The Underrated Gut Microbiota Helminths, Bacteriophages, Fungi, and Archaea
Abstract
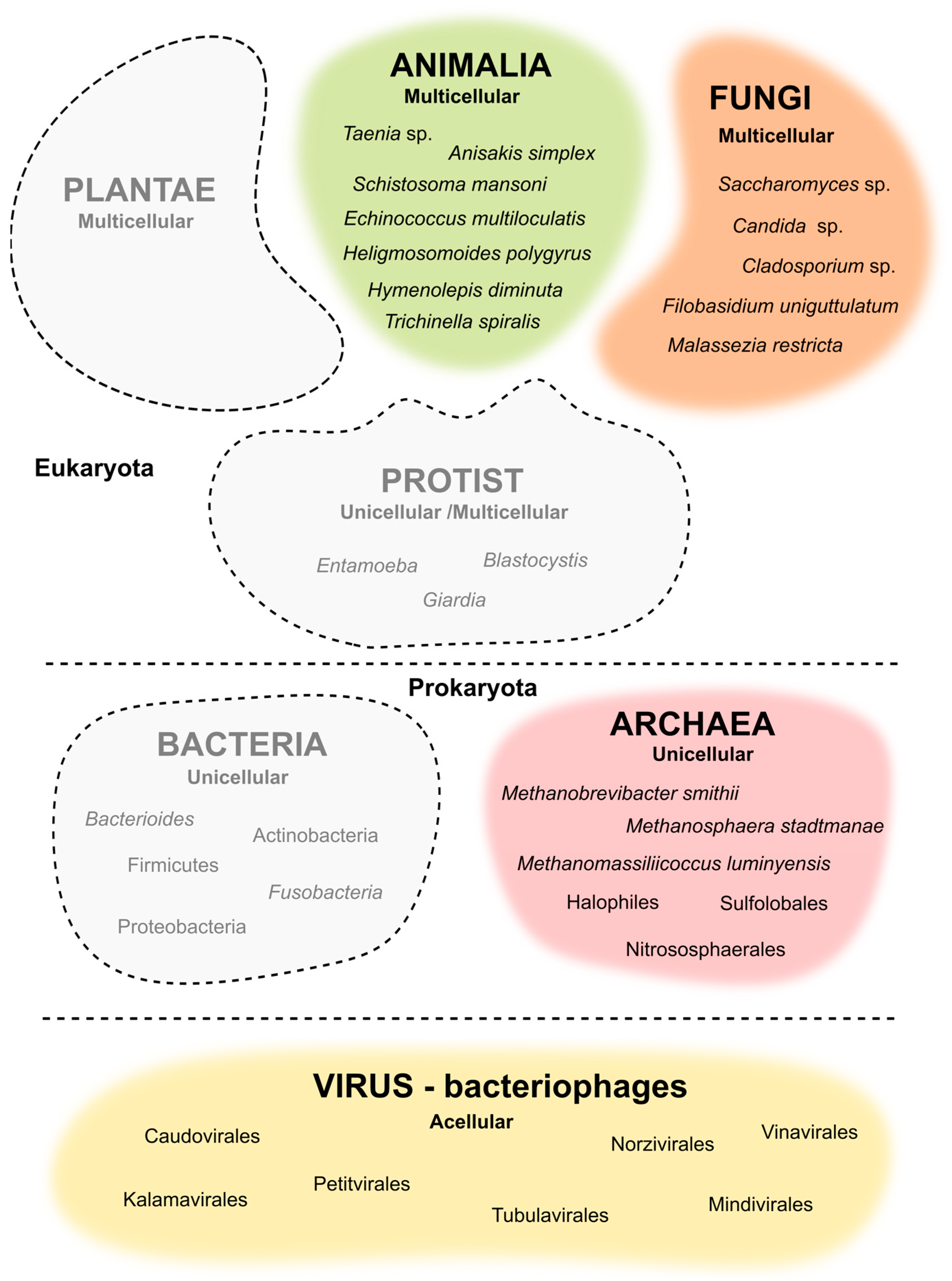
:1. Introduction
2. Helminths
3. Bacteriophages
4. Fungi
5. Archaea
6. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Butler, M.I.; Bastiaanssen, T.F.S.; Long-Smith, C.; Morkl, S.; Berding, K.; Ritz, N.L.; Strain, C.; Patangia, D.; Patel, S.; Stanton, C.; et al. The Gut Microbiome in Social Anxiety Disorder: Evidence of Altered Composition and Function. Transl. Psychiatry 2023, 13, 95. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Tsolis, R.M.; Bäumler, A.J. The Microbiome and Gut Homeostasis. Science 2022, 377, eabp9960. [Google Scholar] [CrossRef] [PubMed]
- Daniel, H. Gut Physiology Meets Microbiome Science. Gut Microbiome 2023, 4, e1. [Google Scholar] [CrossRef]
- Grasa, L.; Abecia, L.; Forcén, R.; Castro, M.; de Jalón, J.A.G.; Latorre, E.; Alcalde, A.I.; Murillo, M.D. Antibiotic-Induced Depletion of Murine Microbiota Induces Mild Inflammation and Changes in Toll-Like Receptor Patterns and Intestinal Motility. Microb. Ecol. 2015, 70, 835–848. [Google Scholar] [CrossRef]
- Layunta, E.; Jäverfelt, S.; Dolan, B.; Arike, L.; Pelaseyed, T. Il-22 Promotes the Formation of a Muc17 Glycocalyx Barrier in the Postnatal Small Intestine During Weaning. Cell Rep. 2021, 34, 108757. [Google Scholar] [CrossRef]
- Suriano, F.; Nyström, E.E.L.; Sergi, D.; Gustafsson, J.K. Diet, Microbiota, and the Mucus Layer: The Guardians of Our Health. Front. Immunol. 2022, 13, 953196. [Google Scholar] [CrossRef]
- de Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut Microbiome and Health: Mechanistic Insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human Nutrition, the Gut Microbiome and the Immune System. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef]
- Mafra, D.; Ribeiro, M.; Fonseca, L.; Regis, B.; Cardozo, L.; Santos, H.F.D.; de Jesus, H.E.; Schultz, J.; Shiels, P.G.; Stenvinkel, P.; et al. Archaea from the Gut Microbiota of Humans: Could Be Linked to Chronic Diseases? Anaerobe 2022, 77, 102629. [Google Scholar] [CrossRef] [PubMed]
- Stevens, E.J.; Bates, K.A.; King, K.C. Host Microbiota Can Facilitate Pathogen Infection. PLoS Pathog. 2021, 17, e1009514. [Google Scholar] [CrossRef]
- Dridi, B. Laboratory Tools for Detection of Archaea in Humans. Clin. Microbiol. Infect. 2012, 18, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Houpikian, P.; Raoult, D. Traditional and Molecular Techniques for the Study of Emerging Bacterial Diseases: One Laboratory’s Perspective. Emerg. Infect. Dis. 2002, 8, 122–131. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List; Bolchacova, E.; et al. Nuclear Ribosomal Internal Transcribed Spacer (Its) Region as a Universal DNA Barcode Marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Nkamga, V.D.; Henrissat, B.; Drancourt, M. Archaea: Essential Inhabitants of the Human Digestive Microbiota. Hum. Microbiome J. 2016, 3, 1–8. [Google Scholar] [CrossRef]
- Scanlan, P.D.; Shanahan, F.; Marchesi, J.R. Human Methanogen Diversity and Incidence in Healthy and Diseased Colonic Groups Using Mcra Gene Analysis. BMC Microbiol. 2008, 8, 79. [Google Scholar] [CrossRef]
- Cassedy, A.; Parle-McDermott, A.; O’kennedy, R. Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods. Front. Mol. Biosci. 2021, 8, 637559. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, Y.; Hu, H.; Tan, Y.; Ma, Y. Metagenomic Analysis Reveals Unexplored Diversity of Archaeal Virome in the Human Gut. Nat. Commun. 2022, 13, 7978. [Google Scholar] [CrossRef]
- Fitzpatrick, A.H.; Rupnik, A.; O’Shea, H.; Crispie, F.; Keaveney, S.; Cotter, P. High Throughput Sequencing for the Detection and Characterization of Rna Viruses. Front. Microbiol. 2021, 12, 621719. [Google Scholar] [CrossRef]
- Quer, J.; Colomer-Castell, S.; Campos, C.; Andrés, C.; Piñana, M.; Cortese, M.F.; González-Sánchez, A.; Garcia-Cehic, D.; Ibáñez, M.; Pumarola, T.; et al. Next-Generation Sequencing for Confronting Virus Pandemics. Viruses 2022, 14, 600. [Google Scholar] [CrossRef]
- Charlier, J.; Rinaldi, L.; Musella, V.; Ploeger, H.; Chartier, C.; Vineer, H.R.; Hinney, B.; von Samson-Himmelstjerna, G.; Băcescu, B.; Mickiewicz, M.; et al. Initial Assessment of the Economic Burden of Major Parasitic Helminth Infections to the Ruminant Livestock Industry in Europe. Prev. Veter. Med. 2020, 182, 105103. [Google Scholar] [CrossRef]
- Taraschewski, H. Host-Parasite Interactions in Acanthocephala: A Morphological Approach. Adv. Parasitol. 2000, 46, 1–179. [Google Scholar] [PubMed]
