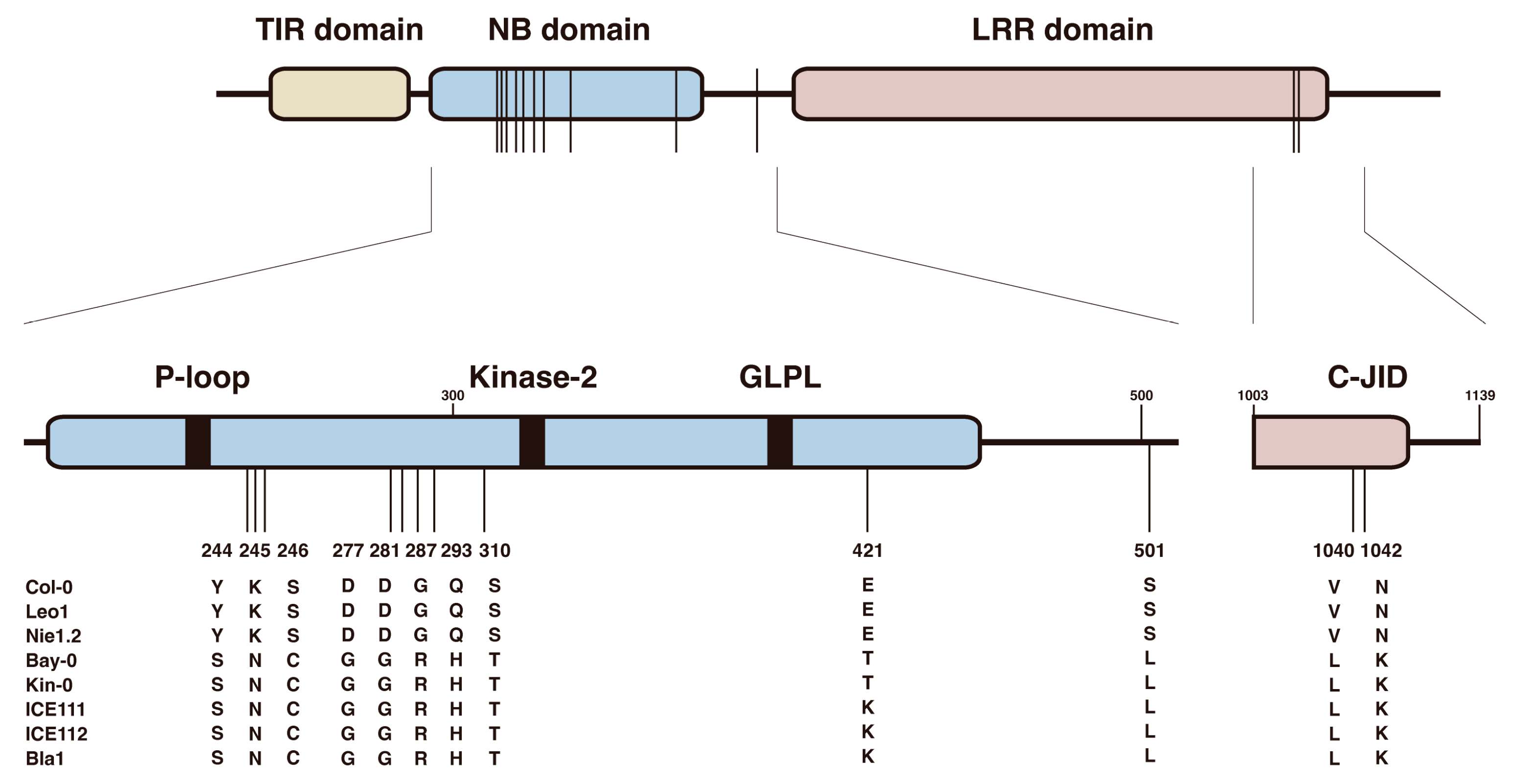
3.5. Twelve Natural Variation Sites in the TNL Receptor VICTR Are Required for Triggering the DFPM-Induced Root Growth Arrest
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lepri, A.; Longo, C.; Messore, A.; Kazmi, H.; Madia, V.N.; Di Santo, R.; Costi, R.; Vittorioso, P. Plants and Small Molecules: An Up-and-Coming Synergy. Plants 2023, 12, 1729. [Google Scholar] [CrossRef] [PubMed]
- Pasquer, Q.T.L.; Tsakoumagkos, I.A.; Hoogendoorn, S. From Phenotypic Hit to Chemical Probe: Chemical Biology Approaches to Elucidate Small Molecule Action in Complex Biological Systems. Molecules 2020, 25, 5702. [Google Scholar] [CrossRef] [PubMed]
- Dejonghe, W.; Russinova, E. Plant Chemical Genetics: From Phenotype-Based Screens to Synthetic Biology. Plant Physiol. 2017, 174, 5–20. [Google Scholar] [CrossRef]
- Xuan, W.; Murphy, E.; Beeckman, T.; Audenaert, D.; De Smet, I. Synthetic molecules: Helping to unravel plant signal transduction. J. Chem. Biol. 2013, 6, 43–50. [Google Scholar] [CrossRef][Green Version]
- Hewage, K.A.H.; Yang, J.F.; Wang, D.; Hao, G.F.; Yang, G.F.; Zhu, J.K. Chemical Manipulation of Abscisic Acid Signaling: A New Approach to Abiotic and Biotic Stress Management in Agriculture. Adv. Sci. 2020, 7, 2001265. [Google Scholar] [CrossRef]
- Hu, D.; Wei, L.; Liao, W. Brassinosteroids in Plants: Crosstalk with Small-Molecule Compounds. Biomolecules 2021, 11, 1800. [Google Scholar] [CrossRef]
- Chini, A.; Monte, I.; Fernandez-Barbero, G.; Boter, M.; Hicks, G.; Raikhel, N.; Solano, R. A small molecule antagonizes jasmonic acid perception and auxin responses in vascular and nonvascular plants. Plant Physiol. 2021, 187, 1399–1413. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Asami, T. Chemical regulators of plant hormones and their applications in basic research and agriculture. Biosci. Biotechnol. Biochem. 2018, 82, 1265–1300. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Ng, L.M.; Melcher, K.; Teh, B.T.; Xu, H.E. Abscisic acid perception and signaling: Structural mechanisms and applications. Acta Pharmacol. Sin. 2014, 35, 567–584. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Kim, T.H.; Hauser, F.; Ha, T.; Xue, S.; Bohmer, M.; Nishimura, N.; Munemasa, S.; Hubbard, K.; Peine, N.; Lee, B.H.; et al. Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr. Biol. 2011, 21, 990–997. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef]
