Pathways of Hypoxia-Inducible Factor (HIF) in the Orchestration of Uterine Fibroids Development
Abstract
1. Introduction
2. Methodology
3. UF in Terms of Hypoxia Response
4. Basic Biology of HIF
5. The Role of Hypoxia-Inducible Factor in Uterine Fibroids
6. HIF and GWAS-Identified Genes
7. Future Perspectives
8. Limitations
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| UF | uterine fibroids |
| NM | normal myometrium |
| HIF | hypoxia-inducible factor |
| EPO | erythropoietin |
| VHL | von Hippel–Lindau |
| CUL2 | cullin 2 |
| VEGF | vascular endothelial growing factor |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| ANG-1 | angiopoietin-1 |
| VEGFR-1 | vascular endothelial growing factor receptor 1 |
| EGF | epidermal growing factor |
| SERPINE1 (PAI1) | plasminogen activator inhibitor 1 |
| TEK | endothelial tyrosine kinase |
| TIMP1 | tissue inhibitor of metalloproteinases 1 |
| HMOX1 | heme oxygenase 1 |
| EDN1 | endothelin 1 |
| iNOS | inducible nitrogen oxide synthase |
| cNOS (eNOS) | constitutive (endothelial) nitric oxide synthase |
| GLUT-1 | glucose transporter type 1 |
| HK | hexokinase |
| ALDOA | aldolase, fructose-bisphosphate A |
| ENO1 | enolase 1 |
| PGK1 | phosphoglycerate kinase 1 |
| PFK/FBPase 3 | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 |
| LDHA | lactate dehydrogenase A |
| HLRCC | hereditary leiomyomatosis and renal cell cancer |
| FH | fumarate hydratase |
| F2,6BP | fructose-2,6-bisphosphate |
| GWAS | genome-wide association studies |
| Bcl-2 | B-cell lymphoma 2 |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
| ROS | reactive oxygen species |
| PPI | protein–protein interactions |
| ATM | ataxia telangiectasia mutated |
| CDC42 | cell division control protein 42 homolog |
| FOXO1 | forkhead box protein O1 |
| IGF1 | insulin-like growth factor I |
| RBMS1 | RNA-binding motif, single-stranded-interacting protein 1 |
| SIM2 | single-minded homolog 2 |
| SIRT3 | NAD-dependent protein deacetylase sirtuin-3, mitochondrial |
| TERT | telomerase reverse transcriptase |
| THRB | thyroid hormone receptor beta |
| TP53 | tumor protein P53 |
| WT1 | Wilms tumor 1 |
| ZEB1 | zinc finger e-box binding homeobox 1 |
References
- Giuliani, E.; As-Sanie, S.; Marsh, E.E. Epidemiology and Management of Uterine Fibroids. Int. J. Gynaecol. Obstet. 2020, 149, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Al Ansari, A.A.; Al Hail, F.A.; Abboud, E. Malignant Transformation of Uterine Leiomyoma. Qatar Med. J. 2012, 2012, 71–74. [Google Scholar] [CrossRef][Green Version]
- Freytag, D.; Günther, V.; Maass, N.; Alkatout, I. Uterine Fibroids and Infertility. Diagnostics 2021, 11, 1455. [Google Scholar] [CrossRef]
- Parazzini, F.; Tozzi, L.; Bianchi, S. Pregnancy Outcome and Uterine Fibroids. Best. Pract. Res. Clin. Obstet. Gynaecol. 2016, 34, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Marjoribanks, J.; Lethaby, A.; Farquhar, C. Surgery versus Medical Therapy for Heavy Menstrual Bleeding. Cochrane Database Syst. Rev. 2016, 2016, CD003855. [Google Scholar] [CrossRef]
- Surrey, E.S.; Hornstein, M.D. Prolonged GnRH Agonist and Add-Back Therapy for Symptomatic Endometriosis: Long-Term Follow-Up. Obstet. Gynecol. 2002, 99 Pt 1, 709–719. [Google Scholar] [CrossRef]
- Lewis, T.D.; Malik, M.; Britten, J.; San Pablo, A.M.; Catherino, W.H. A Comprehensive Review of the Pharmacologic Management of Uterine Leiomyoma. Biomed. Res. Int. 2018, 2018, 2414609. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Schmidberger, H.; Mayer, A. The Warburg Effect: Essential Part of Metabolic Reprogramming and Central Contributor to Cancer Progression. Int. J. Radiat. Biol. 2019, 95, 912–919. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does It Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Islam, M.S.; Ciavattini, A.; Petraglia, F.; Castellucci, M.; Ciarmela, P. Extracellular Matrix in Uterine Leiomyoma Pathogenesis: A Potential Target for Future Therapeutics. Hum. Reprod. Update 2018, 24, 59–85. [Google Scholar] [CrossRef]
- Lee, P.; Chandel, N.S.; Simon, M.C. Cellular Adaptation to Hypoxia through Hypoxia Inducible Factors and Beyond. Nat. Rev. Mol. Cell Biol. 2020, 21, 268–283. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Kim, J.J.; Li, Y.; Xie, J.; Shao, C.; Wei, J.-J. Oxidative Stress-Induced MiRNAs Modulate AKT Signaling and Promote Cellular Senescence in Uterine Leiomyoma. J. Mol. Med. 2018, 96, 1095–1106. [Google Scholar] [CrossRef]
- Smith, K.A.; Waypa, G.B.; Schumacker, P.T. Redox Signaling during Hypoxia in Mammalian Cells. Redox Biol. 2017, 13, 228–234. [Google Scholar] [CrossRef]
- Soldatova, V.A.; Demidenko, A.N.; Soldatov, V.; Deykin, A.; Bushueva, O.; Puchenkova, O.A. Hypoxia-Inducible Factor: Basic Biology and Involvement in Cardiovascular Pathology. Asian J. Pharm. 2018, 12, S1173–S1178. [Google Scholar]