- Mathison, B.A.; Mehta, N.; Couturier, M.R. Human Acanthocephaliasis: A Thorn in the Side of Parasite Diagnostics. J. Clin. Microbiol. 2021, 59, e0269120. [Google Scholar] [CrossRef] [PubMed]
- Pullan, R.L.; Smith, J.L.; Jasrasaria, R.; Brooker, S.J. Global Numbers of Infection and Disease Burden of Soil Transmitted Helminth Infections in 2010. Parasites Vectors 2014, 7, 37. [Google Scholar] [CrossRef]
- Bourke, C.D.; Maizels, R.M.; Mutapi, F. Acquired Immune Heterogeneity and Its Sources in Human Helminth Infection. Parasitology 2011, 138, 139–159. [Google Scholar] [CrossRef]
- Lee, S.C.; Tang, M.S.; Lim, Y.A.L.; Choy, S.H.; Kurtz, Z.D.; Cox, L.M.; Gundra, U.M.; Cho, I.; Bonneau, R.; Blaser, M.J.; et al. Helminth Colonization Is Associated with Increased Diversity of the Gut Microbiota. PLoS Neglected Trop. Dis. 2014, 8, e2880. [Google Scholar] [CrossRef]
- Zaiss, M.M.; Rapin, A.; Lebon, L.; Dubey, L.K.; Mosconi, I.; Sarter, K.; Piersigilli, A.; Menin, L.; Walker, A.W.; Rougemont, J.; et al. The Intestinal Microbiota Contributes to the Ability of Helminths to Modulate Allergic Inflammation. Immunity 2015, 43, 998–1010. [Google Scholar] [CrossRef]
- Kim, C.H. Control of Lymphocyte Functions by Gut Microbiota-Derived Short-Chain Fatty Acids. Cell. Mol. Immunol. 2021, 18, 1161–1171. [Google Scholar] [CrossRef]
- Rausch, S.; Midha, A.; Kuhring, M.; Affinass, N.; Radonic, A.; Kühl, A.A.; Bleich, A.; Renard, B.Y.; Hartmann, S. Parasitic Nematodes Exert Antimicrobial Activity and Benefit from Microbiota-Driven Support for Host Immune Regulation. Front. Immunol. 2018, 9, 2282. [Google Scholar] [CrossRef]
- Robinson, M.W.; Hutchinson, A.T.; Dalton, J.P.; Donnelly, S. Peroxiredoxin: A Central Player in Immune Modulation. Parasite Immunol. 2010, 32, 305–313. [Google Scholar] [CrossRef]
- Zaiss, M.M.; Harris, N.L. Interactions between the Intestinal Microbiome and Helminth Parasites. Parasite Immunol. 2016, 38, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Moyat, M.; Lebon, L.; Perdijk, O.; Wickramasinghe, L.C.; Zaiss, M.M.; Mosconi, I.; Volpe, B.; Guenat, N.; Shah, K.; Coakley, G.; et al. Microbial Regulation of Intestinal Motility Provides Resistance against Helminth Infection. Mucosal Immunol. 2022, 15, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Llinas-Caballero, K.; Caraballo, L. Helminths and Bacterial Microbiota: The Interactions of Two of Humans’ Old Friends. Int. J. Mol. Sci. 2022, 23, 13358. [Google Scholar] [CrossRef] [PubMed]
- Hayes, K.S.; Bancroft, A.J.; Goldrick, M.; Portsmouth, C.; Roberts, I.S.; Grencis, R.K. Exploitation of the Intestinal Microflora by the Parasitic Nematode Trichuris Muris. Science 2010, 328, 1391–1394. [Google Scholar] [CrossRef]
- Jin, X.; Liu, Y.; Wang, J.; Wang, X.; Tang, B.; Liu, M.; Liu, X. Beta-Glucan-Triggered Akkermansia Muciniphila Expansion Facilitates the Expulsion of Intestinal Helminth Via Tlr2 in Mice. Carbohydr. Polym. 2022, 275, 118719. [Google Scholar] [CrossRef]
- Rajamanickam, A.; Munisankar, S.; Menon, P.A.; Dolla, C.; Nutman, T.B.; Babu, S. Helminth Mediated Attenuation of Systemic Inflammation and Microbial Translocation in Helminth-Diabetes Comorbidity. Front. Cell. Infect. Microbiol. 2020, 10, 431. [Google Scholar] [CrossRef]
- Horsnell, W.G.C.; Oudhoff, M.J. Helminths Are Positively Amping up Gut De-Bugging. Cell Host Microbe 2022, 30, 1–2. [Google Scholar] [CrossRef]
- Popov, J.; Caputi, V.; Nandeesha, N.; Rodriguez, D.A.; Pai, N. Microbiota-Immune Interactions in Ulcerative Colitis and Colitis Associated Cancer and Emerging Microbiota-Based Therapies. Int. J. Mol. Sci. 2021, 22, 11365. [Google Scholar] [CrossRef]
- Maruszewska-Cheruiyot, M.; Donskow-Łysoniewska, K.; Doligalska, M. Helminth Therapy: Advances in the Use of Parasitic Worms against Inflammatory Bowel Diseases and Its Challenges. Helminthologia 2018, 55, 1–11. [Google Scholar] [CrossRef] [PubMed]
- León-Cabrera, S.; Callejas, B.E.; Ledesma-Soto, Y.; Coronel, J.; Pérez-Plasencia, C.; Gutiérrez-Cirlos, E.B.; Ávila-Moreno, F.; Rodríguez-Sosa, M.; Hernández-Pando, R.; Marquina-Castillo, B.; et al. Extraintestinal Helminth Infection Reduces the Development of Colitis-Associated Tumorigenesis. Int. J. Biol. Sci. 2014, 10, 948–956. [Google Scholar] [CrossRef]
- Chu, K.M.; Watermeyer, G.; Shelly, L.; Janssen, J.; May, T.D.; Brink, K.; Benefeld, G.; Li, X. Childhood Helminth Exposure Is Protective against Inflammatory Bowel Disease: A Case Control Study in South Africa. Inflamm. Bowel Dis. 2013, 19, 614–620. [Google Scholar] [CrossRef]