- Wu, L.; Chen, H.; Curtis, C.; Fu, Z.Q. Go in for the kill: How plants deploy effector-triggered immunity to combat pathogens. Virulence 2014, 5, 710–721. [Google Scholar] [CrossRef] [PubMed]
- van Wersch, S.; Tian, L.; Hoy, R.; Li, X. Plant NLRs: The Whistleblowers of Plant Immunity. Plant Commun. 2020, 1, 100016. [Google Scholar] [CrossRef] [PubMed]
- Eitas, T.K.; Dangl, J.L. NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 2010, 13, 472–477. [Google Scholar] [CrossRef]
- Cesari, S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2018, 219, 17–24. [Google Scholar] [CrossRef]
- Shao, Z.Q.; Xue, J.Y.; Wu, P.; Zhang, Y.M.; Wu, Y.; Hang, Y.Y.; Wang, B.; Chen, J.Q. Large-Scale Analyses of Angiosperm Nucleotide-Binding Site-Leucine-Rich Repeat Genes Reveal Three Anciently Diverged Classes with Distinct Evolutionary Patterns. Plant Physiol. 2016, 170, 2095–2109. [Google Scholar] [CrossRef]
- Jia, A.; Huang, S.; Ma, S.; Chang, X.; Han, Z.; Chai, J. TIR-catalyzed nucleotide signaling molecules in plant defense. Curr. Opin. Plant Biol. 2023, 73, 102334. [Google Scholar] [CrossRef]
- Essuman, K.; Milbrandt, J.; Dangl, J.L.; Nishimura, M.T. Shared TIR enzymatic functions regulate cell death and immunity across the tree of life. Science 2022, 377, eabo0001. [Google Scholar] [CrossRef] [PubMed]
- Lapin, D.; Johanndrees, O.; Wu, Z.; Li, X.; Parker, J.E. Molecular innovations in plant TIR-based immunity signaling. Plant Cell 2022, 34, 1479–1496. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, W.; Zhang, T.; Gong, Z.; Zhao, H.; Han, G.Z. Out of Water: The Origin and Early Diversification of Plant R-Genes. Plant Physiol. 2018, 177, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Barragan, A.C.; Weigel, D. Plant NLR diversity: The known unknowns of pan-NLRomes. Plant Cell 2021, 33, 814–831. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef]
- Van de Weyer, A.L.; Monteiro, F.; Furzer, O.J.; Nishimura, M.T.; Cevik, V.; Witek, K.; Jones, J.D.G.; Dangl, J.L.; Weigel, D.; Bemm, F. A Species-Wide Inventory of NLR Genes and Alleles in Arabidopsis thaliana. Cell 2019, 178, 1260–1272. [Google Scholar] [CrossRef]
- Gu, L.; Si, W.; Zhao, L.; Yang, S.; Zhang, X. Dynamic evolution of NBS-LRR genes in bread wheat and its progenitors. Mol. Genet. Genom. 2015, 290, 727–738. [Google Scholar] [CrossRef]