- Semenza, G.L.; Nejfelt, M.K.; Chi, S.M.; Antonarakis, S.E. Hypoxia-Inducible Nuclear Factors Bind to an Enhancer Element Located 3′ to the Human Erythropoietin Gene. Proc. Natl. Acad. Sci. USA 1991, 88, 5680–5684. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Pugh, C.W.; Ratcliffe, P.J. Inducible Operation of the Erythropoietin 3′ Enhancer in Multiple Cell Lines: Evidence for a Widespread Oxygen-Sensing Mechanism. Proc. Natl. Acad. Sci. USA 1993, 90, 2423–2427. [Google Scholar] [CrossRef]
- Wang, G.L.; Semenza, G.L. Purification and Characterization of Hypoxia-Inducible Factor 1. J. Biol. Chem. 1995, 270, 1230–1237. [Google Scholar] [CrossRef]
- Kibel, A.; Iliopoulos, O.; DeCaprio, J.A.; Kaelin, W.G. Binding of the von Hippel-Lindau Tumor Suppressor Protein to Elongin B and C. Science 1995, 269, 1444–1446. [Google Scholar] [CrossRef] [PubMed]
- Lonergan, K.M.; Iliopoulos, O.; Ohh, M.; Kamura, T.; Conaway, R.C.; Conaway, J.W.; Kaelin, W.G. Regulation of Hypoxia-Inducible MRNAs by the von Hippel-Lindau Tumor Suppressor Protein Requires Binding to Complexes Containing Elongins B/C and Cul2. Mol. Cell. Biol. 1998, 18, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, J.A.; Jiang, B.H.; Iyer, N.V.; Agani, F.; Leung, S.W.; Koos, R.D.; Semenza, G.L. Activation of Vascular Endothelial Growth Factor Gene Transcription by Hypoxia-Inducible Factor 1. Mol. Cell. Biol. 1996, 16, 4604–4613. [Google Scholar] [CrossRef] [PubMed]
- Ivan, M.; Kondo, K.; Yang, H.; Kim, W.; Valiando, J.; Ohh, M.; Salic, A.; Asara, J.M.; Lane, W.S.; Kaelin, W.G. HIFalpha Targeted for VHL-Mediated Destruction by Proline Hydroxylation: Implications for O2 Sensing. Science 2001, 292, 464–468. [Google Scholar] [CrossRef]
- Mahon, P.C.; Hirota, K.; Semenza, G.L. FIH-1: A Novel Protein That Interacts with HIF-1alpha and VHL to Mediate Repression of HIF-1 Transcriptional Activity. Genes. Dev. 2001, 15, 2675–2686. [Google Scholar] [CrossRef]
- Lando, D.; Peet, D.J.; Gorman, J.J.; Whelan, D.A.; Whitelaw, M.L.; Bruick, R.K. FIH-1 Is an Asparaginyl Hydroxylase Enzyme That Regulates the Transcriptional Activity of Hypoxia-Inducible Factor. Genes. Dev. 2002, 16, 1466–1471. [Google Scholar] [CrossRef] [PubMed]
- Jaskiewicz, M.; Moszynska, A.; Serocki, M.; Króliczewski, J.; Bartoszewska, S.; Collawn, J.F.; Bartoszewski, R. Hypoxia-Inducible Factor (HIF)-3a2 Serves as an Endothelial Cell Fate Executor during Chronic Hypoxia. EXCLI J. 2022, 21, 454–469. [Google Scholar] [CrossRef] [PubMed]
- Serocki, M.; Bartoszewska, S.; Janaszak-Jasiecka, A.; Ochocka, R.J.; Collawn, J.F.; Bartoszewski, R. MiRNAs Regulate the HIF Switch during Hypoxia: A Novel Therapeutic Target. Angiogenesis 2018, 21, 183–202. [Google Scholar] [CrossRef] [PubMed]
- Corrado, C.; Fontana, S. Hypoxia and HIF Signaling: One Axis with Divergent Effects. Int. J. Mol. Sci. 2020, 21, 5611. [Google Scholar] [CrossRef]
- Albogami, S.M.; Al-Kuraishy, H.M.; Al-Maiahy, T.J.; Al-Buhadily, A.K.; Al-Gareeb, A.I.; Alorabi, M.; Alotaibi, S.S.; De Waard, M.; Sabatier, J.-M.; Saad, H.M.; et al. Hypoxia-Inducible Factor 1 and Preeclampsia: A New Perspective. Curr. Hypertens. Rep. 2022, 24, 687–692. [Google Scholar] [CrossRef]
- Fu, X.; Shi, L.; Liu, P.; Jiao, Y.; Guo, S.; Chen, Q.; Zheng, Q.; Chen, X.; Wang, Y. Expression and Clinical Significance of HIF-1α in Follicular Fluid and Granulosa Cells in Infertile PCOS Patients. Reprod. Sci. 2023, 30, 2263–2274. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Chen, Q.; Xiao, J.; Yao, T.; Bian, L.; Liu, C.; Lin, Z. Overexpression of Hypoxia-Inducible Factor-1α Is a Predictor of Poor Prognosis in Cervical Cancer: A Clinicopathologic Study and a Meta-Analysis. Int. J. Gynecol. Cancer 2014, 24, 1054–1064. [Google Scholar] [CrossRef]
- Zhan, L.; Wang, W.; Zhang, Y.; Song, E.; Fan, Y.; Wei, B. Hypoxia-Inducible Factor-1alpha: A Promising Therapeutic Target in Endometriosis. Biochimie 2016, 123, 130–137. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, D.; Lu, X.; Zhang, Q.; Gu, R.; Sun, B.; Sun, Y. Hypoxia and Its Possible Relationship with Endometrial Receptivity in Adenomyosis: A Preliminary Study. Reprod. Biol. Endocrinol. 2021, 19, 7. [Google Scholar] [CrossRef] [PubMed]
- Seeber, L.M.S.; Horrée, N.; Vooijs, M.A.G.G.; Heintz, A.P.M.; van der Wall, E.; Verheijen, R.H.M.; van Diest, P.J. The Role of Hypoxia Inducible Factor-1alpha in Gynecological Cancer. Crit. Rev. Oncol. Hematol. 2011, 78, 173–184. [Google Scholar] [CrossRef]
- Miyashita-Ishiwata, M.; El Sabeh, M.; Reschke, L.D.; Afrin, S.; Borahay, M.A. Differential Response to Hypoxia in Leiomyoma and Myometrial Cells. Life Sci. 2022, 290, 120238. [Google Scholar] [CrossRef]