- Al Nabhani, Z.; Eberl, G. Imprinting of the Immune System by the Microbiota Early in Life. Mucosal Immunol. 2020, 13, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Long, S.R.; Liu, R.D.; Kumar, D.V.; Wang, Z.Q.; Su, C.-W. Immune Protection of a Helminth Protein in the Dss-Induced Colitis Model in Mice. Front. Immunol. 2021, 12, 664998. [Google Scholar] [CrossRef] [PubMed]
- Bager, P.; Hansen, A.V.; Wohlfahrt, J.; Melbye, M. Helminth Infection Does Not Reduce Risk for Chronic Inflammatory Disease in a Population-Based Cohort Study. Gastroenterology 2012, 142, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Cortes, A.; Munoz-Antoli, C.; Esteban, J.G.; Toledo, R. Th2 and Th1 Responses: Clear and Hidden Sides of Immunity against Intestinal Helminths. Trends Parasitol. 2017, 33, 678–693. [Google Scholar] [CrossRef]
- Cho, M.K.; Park, M.K.; Kang, S.A.; Choi, S.H.; Ahn, S.C.; Yu, H.S. Trichinella spiralis Infection Suppressed Gut Inflammation with Cd4+Cd25+Foxp3+ T Cell Recruitment. Korean J. Parasitol. 2012, 50, 385–390. [Google Scholar] [CrossRef]
- Mishra, P.K.; Palma, M.; Bleich, D.; Loke, P.; Gause, W.C. Systemic Impact of Intestinal Helminth Infections. Mucosal Immunol. 2014, 7, 753–762. [Google Scholar] [CrossRef]
- Briggs, N.; Weatherhead, J.; Sastry, K.J.; Hotez, P.J. The Hygiene Hypothesis and Its Inconvenient Truths About Helminth Infections. PLoS Neglected Trop. Dis. 2016, 10, e0004944. [Google Scholar] [CrossRef]
- Santiago, H.C.; Nutman, T.B. Human Helminths and Allergic Disease: The Hygiene Hypothesis and Beyond. Am. J. Trop. Med. Hyg. 2016, 95, 746–753. [Google Scholar] [CrossRef]
- Jain, N. The Early Life Education of the Immune System: Moms, Microbes and (Missed) Opportunities. Gut Microbes 2020, 12, 1824564. [Google Scholar] [CrossRef]
- Laforest-Lapointe, I.; Arrieta, M.-C. Patterns of Early-Life Gut Microbial Colonization During Human Immune Development: An Ecological Perspective. Front. Immunol. 2017, 8, 788. [Google Scholar] [CrossRef]
- Saluzzo, S.; Gorki, A.-D.; Rana, B.M.; Martins, R.; Scanlon, S.; Starkl, P.; Lakovits, K.; Hladik, A.; Korosec, A.; Sharif, O.; et al. First-Breath-Induced Type 2 Pathways Shape the Lung Immune Environment. Cell Rep. 2017, 18, 1893–1905. [Google Scholar] [CrossRef] [PubMed]
- Djuardi, Y.; Supali, T.; Wibowo, H.; Kruize, Y.C.; Versteeg, S.A.; van Ree, R.; Sartono, E.; Yazdanbakhsh, M. The Development of Th2 Responses from Infancy to 4 Years of Age and Atopic Sensitization in Areas Endemic for Helminth Infections. Allergy Asthma Clin. Immunol. 2013, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Toro-Londono, M.A.; Bedoya-Urrego, K.; Garcia-Montoya, G.M.; Galvan-Diaz, A.L.; Alzate, J.F. Intestinal Parasitic Infection Alters Bacterial Gut Microbiota in Children. PeerJ 2019, 7, e6200. [Google Scholar] [CrossRef] [PubMed]
- Twort, F.W. An Investigation on the Nature of Ultra-Microscopic Viruses. Lancet 1915, 186, 1241–1243. [Google Scholar] [CrossRef]
- Ackermann, H.W.; Dubow, M.S.; Gershman, M.; Karska-Wysocki, B.; Kasatiya, S.S.; Loessner, M.J.; Mamet-Bratley, M.D.; Regué, M. Taxonomic Changes in Tailed Phages of Enterobacteria. Arch. Virol. 1997, 142, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, R.W. Bacteriophages. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 2384–2417. [Google Scholar]
- Gregory, A.C.; Zablocki, O.; Zayed, A.A.; Howell, A.; Bolduc, B.; Sullivan, M.B. The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut. Cell Host Microbe 2020, 28, 724–740.e8. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Maurice, C.F. Menage a Trois in the Human Gut: Interactions between Host, Bacteria and Phages. Nat. Rev. Microbiol. 2017, 15, 397–408. [Google Scholar] [CrossRef]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage Diversity, Genomics and Phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef]
- Sausset, R.; Petit, M.A.; Gaboriau-Routhiau, V.; De Paepe, M. New Insights into Intestinal Phages. Mucosal Immunol. 2020, 13, 205–215. [Google Scholar] [CrossRef]
- Dutilh, B.E.; Cassman, N.; McNair, K.; Sanchez, S.E.; Silva, G.G.; Boling, L.; Barr, J.J.; Speth, D.R.; Seguritan, V.; Aziz, R.K.; et al. A Highly Abundant Bacteriophage Discovered in the Unknown Sequences of Human Faecal Metagenomes. Nat. Commun. 2014, 5, 4498. [Google Scholar] [CrossRef]
- Camarillo-Guerrero, L.F.; Almeida, A.; Rangel-Pineros, G.; Finn, R.D.; Lawley, T.D. Massive Expansion of Human Gut Bacteriophage Diversity. Cell 2021, 184, 1098–1109.e9. [Google Scholar] [CrossRef]