- van Wersch, S.; Li, X. Stronger When Together: Clustering of Plant NLR Disease resistance Genes. Trends Plant Sci. 2019, 24, 688–699. [Google Scholar] [CrossRef]
- Kim, T.H.; Kunz, H.H.; Bhattacharjee, S.; Hauser, F.; Park, J.; Engineer, C.; Liu, A.; Ha, T.; Parker, J.E.; Gassmann, W.; et al. Natural variation in small molecule-induced TIR-NB-LRR signaling induces root growth arrest via EDS1- and PAD4-complexed R protein VICTR in Arabidopsis. Plant Cell 2012, 24, 5177–5192. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, J.I. An Efficient Synthesis of N, N-Dialkyl-5-(chlorophenyl)-2-furancarbothioamides from 2-Furoic Acid. J. Korean Chem. Soc. 2016, 60, 457–461. [Google Scholar] [CrossRef][Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Mahanta, N.; Szantai-Kis, D.M.; Petersson, E.J.; Mitchell, D.A. Biosynthesis and Chemical Applications of Thioamides. ACS Chem. Biol. 2019, 14, 142–163. [Google Scholar] [CrossRef] [PubMed]
- Aulakh, J.K.; Lobana, T.S.; Sood, H.; Arora, D.S.; Kaur, R.; Singh, J.; Garcia-Santos, I.; Kaur, M.; Jasinski, J.P. Silver derivatives of multi-donor heterocyclic thioamides as antimicrobial/anticancer agents: Unusual bio-activity against methicillin resistant S. aureus, S. epidermidis, and E. faecalis and human bone cancer MG63 cell line. RSC Adv. 2019, 9, 15470–15487. [Google Scholar] [CrossRef]
- Chen, X.; Mietlicki-Baase, E.G.; Barrett, T.M.; McGrath, L.E.; Koch-Laskowski, K.; Ferrie, J.J.; Hayes, M.R.; Petersson, E.J. Thioamide Substitution Selectively Modulates Proteolysis and Receptor Activity of Therapeutic Peptide Hormones. J. Am. Chem. Soc. 2017, 139, 16688–16695. [Google Scholar] [CrossRef] [PubMed]
- Mikhailovskii, A.G.; Yusov, A.S.; Makhmudov, R.R.; Starkova, A.V.; Rudakova, I.P. Synthesis and Analgesic, Anthelmintic, and Insecticidal Activity of 3,3-Dialkyl-1-(2-Phenylamino-2-Thioxoethyl)-3,4-Dihydroisoquinolinium Chlorides. Pharm. Chem. J. 2018, 52, 716–720. [Google Scholar] [CrossRef]
- Nishida, C.R.; Ortiz de Montellano, P.R. Bioactivation of antituberculosis thioamide and thiourea prodrugs by bacterial and mammalian flavin monooxygenases. Chem. Biol. Interact. 2011, 192, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Khatri, B.; Raghunathan, S.; Chakraborti, S.; Rahisuddin, R.; Kumaran, S.; Tadala, R.; Wagh, P.; Priyakumar, U.D.; Chatterjee, J. Desolvation of Peptide Bond by O to S Substitution Impacts Protein Stability. Angew. Chem. Int. Ed. Engl. 2021, 60, 24870–24874. [Google Scholar] [CrossRef]