- Hou, P.; Zhao, L.; Li, Y.; Luo, F.; Wang, S.; Song, J.; Bai, J. Comparative Expression of Thioredoxin-1 in Uterine Leiomyomas and Myometrium. Mol. Hum. Reprod. 2014, 20, 148–154. [Google Scholar] [CrossRef]
- Ishikawa, H.; Xu, L.; Sone, K.; Kobayashi, T.; Wang, G.; Shozu, M. Hypoxia Induces Hypoxia-Inducible Factor 1α and Potential HIF-Responsive Gene Expression in Uterine Leiomyoma. Reprod. Sci. 2019, 26, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Shi, X.; Sheng, K.; Han, G.; Li, W.; Zhao, Q.; Jiang, B.; Feng, J.; Li, J.; Gu, Y. PI3K/Akt Signaling Transduction Pathway, Erythropoiesis and Glycolysis in Hypoxia (Review). Mol. Med. Rep. 2019, 19, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Melincovici, C.S.; Boşca, A.B.; Şuşman, S.; Mărginean, M.; Mihu, C.; Istrate, M.; Moldovan, I.M.; Roman, A.L.; Mihu, C.M. Vascular Endothelial Growth Factor (VEGF)—Key Factor in Normal and Pathological Angiogenesis. Rom. J. Morphol. Embryol. 2018, 59, 455–467. [Google Scholar]
- Krüger-Genge, A.; Blocki, A.; Franke, R.-P.; Jung, F. Vascular Endothelial Cell Biology: An Update. Int. J. Mol. Sci. 2019, 20, 4411. [Google Scholar] [CrossRef]
- Fletcher, N.M.; Abusamaan, M.S.; Memaj, I.; Saed, M.G.; Al-Hendy, A.; Diamond, M.P.; Saed, G.M. Oxidative Stress: A Key Regulator of Leiomyoma Cell Survival. Fertil. Steril. 2017, 107, 1387–1394.e1. [Google Scholar] [CrossRef]
- Tirpe, A.A.; Gulei, D.; Ciortea, S.M.; Crivii, C.; Berindan-Neagoe, I. Hypoxia: Overview on Hypoxia-Mediated Mechanisms with a Focus on the Role of HIF Genes. Int. J. Mol. Sci. 2019, 20, 6140. [Google Scholar] [CrossRef]
- Hague, S.; Zhang, L.; Oehler, M.K.; Manek, S.; MacKenzie, I.Z.; Bicknell, R.; Rees, M.C. Expression of the Hypoxically Regulated Angiogenic Factor Adrenomedullin Correlates with Uterine Leiomyoma Vascular Density. Clin. Cancer Res. 2000, 6, 2808–2814. [Google Scholar] [PubMed]
- Korompelis, P.; Piperi, C.; Adamopoulos, C.; Dalagiorgou, G.; Korkolopoulou, P.; Sepsa, A.; Antsaklis, A.; Papavassiliou, A.G. Expression of Vascular Endothelial Factor-A, Gelatinases (MMP-2, MMP-9) and TIMP-1 in Uterine Leiomyomas. Clin. Chem. Lab. Med. 2015, 53, 1415–1424. [Google Scholar] [CrossRef]
- Wei, J.-J.; Zhang, X.-M.; Chiriboga, L.; Yee, H.; Perle, M.A.; Mittal, K. Spatial Differences in Biologic Activity of Large Uterine Leiomyomata. Fertil. Steril. 2006, 85, 179–187. [Google Scholar] [CrossRef]
- Asano, R.; Asai-Sato, M.; Matsukuma, S.; Mizushima, T.; Taguri, M.; Yoshihara, M.; Inada, M.; Fukui, A.; Suzuki, Y.; Miyagi, Y.; et al. Expression of Erythropoietin Messenger Ribonucleic Acid in Wild-Type MED12 Uterine Leiomyomas under Estrogenic Influence: New Insights into Related Growth Disparities. Fertil. Steril. 2019, 111, 178–185. [Google Scholar] [CrossRef]
- Joo, B.S.; Park, M.J.; Kim, C.-W.; Lee, K.S.; Joo, J.K. Differential Expression of Visfatin, Leptin, Stromal Cell Derived Factor-1α, Endothelial Nitric Oxide Synthase, and Vascular Endothelial Growth Factor in Human Leiomyomas. Gynecol. Endocrinol. 2017, 33, 306–310. [Google Scholar] [CrossRef]
- Sanci, M.; Dikis, C.; Inan, S.; Turkoz, E.; Dicle, N.; Ispahi, C. Immunolocalization of VEGF, VEGF Receptors, EGF-R and Ki-67 in Leiomyoma, Cellular Leiomyoma and Leiomyosarcoma. Acta Histochem. 2011, 113, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Tsiligiannis, S.E.; Zaitseva, M.; Coombs, P.R.; Shekleton, P.; Olshansky, M.; Hickey, M.; Vollenhoven, B.; Rogers, P.A.W. Fibroid-Associated Heavy Menstrual Bleeding: Correlation between Clinical Features, Doppler Ultrasound Assessment of Vasculature, and Tissue Gene Expression Profiles. Reprod. Sci. 2013, 20, 361–370. [Google Scholar] [CrossRef]
- Dixon, D.; He, H.; Haseman, J.K. Immunohistochemical Localization of Growth Factors and Their Receptors in Uterine Leiomyomas and Matched Myometrium. Environ. Health Perspect. 2000, 108 (Suppl. S5), 795–802. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Chae, B.; Kim, M.-R. The Potential of Transforming Growth Factor-Beta Inhibitor and Vascular Endothelial Growth Factor Inhibitor as Therapeutic Agents for Uterine Leiomyoma. Int. J. Med. Sci. 2022, 19, 1779–1786. [Google Scholar] [CrossRef]
- Sourla, A.; Polychronakos, C.; Zeng, W.R.; Nepveu, A.; Kukuvitis, A.; Naud, F.; Koutsilieris, M. Plasminogen Activator Inhibitor 1 Messenger RNA Expression and Molecular Evidence for Del(7)(Q22) in Uterine Leiomyomas. Cancer Res. 1996, 56, 3123–3128. [Google Scholar]
- Cheng, Z.; Xie, Y.; Dai, H.; Hu, L.; Zhu, Y.; Gong, J. Unequal Tissue Expression of Proteins from the PA/PAI System, Myoma Necrosis, and Uterus Survival after Uterine Artery Occlusion. Int. J. Gynaecol. Obstet. 2008, 102, 55–59. [Google Scholar] [CrossRef]