- Park, G.W.; Ng, T.F.F.; Freeland, A.L.; Marconi, V.C.; Boom, J.A.; Staat, M.A.; Montmayeur, A.M.; Browne, H.; Narayanan, J.; Payne, D.C.; et al. Crassphage as a Novel Tool to Detect Human Fecal Contamination on Environmental Surfaces and Hands. Emerg. Infect. Dis. 2020, 26, 1731–1739. [Google Scholar] [CrossRef]
- Coughlan, S.; Das, A.; O’Herlihy, E.; Shanahan, F.; O’Toole, P.W.; Jeffery, I.B. The Gut Virome in Irritable Bowel Syndrome Differs from That of Controls. Gut Microbes 2021, 13, 1887719. [Google Scholar] [CrossRef]
- Reyes, A.; Blanton, L.V.; Cao, S.; Zhao, G.; Manary, M.; Trehan, I.; Smith, M.I.; Wang, D.; Virgin, H.W.; Rohwer, F.; et al. Gut DNA Viromes of Malawian Twins Discordant for Severe Acute Malnutrition. Proc. Natl. Acad. Sci. USA 2015, 112, 11941–11946. [Google Scholar] [CrossRef]
- Norman, J.M.; Handley, S.A.; Baldridge, M.T.; Droit, L.; Liu, C.Y.; Keller, B.C.; Kambal, A.; Monaco, C.L.; Zhao, G.; Fleshner, P.; et al. Disease-Specific Alterations in the Enteric Virome in Inflammatory Bowel Disease. Cell 2015, 160, 447–460. [Google Scholar] [CrossRef]
- Clooney, A.G.; Sutton, T.D.; Shkoporov, A.N.; Holohan, R.K.; Daly, K.M.; O’regan, O.; Ryan, F.J.; Draper, L.A.; Plevy, S.E.; Ross, R.P.; et al. Whole-Virome Analysis Sheds Light on Viral Dark Matter in Inflammatory Bowel Disease. Cell Host Microbe 2019, 26, 764–778.e5. [Google Scholar] [CrossRef]
- Emlet, C.; Ruffin, M.; Lamendella, R. Enteric Virome and Carcinogenesis in the Gut. Dig. Dis. Sci. 2020, 65, 852–864. [Google Scholar] [CrossRef]
- Tiamani, K.; Luo, S.; Schulz, S.; Xue, J.; Costa, R.; Mirzaei, M.K.; Deng, L. The Role of Virome in the Gastrointestinal Tract and Beyond. FEMS Microbiol. Rev. 2022, 46, fuac027. [Google Scholar] [CrossRef]
- Liang, G.; Zhao, C.; Zhang, H.; Mattei, L.; Sherrill-Mix, S.; Bittinger, K.; Kessler, L.R.; Wu, G.D.; Baldassano, R.N.; DeRusso, P.; et al. The Stepwise Assembly of the Neonatal Virome Is Modulated by Breastfeeding. Nature 2020, 581, 470–474. [Google Scholar] [CrossRef]
- Lim, E.S.; Zhou, Y.; Zhao, G.; Bauer, I.K.; Droit, L.; Ndao, I.M.; Warner, B.B.; Tarr, P.I.; Wang, D.; Holtz, L.R. Early Life Dynamics of the Human Gut Virome and Bacterial Microbiome in Infants. Nat. Med. 2015, 21, 1228–1234. [Google Scholar] [CrossRef]
- Maqsood, R.; Rodgers, R.; Rodriguez, C.; Handley, S.A.; Ndao, I.M.; Tarr, P.I.; Warner, B.B.; Lim, E.S.; Holtz, L.R. Discordant Transmission of Bacteria and Viruses from Mothers to Babies at Birth. Microbiome 2019, 7, 156. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Clooney, A.G.; Sutton, T.D.S.; Ryan, F.J.; Daly, K.M.; Nolan, J.A.; McDonnell, S.A.; Khokhlova, E.V.; Draper, L.A.; Forde, A.; et al. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell Host Microbe 2019, 26, 527–541.e5. [Google Scholar] [CrossRef] [PubMed]
- Baumann-Dudenhoeffer, A.M.; D’Souza, A.W.; Tarr, P.I.; Warner, B.B.; Dantas, G. Infant Diet and Maternal Gestational Weight Gain Predict Early Metabolic Maturation of Gut Microbiomes. Nat. Med. 2018, 24, 1822–1829. [Google Scholar] [CrossRef] [PubMed]
- Duranti, S.; Lugli, G.A.; Mancabelli, L.; Armanini, F.; Turroni, F.; James, K.; Ferretti, P.; Gorfer, V.; Ferrario, C.; Milani, C.; et al. Maternal Inheritance of Bifidobacterial Communities and Bifidophages in Infants through Vertical Transmission. Microbiome 2017, 5, 66. [Google Scholar] [CrossRef]
- Gregory, K.E.; LaPlante, R.D.; Shan, G.; Kumar, D.V.; Gregas, M. Mode of Birth Influences Preterm Infant Intestinal Colonization with Bacteroides over the Early Neonatal Period. Adv. Neonatal Care 2015, 15, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Van Belleghem, J.D.; Clement, F.; Merabishvili, M.; Lavigne, R.; Vaneechoutte, M. Pro- and Anti-Inflammatory Responses of Peripheral Blood Mononuclear Cells Induced by Staphylococcus Aureus and Pseudomonas Aeruginosa Phages. Sci. Rep. 2017, 7, 8004. [Google Scholar] [CrossRef]
- Gogokhia, L.; Buhrke, K.; Bell, R.; Hoffman, B.; Brown, D.G.; Hanke-Gogokhia, C.; Ajami, N.J.; Wong, M.C.; Ghazaryan, A.; Valentine, J.F.; et al. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host Microbe 2019, 25, 285–299.e8. [Google Scholar] [CrossRef] [PubMed]
- Miedzybrodzki, R.; Switala-Jelen, K.; Fortuna, W.; Weber-Dabrowska, B.; Przerwa, A.; Lusiak-Szelachowska, M.; Dabrowska, K.; Kurzepa, A.; Boratynski, J.; Syper, D.; et al. Bacteriophage Preparation Inhibition of Reactive Oxygen Species Generation by Endotoxin-Stimulated Polymorphonuclear Leukocytes. Virus Res. 2008, 131, 233–242. [Google Scholar] [CrossRef]