- Dubey, N.; Singh, K. Role of NBS-LRR proteins in plant defense. In Molecular Aspects of Plant-Pathogen Interaction; Singh, A., Singh, I.K., Eds.; Springer: Singapore, 2018; pp. 115–138. [Google Scholar]
- Chen, J.; Zhang, X.; Rathjen, J.P.; Dodds, P.N. Direct recognition of pathogen effectors by plant NLR immune receptors and downstream signalling. Essays Biochem. 2022, 66, 471–483. [Google Scholar] [CrossRef]
- Maruta, N.; Burdett, H.; Lim, B.Y.J.; Hu, X.; Desa, S.; Manik, M.K.; Kobe, B. Structural basis of NLR activation and innate immune signalling in plants. Immunogenetics 2022, 74, 5–26. [Google Scholar] [CrossRef]
- Martin, R.; Qi, T.; Zhang, H.; Liu, F.; King, M.; Toth, C.; Nogales, E.; Staskawicz, B.J. Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science 2020, 370, 1185. [Google Scholar] [CrossRef]
- Saucet, S.B.; Esmenjaud, D.; Van Ghelder, C. Integrity of the post-LRR domain is required for TIR-NB-LRR function. Mol. Plant-Microbe Interact. 2021, 34, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Lapin, D.; Liu, L.; Sun, Y.; Song, W.; Zhang, X.; Logemann, E.; Yu, D.; Wang, J.; Jirschitzka, J.; et al. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 2020, 370, 1184. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Cho, M.; Kim, T.-H. Novel Compounds Derived from DFPM Induce Root Growth Arrest through the Specific VICTR Alleles of Arabidopsis Accessions. Life 2023, 13, 1797. https://doi.org/10.3390/life13091797
Kim S, Cho M, Kim T-H. Novel Compounds Derived from DFPM Induce Root Growth Arrest through the Specific VICTR Alleles of Arabidopsis Accessions. Life. 2023; 13(9):1797. https://doi.org/10.3390/life13091797
Chicago/Turabian StyleKim, Seojung, Miri Cho, and Tae-Houn Kim. 2023. "Novel Compounds Derived from DFPM Induce Root Growth Arrest through the Specific VICTR Alleles of Arabidopsis Accessions" Life 13, no. 9: 1797. https://doi.org/10.3390/life13091797
APA StyleKim, S., Cho, M., & Kim, T.-H. (2023). Novel Compounds Derived from DFPM Induce Root Growth Arrest through the Specific VICTR Alleles of Arabidopsis Accessions. Life, 13(9), 1797. https://doi.org/10.3390/life13091797