- Bogusiewicz, M.; Stryjecka-Zimmer, M.; Postawski, K.; Jakimiuk, A.J.; Rechberger, T. Activity of Matrix Metalloproteinase-2 and -9 and Contents of Their Tissue Inhibitors in Uterine Leiomyoma and Corresponding Myometrium. Gynecol. Endocrinol. 2007, 23, 541–546. [Google Scholar] [CrossRef]
- Governini, L.; Marrocco, C.; Semplici, B.; Pavone, V.; Belmonte, G.; Luisi, S.; Petraglia, F.; Luddi, A.; Piomboni, P. Extracellular Matrix Remodeling and Inflammatory Pathway in Human Endometrium: Insights from Uterine Leiomyomas. Fertil. Steril. 2021, 116, 1404–1414. [Google Scholar] [CrossRef]
- Pekonen, F.; Nyman, T.; Rutanen, E.M. Differential Expression of MRNAs for Endothelin-Related Proteins in Human Endometrium, Myometrium and Leiomyoma. Mol. Cell. Endocrinol. 1994, 103, 165–170. [Google Scholar] [CrossRef]
- Wallace, K.; Chatman, K.; Porter, J.; Scott, J.; Johnson, V.; Moseley, J.; LaMarca, B. Enodthelin 1 Is Elevated in Plasma and Explants from Patients Having Uterine Leiomyomas. Reprod. Sci. 2014, 21, 1196–1205. [Google Scholar] [CrossRef]
- Plewka, A.; Madej, P.; Plewka, D.; Kowalczyk, A.; Miskiewicz, A.; Wittek, P.; Leks, T.; Bilski, R. Immunohistochemical Localization of Selected Pro-Inflammatory Factors in Uterine Myomas and Myometrium in Women of Various Ages. Folia Histochem. Cytobiol. 2013, 51, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Oh, N.-J.; Ryu, K.-Y.; Jung, C.-N.; Yi, S.Y.; Kim, S.-R. Expression of Endothelial Nitric Oxide Synthase in the Uterus of Patients with Leiomyoma or Adenomyosis. J. Obstet. Gynaecol. Res. 2013, 39, 536–542. [Google Scholar] [CrossRef]
- Gokdeniz, R.; Mizrak, B.; Ozen, S.; Bazoglu, N. Endothelial Nitric Oxide Synthase Expression in Leiomyoma and Parental Myometrium. Gynecol. Obstet. Investig. 2000, 49, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Asano, R.; Asai-Sato, M.; Miyagi, Y.; Mizushima, T.; Koyama-Sato, M.; Nagashima, Y.; Taguri, M.; Sakakibara, H.; Hirahara, F.; Miyagi, E. Aberrant Expression of Erythropoietin in Uterine Leiomyoma: Implications in Tumor Growth. Am. J. Obstet. Gynecol. 2015, 213, e1–e8. [Google Scholar] [CrossRef]
- Knapp, P.; Chabowski, A.; Posmyk, R.; Górski, J. Expression of the Energy Substrate Transporters in Uterine Fibroids. Prostaglandins Other Lipid Mediat. 2016, 123, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.-H.; Cho, C.-H.; Cha, S.-D.; Back, W.-K.; Kim, M.-K.; Kim, J.-C. Gene Expression Analysis between Uterine Leiomyoma and Normal Myometrial Tissues by DNA Chip. Korean J. Obstet. Gynecol. 2003, 46, 701–706. [Google Scholar]
- Catherino, W.H.; Mayers, C.M.; Mantzouris, T.; Armstrong, A.Y.; Linehan, W.M.; Segars, J.H. Compensatory Alterations in Energy Homeostasis Characterized in Uterine Tumors from Hereditary Leiomyomatosis and Renal Cell Cancer. Fertil. Steril. 2007, 88 (Suppl. S4), 1039–1048. [Google Scholar] [CrossRef]
- Vanharanta, S.; Pollard, P.J.; Lehtonen, H.J.; Laiho, P.; Sjöberg, J.; Leminen, A.; Aittomäki, K.; Arola, J.; Kruhoffer, M.; Orntoft, T.F.; et al. Distinct Expression Profile in Fumarate-Hydratase-Deficient Uterine Fibroids. Hum. Mol. Genet. 2006, 15, 97–103. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Zhou, J. Expression of bcl-2 and bax protein in uterine leiomyosarcomas and leiomyomas. J. Cent. South University Med. Sci. 2005, 30, 183–186. [Google Scholar]
- Wu, X.; Blanck, A.; Olovsson, M.; Henriksen, R.; Lindblom, B. Expression of Bcl-2, Bcl-x, Mcl-1, Bax and Bak in Human Uterine Leiomyomas and Myometrium during the Menstrual Cycle and after Menopause. J. Steroid Biochem. Mol. Biol. 2002, 80, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Kovács, K.A.; Lengyel, F.; Környei, J.L.; Vértes, Z.; Szabó, I.; Sümegi, B.; Vértes, M. Differential Expression of Akt/Protein Kinase B, Bcl-2 and Bax Proteins in Human Leiomyoma and Myometrium. J. Steroid Biochem. Mol. Biol. 2003, 87, 233–240. [Google Scholar] [CrossRef]
- Csatlós, É.; Máté, S.; Laky, M.; Rigó, J.; Joó, J.G. Role of Apoptosis in the Development of Uterine Leiomyoma: Analysis of Expression Patterns of Bcl-2 and Bax in Human Leiomyoma Tissue With Clinical Correlations. Int. J. Gynecol. Pathol. 2015, 34, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Salimi, S.; Hajizadeh, A.; Yaghmaei, M.; Rezaie, S.; Shahrakypour, M.; Teimoori, B.; Parache, M.; Naghavi, A.; Mokhtari, M. The Effects of P21 Gene C98A Polymorphism on Development of Uterine Leiomyoma in Southeast Iranian Women. Tumour Biol. 2016, 37, 12497–12502. [Google Scholar] [CrossRef]
- Polonikov, A.V.; Samgina, T.A.; Nazarenko, P.M.; Bushueva, O.Y.; Ivanov, V.P. Alcohol Consumption and Cigarette Smoking Are Important Modifiers of the Association Between Acute Pancreatitis and the PRSS1–PRSS2 Locus in Men. Pancreas 2017, 46, 230–236. [Google Scholar] [CrossRef]