- Górski, A.; Kniotek, M.; Perkowska-Ptasińska, A.; Mróz, A.; Przerwa, A.; Gorczyca, W.; Dąbrowska, K.; Weber-Dąbrowska, B.; Nowaczyk, M. Bacteriophages and Transplantation Tolerance. Transplant. Proc. 2006, 38, 331–333. [Google Scholar] [CrossRef]
- d’Hérelle, F.; Smith, G.H. The Bacteriophage and Its Behavior; The Williams & Wilkins Company: Baltimore, MD, USA, 1926. [Google Scholar]
- Duan, Y.; Young, R.; Schnabl, B. Bacteriophages and Their Potential for Treatment of Gastrointestinal Diseases. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 135–144. [Google Scholar] [CrossRef]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails to Treat a Patient with a Disseminated Resistant Acinetobacter Baumannii Infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef]
- Corbellino, M.; Kieffer, N.; Kutateladze, M.; Balarjishvili, N.; Leshkasheli, L.; Askilashvili, L.; Tsertsvadze, G.; Rimoldi, S.G.; Nizharadze, D.; Hoyle, N.; et al. Eradication of a Multidrug-Resistant, Carbapenemase-Producing Klebsiella Pneumoniae Isolate Following Oral and Intra-Rectal Therapy with a Custom Made, Lytic Bacteriophage Preparation. Clin. Infect. Dis. 2020, 70, 1998–2001. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, Y.; Shen, S.; Hou, Y.; Chen, Y.; Wang, T. The Mycobiota of the Human Body: A Spark Can Start a Prairie Fire. Gut Microbes 2020, 11, 655–679. [Google Scholar] [CrossRef]
- Underhill, D.M.; Iliev, I.D. The Mycobiota: Interactions between Commensal Fungi and the Host Immune System. Nat. Rev. Immunol. 2014, 14, 405–416. [Google Scholar] [CrossRef]
- Richard, M.L.; Sokol, H. The Gut Mycobiota: Insights into Analysis, Environmental Interactions and Role in Gastrointestinal Diseases. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 331–345. [Google Scholar] [CrossRef]
- Wu, X.; Xia, Y.; He, F.; Zhu, C.; Ren, W. Intestinal Mycobiota in Health and Diseases: From a Disrupted Equilibrium to Clinical Opportunities. Microbiome 2021, 9, 60. [Google Scholar] [CrossRef]
- Li, X.V.; Leonardi, I.; Iliev, I.D. Gut Mycobiota in Immunity and Inflammatory Disease. Immunity 2019, 50, 1365–1379. [Google Scholar] [CrossRef]
- Hall, R.A.; Noverr, M.C. Fungal Interactions with the Human Host: Exploring the Spectrum of Symbiosis. Curr. Opin. Microbiol. 2017, 40, 58–64. [Google Scholar] [CrossRef]
- Hatoum, R.; Labrie, S.; Fliss, I. Antimicrobial and Probiotic Properties of Yeasts: From Fundamental to Novel Applications. Front. Microbiol. 2012, 3, 421. [Google Scholar] [CrossRef]
- Kelesidis, T.; Pothoulakis, C. Efficacy and Safety of the Probiotic Saccharomyces Boulardii for the Prevention and Therapy of Gastrointestinal Disorders. Ther. Adv. Gastroenterol. 2012, 5, 111–125. [Google Scholar] [CrossRef]
- Sovran, B.; Planchais, J.; Jegou, S.; Straube, M.; Lamas, B.; Natividad, J.M.; Agus, A.; Dupraz, L.; Glodt, J.; Da Costa, G.; et al. Enterobacteriaceae Are Essential for the Modulation of Colitis Severity by Fungi. Microbiome 2018, 6, 152. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut Microbiota, Metabolites and Host Immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Strati, F.; Di Paola, M.; Stefanini, I.; Albanese, D.; Rizzetto, L.; Lionetti, P.; Calabrò, A.; Jousson, O.; Donati, C.; Cavalieri, D.; et al. Age and Gender Affect the Composition of Fungal Population of the Human Gastrointestinal Tract. Front. Microbiol. 2016, 7, 1227. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal Microbiota Dysbiosis in Ibd. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef]
- Liguori, G.; Lamas, B.; Richard, M.L.; Brandi, G.; Da Costa, G.; Hoffmann, T.W.; Di Simone, M.P.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Fungal Dysbiosis in Mucosa-Associated Microbiota of Crohn’s Disease Patients. J. Crohn’s Colitis 2016, 10, 296–305. [Google Scholar] [CrossRef]
- Limon, J.J.; Tang, J.; Li, D.; Wolf, A.J.; Michelsen, K.S.; Funari, V.; Gargus, M.; Nguyen, C.; Sharma, P.; Maymi, V.I.; et al. Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 2019, 25, 377–388.e6. [Google Scholar] [CrossRef]
- Romani, L. Immunity to Fungal Infections. Nat. Rev. Immunol. 2004, 4, 11–24. [Google Scholar] [CrossRef]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef]
- Naglik, J.R.; Fidel, P.L., Jr.; Odds, F.C. Animal Models of Mucosal Candida Infection. FEMS Microbiol. Lett. 2008, 283, 129–139. [Google Scholar] [CrossRef]
- Noverr, M.C.; Falkowski, N.R.; McDonald, R.A.; McKenzie, A.N.; Huffnagle, G.B. Development of Allergic Airway Disease in Mice Following Antibiotic Therapy and Fungal Microbiota Increase: Role of Host Genetics, Antigen, and Interleukin-13. Infect. Immun. 2005, 73, 30–38. [Google Scholar] [CrossRef]