- Novakov, V.; Novakova, O.; Sorokina, I. Genetic Markers of Knee Osteoarthritis in Women of the Central Chernozem Region of Russia. Res. Results Biomed. 2023, 9, 191–205. [Google Scholar] [CrossRef]
- Abramova, M. Genetic Markers of Severe Preeclampsia. Res. Results Biomed. 2022, 8, 305–316. [Google Scholar] [CrossRef]
- Malashenkova, I.K.; Ushakov, V.I.; Krynskiy, S.A.; Ogurtsov, D.P.; Khailov, N.A. The Association of Inflammatory Status and Immunological Parameters with Single-Nucleotide Polymorphisms of Cytokine and Toll-like Receptor Genes in Patients with Schizophrenia. Res. Results Biomed. 2022, 8, 148–163. [Google Scholar] [CrossRef]
- Rashid, M.; Zadeh, L.R.; Baradaran, B.; Molavi, O.; Ghesmati, Z.; Sabzichi, M.; Ramezani, F. Up-down Regulation of HIF-1α in Cancer Progression. Gene 2021, 798, 145796. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zang, Y.; Zhao, F.; Li, Z.; Zhang, J.; Fang, L.; Li, M.; Xing, L.; Xu, Z.; Yu, J. Inhibition of HIF-1α by PX-478 Suppresses Tumor Growth of Esophageal Squamous Cell Cancer In Vitro and In Vivo. Am. J. Cancer Res. 2017, 7, 1198–1212. [Google Scholar] [PubMed]
- Lu, J.; Wei, H.; Sun, W.; Geng, J.; Liu, K.; Liu, J.; Liu, Z.; Fu, J.; He, Y.; Wang, K. NKT2152: A Highly Potent HIF2α Inhibitor and Its Therapeutic Potential in Solid Tumors beyond CcRCC. Cancer Res. 2022, 82 (Suppl. S12), 6330. [Google Scholar] [CrossRef]
- Guo, X.C.; Segars, J.H. The Impact and Management of Fibroids for Fertility: An Evidence-Based Approach. Obstet. Gynecol. Clin. N. Am. 2012, 39, 521–533. [Google Scholar] [CrossRef]
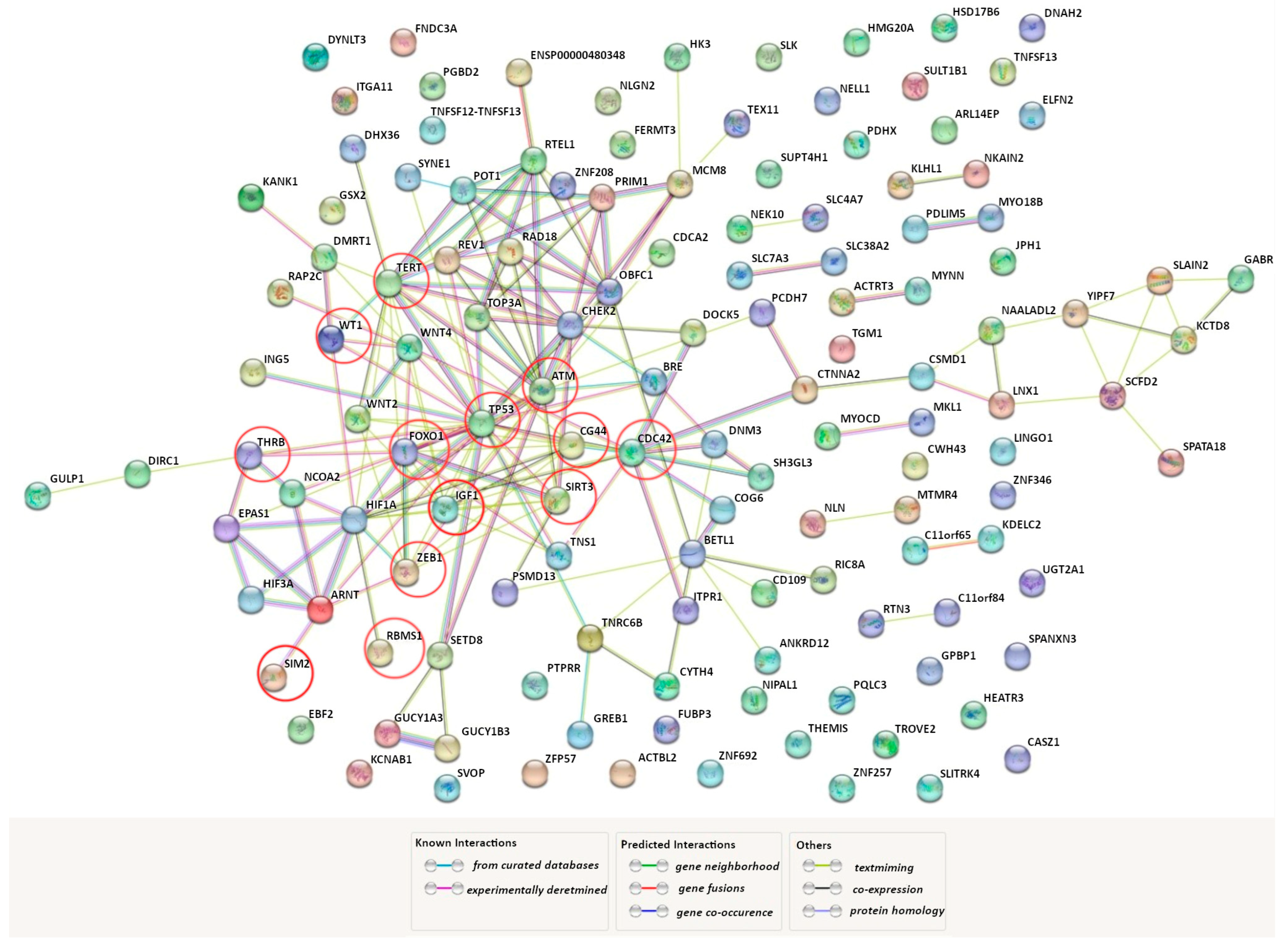
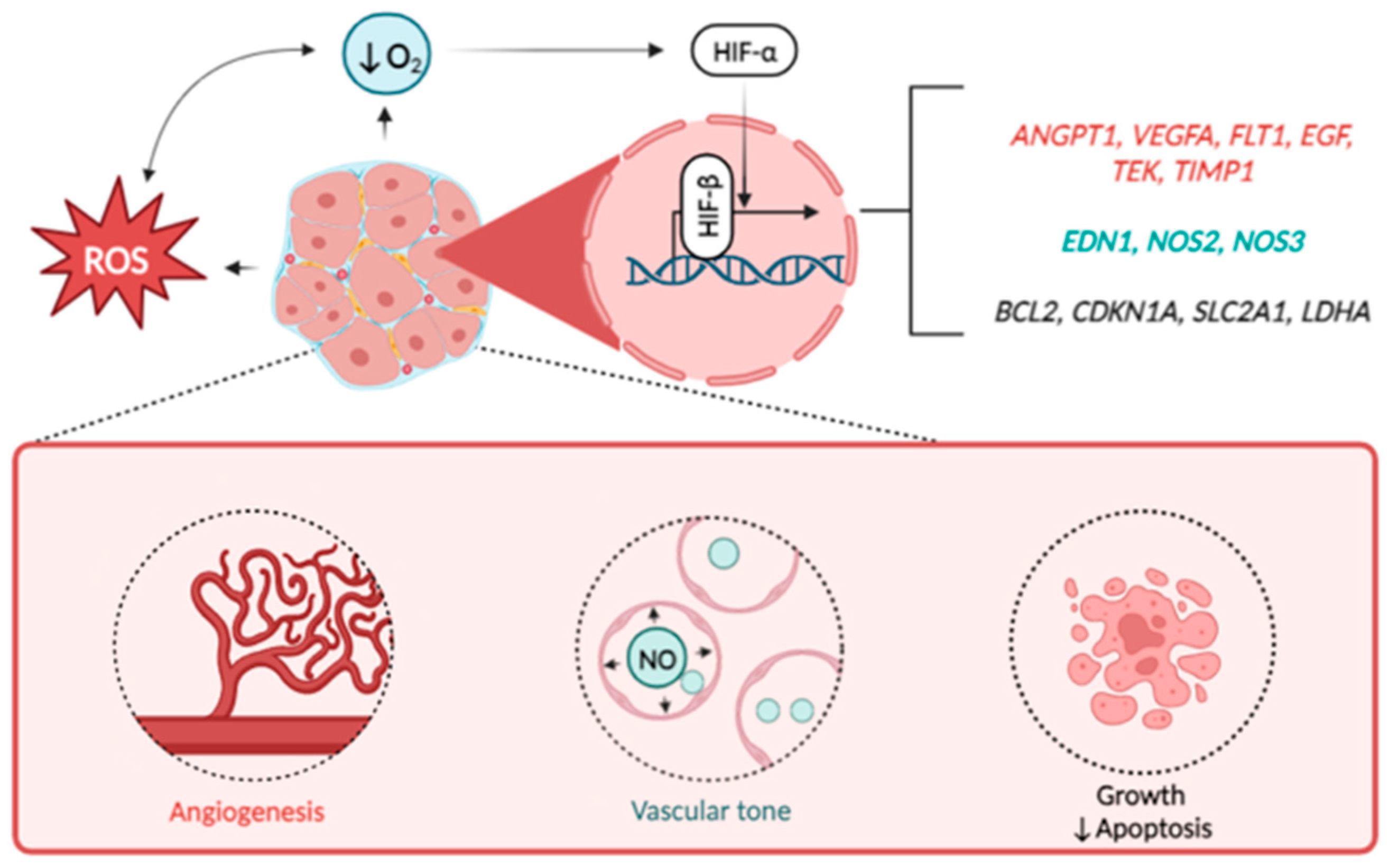
| Factor * | Function ** | Reference | Design of the Study | Main Findings in UF |
|---|---|---|---|---|
| VEGF-A | Promotes the PI3K/Akt pathway, endothelial cell growth, and neo-angiogenesis. | Hague et al. [41] | Comparison of VEGF-A protein levels between women with UF (n = 52) and women without UF (control group, n = 39). | Increased VEGF-A expression was detected in UF compared with controls (p < 0.05). |
| Korompelis et al. [42] | Comparison of the blood levels of VEGF-A protein in women with UF (n = 46) and healthy controls (n = 39). | VEGF-A was significantly elevated in the blood of UF patients compared with controls (p < 0.001). | ||
| Wei et al. [43] | Comparison of VEGF protein in UF tissue (n = 7) and normal myometrium from the same patients (n = 7) Measurements were performed in six zones of UF, from the periphery to the centre. | The level of hypoxia has a linear correlation to VEGF expression levels. VEGF concentrations in UF were gradually increasing from the periphery to the central UF zone (p < 0.05). | ||
| Asano et al. [44] | Comparison of VEGF mRNA in MED12-mutated UF (n = 52) and MED12-wild-type UF (n = 56). | MED12-mutated UF expressed higher VEGF mRNA levels compared with wild-type tumors (p = 0.024). | ||
| Joo et al. [45] | Comparison of VEGF-A mRNA levels in intramural (n = 19) and subserosal (n = 4) UF tissue and normal myometrium (n = 10). | VEGF-A mRNA was significantly higher in both UF types (p < 0.05). | ||
| FLT-1 (VEGFR-1) | Encodes vascular endothelial growth factor receptor 1, which mediates VEGF signaling. | Sanci et al. [46] | Comparison of FLT-1 protein between cellular UF tissue (n = 20) and ordinary UF (n = 20). | Moderate FLT-1 immunostaining intensities in UF and its higher level in cellular UF compared with ordinary UF (p < 0.01). |
| EGF | Induces angiogenesis, cell growth, and differentiation through binding with EGFR. | Tsiligiannis et al. [47] | The level of EGF mRNA was compared between UF (n = 11) and surrounding normal myometrial tissue (n = 11). | An increased level of EGF expression in UF than in NM (p = 0.01–0.05). |
| Dixon et al. [48] | The level of EGF protein expression was studied in UF samples (n = 7) and matched myometrial samples (n = 7). | Lower EGF expression in UF than in myometrium (p < 0.05). | ||
| Park et al. [49] | The level of EGF protein was compared between UF (n = 12) and surrounding normal myometrial tissue (n = 12). | There is no difference in EGF expression between UF and NM (p > 0.05). | ||
| SERPINE1 | Inhibits two ferments: tissue plasminogen activator and urokinase, and hence fibrinolysis. | Sourla et al. [50] | Comparison of SERPINE1 mRNA between UF samples (n = 16) and adjacent myometrium (n = 16). | 11 UF showed higher expression of SERPINE1 mRNA compared with the adjacent myometrium, and 5 UF showed a lower level (p< 0.05). |
| Cheng et al. [51] | Comparison of SERPINE1 mRNA and protein in UF samples (n = 30) and the adjacent myometrial tissue. | Higher expression of SERPINE1 mRNA and protein was investigated in UF compared with the adjacent myometrium (p < 0.05). | ||
| TEK | Angiopoietin-1 receptor. Participates in angiogenesis. | Tsiligiannis et al. [9] | Comparison of TEK mRNA expression between UF (n = 11) and surrounding normal myometrial tissue (n = 11). | Decreased level of TEK expression in UF compared with NM (p < 0.01). |
| TIMP1 | Inhibits proteins from the MMP family. It takes part in extracellular matrix restructuring and slows down endothelial cell migration. | Bogusiewicz et al. [52] | Comparison of TIMP1 protein in UF (n = 20) and the corresponding myometrium (n = 20) of hysterectomized women. | There are no differences between comparable groups (p > 0.05). |
| Tsiligiannis et al. [47] | Comparison of TIMP1 mRNA between UF (n = 11) and surrounding normal myometrial tissue (n = 11). | Higher level of TIMP1 mRNA in UF and proximal myometrium compared with distal myometrium (p > 0.1–0.2). | ||
| Korompelis et al. [42] | Comparison of blood TIMP1 protein level in women with UF (n = 46) and healthy controls (n = 39). | TIMP1 blood level was elevated in UF patients compared with healthy woman (p = 0.003). | ||
| Governini et al. [53] | Comparison of TIMP1 mRNA in women with UF (n = 18) and control women (n = 18). | There were no differences in TIMP1 expression (p > 0.05). | ||
| EDN1 | Strong activator of vasoconstriction. | Pekonen et al. [54] | Comparison of EDN1 mRNA levels in UF (n = 8) and adjacent myometrium (n = 8). | There is no difference between normal myometrium and UF in the abundance of EDN1 mRNA (p > 0.05). |
| Wallace et al. [55] | Comparison of EDN1 protein blood levels in UF (n = 32) and the control group (n = 11). The secretion of ET-1 protein under hypoxia and normoxia was studied using UF samples (n = 32), samples from the adjacent myometrium (n = 32), and samples from control women (n = 11). EDN1 transcript-preproendothelin mRNA (PPET) was quantitated in UF (n = 7) and NM (n = 7) under normoxic and hypoxic conditions. | The circulating EDN1 level was greater in UF patients compared with controls (p < 0.005). Secretion of EDN1 was higher in UF compared with adjacent myometrium (p = 0.025). Hypoxia-induced ET-1 secretion from UF was higher compared to normoxic UF (p = 0.001). EDN1 secretion from UF cultured under normoxia was higher compared with adjacent myometrium (p = 0.02). EDN1 secretion was higher in UF exposed to 24 h of hypoxia (p = 0.005) compared with myometrium explants exposed to hypoxia. Under normoxia, PPET mRNA was increased in UF compared with NM (p = 0.01) and in hypoxic UF compared to hypoxic NM (p = 0.002). | ||
| Miyashita-Ishiwata et al. [33] | Comparison of EDN1 in UF and myometrial cells cultured under normoxic and hypoxic conditions. | Hypoxia induced EDN1 expression in the culture media in leiomyoma but not in myometrial cells. | ||
| iNOS | Inducible enzyme, which produces NO. | Plewka et al. [56] | Comparison of iNOS protein levels in the myometrium of six groups of women with different myoma sizes in perimenopausal and reproductive age. | Increased NO expression in both small and large UF compared to control in women of reproductive and perimenopausal age (p = 0.05). Higher iNOS expression in large myomas compared to small ones (p = 0.05). |
| Fletcher et al. [39] | Comparison of expression of iNOS mRNA and nitrate/nitrite ratio (activity index of iNOS) in UF and NM cell lines exposed to normal (20% O2) and hypoxic (2% O2) conditions for 24 h. | Higher iNOS mRNA expression (p = 0.059) and total nitrate/nitrite ratio (p < 0.04) in UF than in NM under normoxic conditions. Hypoxic conditions led to a more significant increase in iNOS levels in NM (p < 0.001) than in UF (p = 0.268). Under hypoxia, the nitrate/nitrite ratio increased in UF (p = 0.091) and NM (p < 0.001). | ||
| cNOS (eNOS) | Produces NO, the predominant isoform in endothelial cells. | Oh et al. [57] | Comparison of cNOS protein in two groups of perimenopausal women with UF (n = 24) and the control group of healthy patients (n = 8). | Higher level of cNOS expression in the UF women compared with the control (p = 0.05). Inside UF patients, cNOS expression was higher in symptomatic patients (menorrhagia and dysmenorrhea) compared to asymptomatic patients (endometrium p = 0.0029, myometrium p = 0.0276). |
| Gokdeniz et al. [58] | Comparison of cNOS protein levels in UF tissue and normal myometrium (n = 8). | Higher level of cNOS in the smooth muscle cells of UF compared to NM (p < 0.005). | ||
| Joo et al. [45] | Comparison of cNOS mRNA expression in UF patients (n = 23) and normal myometrium (n = 10). | Expressions of cNOS mRNA were higher in intramural and subserosal UF compared with normal myometrium (p < 0.05). Expression of cNOS mRNA was higher in large UF than small UF (p < 0.05). | ||