- Mason, K.L.; Erb Downward, J.R.; Mason, K.D.; Falkowski, N.R.; Eaton, K.A.; Kao, J.Y.; Young, V.B.; Huffnagle, G.B. Candida Albicans and Bacterial Microbiota Interactions in the Cecum During Recolonization Following Broad-Spectrum Antibiotic Therapy. Infect. Immun. 2012, 80, 3371–3380. [Google Scholar] [CrossRef] [PubMed]
- Samonis, G.; Gikas, A.; Anaissie, E.J.; Vrenzos, G.; Maraki, S.; Tselentis, Y.; Bodey, G.P. Prospective Evaluation of Effects of Broad-Spectrum Antibiotics on Gastrointestinal Yeast Colonization of Humans. Antimicrob. Agents Chemother. 1993, 37, 51–53. [Google Scholar] [CrossRef]
- Mulligan, M.E.; Citron, D.M.; McNamara, B.T.; Finegold, S.M. Impact of Cefoperazone Therapy on Fecal Flora. Antimicrob. Agents Chemother. 1982, 22, 226–230. [Google Scholar] [CrossRef] [PubMed]
- van Tilburg Bernardes, E.; Pettersen, V.K.; Gutierrez, M.W.; Laforest-Lapointe, I.; Jendzjowsky, N.G.; Cavin, J.-B.; Vicentini, F.A.; Keenan, C.M.; Ramay, H.R.; Samara, J.; et al. Intestinal Fungi Are Causally Implicated in Microbiome Assembly and Immune Development in Mice. Nat. Commun. 2020, 11, 2577. [Google Scholar] [CrossRef]
- Frey-Klett, P.; Burlinson, P.; Deveau, A.; Barret, M.; Tarkka, M.; Sarniguet, A. Bacterial-Fungal Interactions: Hyphens between Agricultural, Clinical, Environmental, and Food Microbiologists. Microbiol. Mol. Biol. Rev. 2011, 75, 583–609. [Google Scholar] [CrossRef]
- Erb Downward, J.R.; Falkowski, N.R.; Mason, K.L.; Muraglia, R.; Huffnagle, G.B. Modulation of Post-Antibiotic Bacterial Community Reassembly and Host Response by Candida Albicans. Sci. Rep. 2013, 3, 2191. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, C.A.; Gresnigt, M.S.; Hube, B. The Gut, the Bad and the Harmless: Candida Albicans as a Commensal and Opportunistic Pathogen in the Intestine. Curr. Opin. Microbiol. 2020, 56, 7–15. [Google Scholar] [CrossRef]
- Woese, C.R.; Fox, G.E. Phylogenetic Structure of the Prokaryotic Domain: The Primary Kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef]
- Koga, Y.; Morii, H. Biosynthesis of Ether-Type Polar Lipids in Archaea and Evolutionary Considerations. Microbiol. Mol. Biol. Rev. 2007, 71, 97–120. [Google Scholar] [CrossRef]
- Lombard, J.; López-García, P.; Moreira, D. The Early Evolution of Lipid Membranes and the Three Domains of Life. Nat. Rev. Genet. 2012, 10, 507–515. [Google Scholar] [CrossRef]
- Jain, S.; Caforio, A.; Driessen, A.J.M. Biosynthesis of Archaeal Membrane Ether Lipids. Front. Microbiol. 2014, 5, 641. [Google Scholar] [CrossRef]
- Caforio, A.; Driessen, A.J.M. Archaeal Phospholipids: Structural Properties and Biosynthesis. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2017, 1862, 1325–1339. [Google Scholar] [CrossRef]
- Siliakus, M.F.; van der Oost, J.; Kengen, S.W.M. Adaptations of Archaeal and Bacterial Membranes to Variations in Temperature, Ph and Pressure. Extremophiles 2017, 21, 651–670. [Google Scholar] [CrossRef]
- Pereira, S.L.; Reeve, J.N. Histones and Nucleosomes in Archaea and Eukarya: A Comparative Analysis. Extremophiles 1998, 2, 141–148. [Google Scholar] [CrossRef]
- Baker, B.J.; De Anda, V.; Seitz, K.W.; Dombrowski, N.; Santoro, A.E.; Lloyd, K.G. Diversity, Ecology and Evolution of Archaea. Nat. Microbiol. 2020, 5, 887–900. [Google Scholar] [CrossRef]
- Aminov, R.I. Role of Archaea in Human Disease. Front. Cell. Infect. Microbiol. 2013, 3, 42. [Google Scholar] [CrossRef]
- Bang, C.; Schmitz, R.A. Archaea: Forgotten Players in the Microbiome. Emerg. Top. Life Sci. 2018, 2, 459–468. [Google Scholar]
- Lurie-Weinberger, M.N.; Gophna, U. Archaea in and on the Human Body: Health Implications and Future Directions. PLoS Pathog. 2015, 11, e1004833. [Google Scholar] [CrossRef]
- Faveri, M.; Gonçalves, L.F.H.; Feres, M.; Figueiredo, L.C.; Gouveia, L.A.; Shibli, J.A.; Mayer, M.P.A. Prevalence and Microbiological Diversity of Archaea in Peri-Implantitis Subjects by 16s Ribosomal Rna Clonal Analysis. J. Periodontal Res. 2011, 46, 338–344. [Google Scholar] [CrossRef]
- Kim, J.Y.; Whon, T.W.; Lim, M.Y.; Kim, Y.B.; Kim, N.; Kwon, M.-S.; Kim, J.; Lee, S.H.; Choi, H.-J.; Nam, I.-H.; et al. The Human Gut Archaeome: Identification of Diverse Haloarchaea in Korean Subjects. Microbiome 2020, 8, 114. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Pausan, M.R.; Csorba, C.; Singer, G.; Till, H.; Schöpf, V.; Santigli, E.; Klug, B.; Högenauer, C.; Blohs, M.; Moissl-Eichinger, C. Exploring the Archaeome: Detection of Archaeal Signatures in the Human Body. Front. Microbiol. 2019, 10, 2796. [Google Scholar] [CrossRef]