| EPO | Activator of erythropoiesis in the bone marrow. Under hypoxic conditions, it increases the oxygen-carrying capacity of the blood. | Asano et al. [59] | Comparison of EPO mRNA expression in UF patients (n = 114) and NM from these patients (n = 17). | Higher mRNA expression of EPO in the UF than in the NM (p = 0.025). |
| Asano et al. [44] | Comparison of EPO mRNA expression in MED12-mutated UF (n = 52) and MED12-wild-type UF (n = 56). | The EPO mRNA level was threefold higher in UF with wild-type MED12 genes compared to mutated ones (p = 0.01). | ||
| GLUT-1 | One of the glucose transporter families. Activates glucose passing through the blood–brain barrier, the cell membranes of red blood cells, and tumors. | Ishikawa et al. [35] | Comparison of GLUT-1 mRNA expression in cell cultures of UF under hypoxia and normoxia. | Hypoxia induced the GLUT-1 mRNA expression (p < 0.05). |
| Knapp et al. [60] | Comparison of GLUT-1 mRNA expression in NM specimens and UF women (n = 74). | Higher GLUT-1 mRNA expression in UF compared to NM (p < 0.05). | ||
| HK1 | The first enzyme of glycolysis, phosphorylating glucose to glucose-6-phosphate. | Kwon et al. [61] | Comparison of HK1 mRNA derived from UF and corresponding NM were labeled with Cy5 and Cy3 fluorescein (n = 5). | HK1 mRNA was higher in UF compared to NM (p < 0.05). |
| Catherino et al. [62] | Comparison of 3 UF and NM tissue pairs obtained from a patient with the HLRCC (hereditary leiomyomatosis and renal cell cancer) and patients with nonsyndromic or common UF (n = 11). | Similar expression of HK1 mRNA in nonsyndromic UF and HLRCC compared to NM (p > 0.05). | ||
| ALDOA | Glycolytic enzyme which provides the reversible conversion of fructose-1,6-bisphosphate to dihydroxyacetone phosphate and glyceraldehyde 3-phosphate. | Ishikawa et al. [35] | Comparison of mRNA expression of ALDOA in cells cultures of UF under hypoxia and normoxia. | Hypoxia induced the ALDOA mRNA expression (p < 0.05). |
| Catherino et al. [62] | Comparison of 3 UF and NM tissue pairs obtained from a patient with the HLRCC (hereditary leiomyomatosis and renal cell cancer) and patients with nonsyndromic or common UF (n = 11). | HLRCC fibroids overexpressed ALDOA mRNA (p < 0.05). Expression of this gene was not changed in nonsyndromic UF (p < 0.01). | ||
| ENO1 | Glycolytic enzyme, which provides the conversion of 2-phosphoglycerate to phosphoenolpyruvate. This protein involved in allergic reaction, growth and hypoxia tolerance | Vanharanta et al. [63] | Comparison of ENO1 mRNA levels in UF carrying FH mutations (n = 7) and wild-type FH (n = 15). | Overexpression of ENO1 mRNA in FH-mutated tumors compared to nonmutated UF (p < 0.05). |
| Catherino et al. [62] | Comparison of 3 UF and NM tissue pairs obtained from a patient with HLRCC (hereditary leiomyomatosis and renal cell cancer) and patients with nonsyndromic UF (n = 11). | HLRCC fibroids overexpressed ENO1 mRNA (p < 0.01). Expression of this gene was not changed in nonsyndromic UF. | ||
| Ishikawa et al. [35] | Comparison of ENO1 mRNA expression in cell cultures of UF under hypoxia and normoxia. | Hypoxia induced ENO1 mRNA expression (p < 0.05). | ||
| PGK1 | Glycolytic enzyme catalysing the conversion of 1,3-diphosphoglycerate to 3-phosphoglycerate. | Catherino et al. [62] | Comparison of 3 UF and NM tissue pairs obtained from a patient with the HLRCC (hereditary leiomyomatosis and renal cell cancer) and patients with nonsyndromic or common UF (n = 11). | HLRCC fibroids overexpressed PGK1 mRNA (p < 0.01). Expression of this gene was not changed in nonsyndromic UF. |
| PFK/FBPase 3 | Involved in the synthesis of fructose-2,6-bisphosphate (F2,6BP) and the degradation of F2,6BP. | Catherino et al. [62] | Comparison of 3 UF and NM tissue samples obtained from patients with hereditary leiomyomatosis and renal cell cancer and patients with nonsyndromic or common UF (n = 11). | HLRCC fibroids overexpressed PFK/FBPase 3 mRNA (p < 0.01). Expression of this gene was not changed in nonsyndromic UF. |
| Ishikawa et al. [35] | A comparison of PFK/FBPase 3 mRNA expression was evaluated in cell cultures of UF under hypoxia and normoxia. | Hypoxia induced PFK/FBPase 3 mRNA expression (p < 0.05). | ||
| LDHA | The final glycolytic enzyme that converts L-lactate to pyruvate. | Vanharanta et al. [63] | Comparison of LDHA mRNA expression in UF carrying FH mutations (n = 7) and UF with a wild-type FH (n = 15). | Overexpression of LDHA mRNA in FH-mutated tumors compared to nonmutated UF (p < 0.05). |
| Ishikawa et al. [35] | Comparison of LDHA mRNA expression in cell cultures of UF under hypoxia and normoxia. | Hypoxia induced LDHA mRNA expression (p < 0.05). | ||
| Bcl-2 | Antiapoptotic gene from the B-cell lymphoma gene family. | Zhang et al. [64] | Comparison of Bcl-2 protein expression in patients with UF (n = 34), uterine leiomyosarcoma (n = 34) and normal myometrial samples (n = 34). | Higher Bcl-2 expression in UF than in NM and in uterine leiomyosarcoma (p < 0.01). |
| Wu et al. [65] | Comparison of Bcl-2 protein levels in UF tissue (n = 24) and correspondent NM (n = 22) in patients with UF. | Bcl-2 was overexpressed in UF compared to NM only in the proliferative phase of the menstrual cycle (p < 0.05). Bcl-2 was more expressed in UF from fertile women than from menopausal women (p < 0.05). | ||
| Zhang et al. [64] | Comparison of Bcl-2 protein expression in UF and normal myometrium (n = 40). | Bcl-2 was highly expressed in UF during the whole menstrual cycle compared to NM (p < 0.01). The increase rate was higher in the secretory phase of the menstrual cycle than in the proliferative phase (p < 0.01). | ||
| Kovács et al. [66] | Comparison of Bcl-2 protein in normal and UF specimens from cyclic (n = 16) and menopausal (n = 5) women. | The amount of Bcl-2 in the UF was higher than in the NM (p < 0.05) in the proliferative and secretory phases (p < 0.05). The rate of increase was higher in the secretory phase than in the proliferative phase. In menopausal women, no change was detected in UF relative to NM. | ||
| Csatlós et al. [67] | Comparison of Bcl-2 mRNA in UF women (n = 101) and control women (n = 110). | Bcl-2 mRNA overexpression in UF compared to the control group (p = 0.02–0.04). Bcl-2 expression was positively correlated with the number of tumors. | ||
| Fletcher et al. [39] | Comparison of Bcl-2/Bax RNA ratio in UF and NM cell lines exposed to normal (20% O2) and hypoxic (2% O2) conditions for 24 h. | Higher level of Bcl-2/Bax ratio in UF than in NM under normoxic conditions (p < 0.001). Hypoxia increased the Bcl-2/Bax ratio and decreased the level of apoptosis in UF; in contrast, changes in UF were opposite (decreased Bcl-2/Bax, p < 0.04). | ||
| CDKN1A | Regulator of cell cycle progression at the G1 phase. | Salimi et al. [68] | Comparison of CDKN1A 98A allele frequency in genomic DNA extracted from blood samples of 154 women with UF and 197 matched controls. | The frequency of the CDKN1A 98A allele was significantly higher in the UF women compared to controls (p = 0.04). |
| Biological Process | Fold Enrichment | FDR |
|---|---|---|
| Positive regulation of transcription from RNA polymerase II promoter in response to hypoxia | >100 | 2.04 × 10−3 |
| Regulation of transcription from RNA polymerase II promoter in response to oxidative stress | >100 | 2.90 × 10−3 |
| DNA damage response, signal transduction by P53 class mediator resulting in cell cycle arrest | >100 | 1.21 × 10−2 |
| Positive regulation of nitric-oxide synthase activity | >100 | 1.37 × 10−2 |
| Positive regulation of glycolytic process | >100 | 1.36 × 10−2 |
| Regulation of DNA damage response, signal transduction by P53 class mediator | 93.16 | 1.02 × 10−3 |
| Regulation of intrinsic apoptotic signaling pathway by P53 class mediator | 71.24 | 2.56 × 10−2 |
| Positive regulation of vascular associated smooth muscle cell proliferation | 65.47 | 2.87 × 10−2 |
| Positive regulation of smooth muscle cell migration | 65.47 | 2.86 × 10−2 |
| Myoblast differentiation | 62.11 | 3.02 × 10−2 |
| Negative regulation of reactive oxygen species metabolic process | 55.05 | 3.59 × 10−2 |
| Positive regulation of cell growth | 22.15 | 2.23 × 10−2 |
| Mesenchymal cell differentiation | 21.25 | 2.43 × 10−2 |
| Negative regulation of apoptotic signaling pathway | 21.15 | 3.97 × 10−3 |
| Positive regulation of apoptotic process | 9.50 | 3.83 × 10−2 |
| Data were obtained using the bioinformatics tool Gene Ontology (http://geneontology.org/ (accessed on 4 June 2023)) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedotova, M.; Barysheva, E.; Bushueva, O. Pathways of Hypoxia-Inducible Factor (HIF) in the Orchestration of Uterine Fibroids Development. Life 2023, 13, 1740. https://doi.org/10.3390/life13081740
Fedotova M, Barysheva E, Bushueva O. Pathways of Hypoxia-Inducible Factor (HIF) in the Orchestration of Uterine Fibroids Development. Life. 2023; 13(8):1740. https://doi.org/10.3390/life13081740
Chicago/Turabian StyleFedotova, Maria, Ekaterina Barysheva, and Olga Bushueva. 2023. "Pathways of Hypoxia-Inducible Factor (HIF) in the Orchestration of Uterine Fibroids Development" Life 13, no. 8: 1740. https://doi.org/10.3390/life13081740
APA StyleFedotova, M., Barysheva, E., & Bushueva, O. (2023). Pathways of Hypoxia-Inducible Factor (HIF) in the Orchestration of Uterine Fibroids Development. Life, 13(8), 1740. https://doi.org/10.3390/life13081740