- Patra, A.K.; Park, T.; Kim, M.; Yu, Z. Rumen Methanogens and Mitigation of Methane Emission by Anti-Methanogenic Compounds and Substances. J. Anim. Sci. Biotechnol. 2017, 8, 13. [Google Scholar] [CrossRef]
- Guindo, C.O.; Drancourt, M.; Grine, G. Digestive Tract Methanodrome: Physiological Roles of Human Microbiota-Associated Methanogens. Microb. Pathog. 2020, 149, 104425. [Google Scholar] [CrossRef]
- Dridi, B.; Henry, M.; El Khéchine, A.; Raoult, D.; Drancourt, M. High Prevalence of Methanobrevibacter Smithii and Methanosphaera Stadtmanae Detected in the Human Gut Using an Improved DNA Detection Protocol. PLoS ONE 2009, 4, e7063. [Google Scholar] [CrossRef]
- Rieu-Lesme, F.; Delbès, C.; Sollelis, L. Recovery of Partial 16s Rdna Sequences Suggests the Presence of Crenarchaeota in the Human Digestive Ecosystem. Curr. Microbiol. 2005, 51, 317–321. [Google Scholar] [CrossRef]
- Gaci, N.; Borrel, G.; Tottey, W.; O’Toole, P.W.; Brugere, J.F. Archaea and the Human Gut: New Beginning of an Old Story. World J. Gastroenterol. 2014, 20, 16062–16078. [Google Scholar] [CrossRef] [PubMed]
- Million, M.; Alou, M.T.; Khelaifia, S.; Bachar, D.; Lagier, J.-C.; Dione, N.; Brah, S.; Hugon, P.; Lombard, V.; Armougom, F.; et al. Increased Gut Redox and Depletion of Anaerobic and Methanogenic Prokaryotes in Severe Acute Malnutrition. Sci. Rep. 2016, 6, 26051. [Google Scholar] [CrossRef]
- McKay, L.F.; Eastwood, M.A.; Brydon, W.G. Methane Excretion in Man—A Study of Breath, Flatus, and Faeces. Gut 1985, 26, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Bin Waqar, S.H.; Rehan, A. Methane and Constipation-Predominant Irritable Bowel Syndrome: Entwining Pillars of Emerging Neurogastroenterology. Cureus 2019, 11, e4764. [Google Scholar] [CrossRef]
- Attaluri, A.; Jackson, M.; Valestin, J.; Rao, S.S. Methanogenic Flora Is Associated with Altered Colonic Transit but Not Stool Characteristics in Constipation without Ibs. Am. J. Gastroenterol. 2010, 105, 1407–1411. [Google Scholar] [CrossRef]
- Coker, O.O.; Wu, W.K.K.; Wong, S.H.; Sung, J.J.; Yu, J. Altered Gut Archaea Composition and Interaction with Bacteria Are Associated with Colorectal Cancer. Gastroenterology 2020, 159, 1459–1470.e5. [Google Scholar] [CrossRef] [PubMed]
- Barnett, D.J.; Mommers, M.; Penders, J.; Arts, I.C.; Thijs, C. Intestinal Archaea Inversely Associated with Childhood Asthma. J. Allergy Clin. Immunol. 2019, 143, 2305–2307. [Google Scholar] [CrossRef] [PubMed]
- Barcik, W.; Boutin, R.C.T.; Sokolowska, M.; Finlay, B.B. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity 2020, 52, 241–255. [Google Scholar] [CrossRef]
- Bang, C.; Weidenbach, K.; Gutsmann, T.; Heine, H.; Schmitz, R.A. The Intestinal Archaea Methanosphaera Stadtmanae and Methanobrevibacter Smithii Activate Human Dendritic Cells. PLoS ONE 2014, 9, e99411. [Google Scholar] [CrossRef]
- Lecours, P.B.; Duchaine, C.; Taillefer, M.; Tremblay, C.; Veillette, M.; Cormier, Y.; Marsolais, D. Immunogenic Properties of Archaeal Species Found in Bioaerosols. PLoS ONE 2011, 6, e23326. [Google Scholar] [CrossRef]
- Bragin, A.O.; Sokolov, V.S.; Demenkov, P.S.; Ivanisenko, T.V.; Bragina, E.Y.; Matushkin, Y.G.; Ivanisenko, V.A. Prediction of Bacterial and Archaeal Allergenicity with Allpred Program. Mol. Biol. 2018, 52, 279–284. [Google Scholar] [CrossRef]
- Yang, K.; Yang, J.; Zhang, J.; Wei, Q.; Ng, S.C.; Wang, H. IDDF2021-ABS-0130 Dysbiosis of Gut Archaea in Obesity Recovered after Bariatric Surgery. Gut 2021, 70 (Suppl. 2), A46. [Google Scholar]
- Basseri, R.J.; Basseri, B.; Pimentel, M.; Chong, K.; Youdim, A.; Low, K.; Hwang, L.; Soffer, E.; Chang, C.; Mathur, R. Intestinal Methane Production in Obese Individuals Is Associated with a Higher Body Mass Index. Gastroenterol. Hepatol. 2012, 8, 22–28. [Google Scholar]
- Johansson, M.E.V.; Phillipson, M.; Petersson, J.; Velcich, A.; Holm, L.; Hansson, G.C. The Inner of the Two Muc2 Mucin-Dependent Mucus Layers in Colon Is Devoid of Bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 15064–15069. [Google Scholar] [CrossRef]
- Luis, A.S.; Hansson, G.C. Intestinal Mucus and Their Glycans: A Habitat for Thriving Microbiota. Cell Host Microbe 2023, 31, 1087–1100. [Google Scholar] [CrossRef]
- Houghteling, P.D.; Walker, W.A. Why Is Initial Bacterial Colonization of the Intestine Important to Infants’ and Children’s Health? J. Pediatr. Gastroenterol. Nutr. 2015, 60, 294–307. [Google Scholar] [CrossRef]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal Gut Microbiota Associates with Childhood Multisensitized Atopy and T Cell Differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Khan, A.A.; Ghosh, P.; Taranu, Z.E.; Taguer, M.; Ru, J.; Chowdhury, R.; Kabir, M.; Deng, L.; Mondal, D.; et al. Bacteriophages Isolated from Stunted Children Can Regulate Gut Bacterial Communities in an Age-Specific Manner. Cell Host Microbe 2020, 27, 199–212.e5. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.A.; Holtz, L.R. Gut Virome in Early Life: Origins and Implications. Curr. Opin. Virol. 2022, 55, 101233. [Google Scholar] [CrossRef]
- Piazzesi, A.; Putignani, L. Impact of Helminth-Microbiome Interactions on Childhood Health and Development—A Clinical Perspective. Parasite Immunol. 2023, 45, e12949. [Google Scholar] [CrossRef]
- van de Pol, J.A.A.; van Best, N.; Mbakwa, C.A.; Thijs, C.; Savelkoul, P.H.; Arts, I.C.W.; Hornef, M.W.; Mommers, M.; Penders, J. Gut Colonization by Methanogenic Archaea Is Associated with Organic Dairy Consumption in Children. Front. Microbiol. 2017, 8, 355. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Zhang, L.; Han, Y.; Wu, L.; Wang, B. The Development of Early Life Microbiota in Human Health and Disease. Engineering 2022, 12, 101–114. [Google Scholar] [CrossRef]
- Gutiérrez, B.; Domingo-Calap, P. Phage Therapy in Gastrointestinal Diseases. Microorganisms 2020, 8, 1420. [Google Scholar] [CrossRef]
- Matijasic, M.; Mestrovic, T.; Paljetak, H.C.; Peric, M.; Baresic, A.; Verbanac, D. Gut Microbiota Beyond Bacteria-Mycobiome, Virome, Archaeome, and Eukaryotic Parasites in Ibd. Int. J. Mol. Sci. 2020, 21, 2668. [Google Scholar] [CrossRef]
- Medina, R.H.; Kutuzova, S.; Nielsen, K.N.; Johansen, J.; Hansen, L.H.; Nielsen, M.; Rasmussen, S. Machine Learning and Deep Learning Applications in Microbiome Research. ISME Commun. 2022, 2, 98. [Google Scholar] [CrossRef]
| Phylum | Species Example | Symptoms |
|---|---|---|
| Cestodes | Taenia saginata | Usually do not cause symptoms, but infected patients can experience non-specific symptoms such as abdominal discomfort, nausea, vomiting, diarrhea, and weight loss, among others. |
| Taenia solium | ||
| Nematodes | Anisakis simplex | Non-specific symptoms include abdominal pain and general discomfort with uncommon cases of extra-gastrointestinal symptoms such as allergy with angioedema, urticaria, and anaphylaxis. |
| Trematodes | Schistosoma mansoni | Non-specific symptoms include abdominal pain, enlarged liver as well as hematochezia, and hematuria. |
| Order | Family | Characteristics |
|---|---|---|
| Caudovirales | Siphoviridae | dsDNA, long, noncontractile tails |
| Myoviridae | dsDNA, contractile tails | |
| Podoviridae | dsDNA, short, stubby tails | |
| Kalamavirales | Tectiviridae | dsDNA, internal membrane |
| Petitvirales | Microviridae | ssDNA, circular, icosahedral |
| Tubulavirales | Inoviridae | ssDNA filamentous |
| Norzivirales | Fiersviridae | ssRNA, small icosahedral |
| Mindivirales | Cystoviridae | dsRNA, segmented, enveloped |
| Vinavirales | Corticoviridae | dsDNA, circular, internal membrane |
| Unassigned | Plasmaviridae | dsDNA, circular, enveloped |
| Disease | Changes in Phageome |
|---|---|
| Ulcerative colitis | Increased Caudovirales |
| Crohn’s disease | Lytic phages are replaced with lysogenic phages |
| Colorectal cancer | More diverse phageome |
| Coeliac disease | More Enterobacteria phages, fewer Lactococcus and Streptococcus phages |
| Enriched Archaea CRC | Depleted Archaea CRC |
|---|---|
| Halorubrum tropicale | Methanocorpusculum (2 sp.) |
| Halococcus morrhuae | Methanosarcina (3 sp.) |
| Halococcus salifodinae | Methanobrevibacter (3 sp.) |
| Halovenus aranensis | Methanobacterium (3 sp.) |
| Natrinema sps J7-2 | Methanooccus (2 sp.) |
| Methanothrix soehngenii | Methanospaera |
| Methanoculleu marisnigri |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Bonete, M.J.; Rajan, A.; Suriano, F.; Layunta, E. The Underrated Gut Microbiota Helminths, Bacteriophages, Fungi, and Archaea. Life 2023, 13, 1765. https://doi.org/10.3390/life13081765
Garcia-Bonete MJ, Rajan A, Suriano F, Layunta E. The Underrated Gut Microbiota Helminths, Bacteriophages, Fungi, and Archaea. Life. 2023; 13(8):1765. https://doi.org/10.3390/life13081765
Chicago/Turabian StyleGarcia-Bonete, Maria Jose, Anandi Rajan, Francesco Suriano, and Elena Layunta. 2023. "The Underrated Gut Microbiota Helminths, Bacteriophages, Fungi, and Archaea" Life 13, no. 8: 1765. https://doi.org/10.3390/life13081765





