Aligning Cross-Species Interactomes for Studying Complex and Chronic Diseases
Abstract
1. Introduction
2. Related Work
2.1. Network Comparison
2.2. Network Alignment Algorithms
2.3. Semantic Similarity Measures
3. Materials and Methods
3.1. Dataset: AD-Related PPI Networks
3.2. Dataset: PD-Related PPI Networks
3.3. Local Network Alignmnent
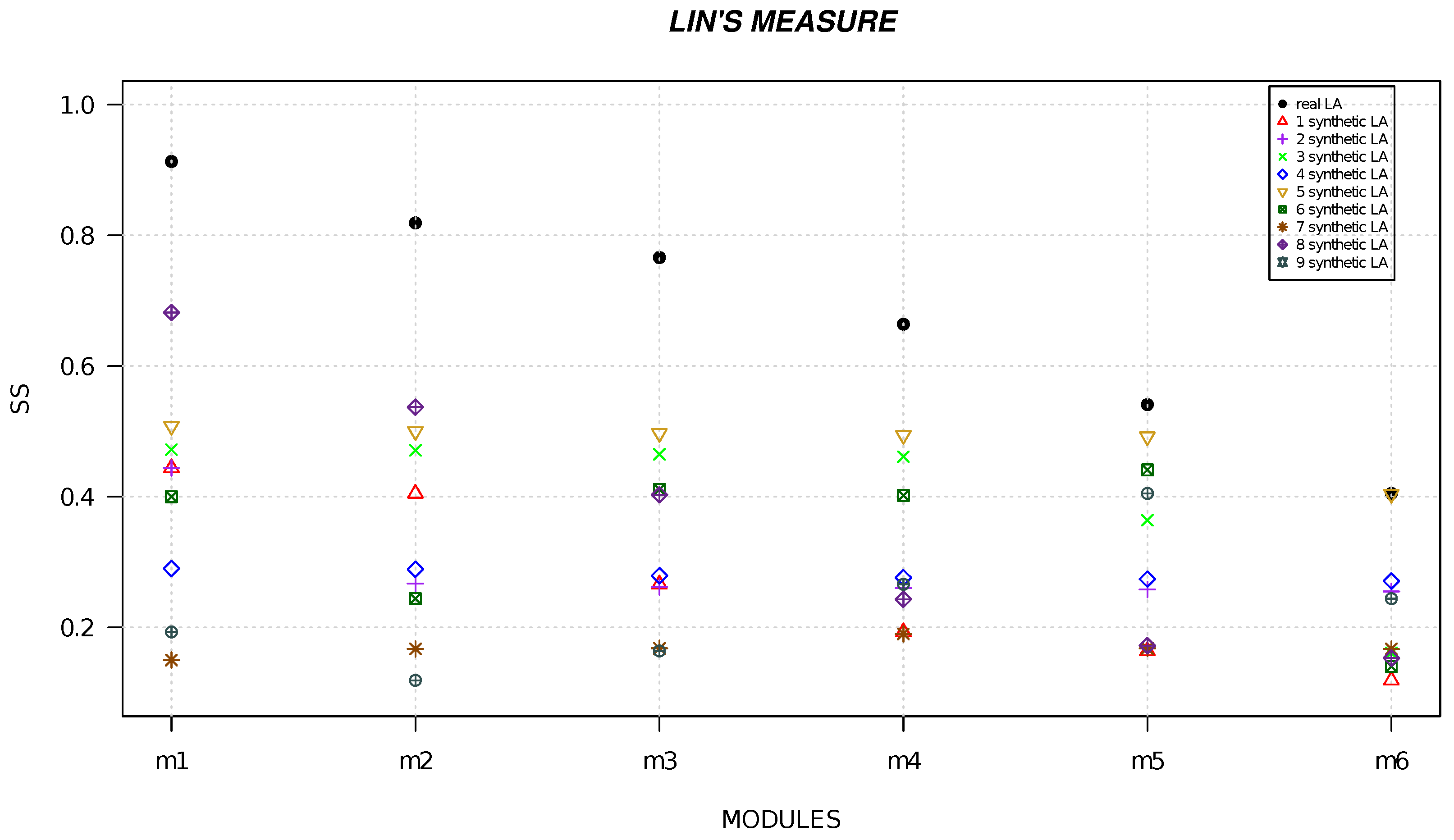
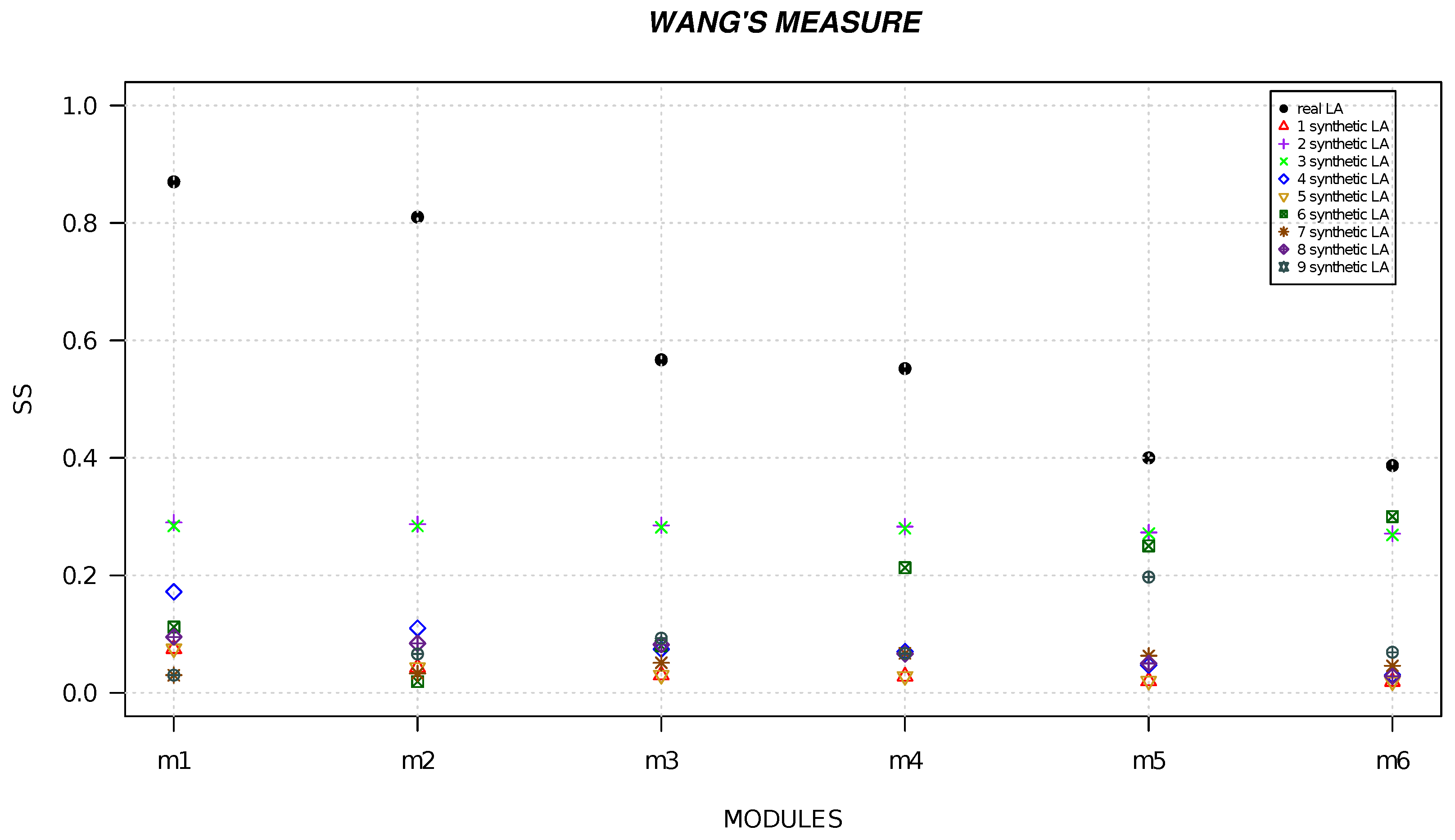
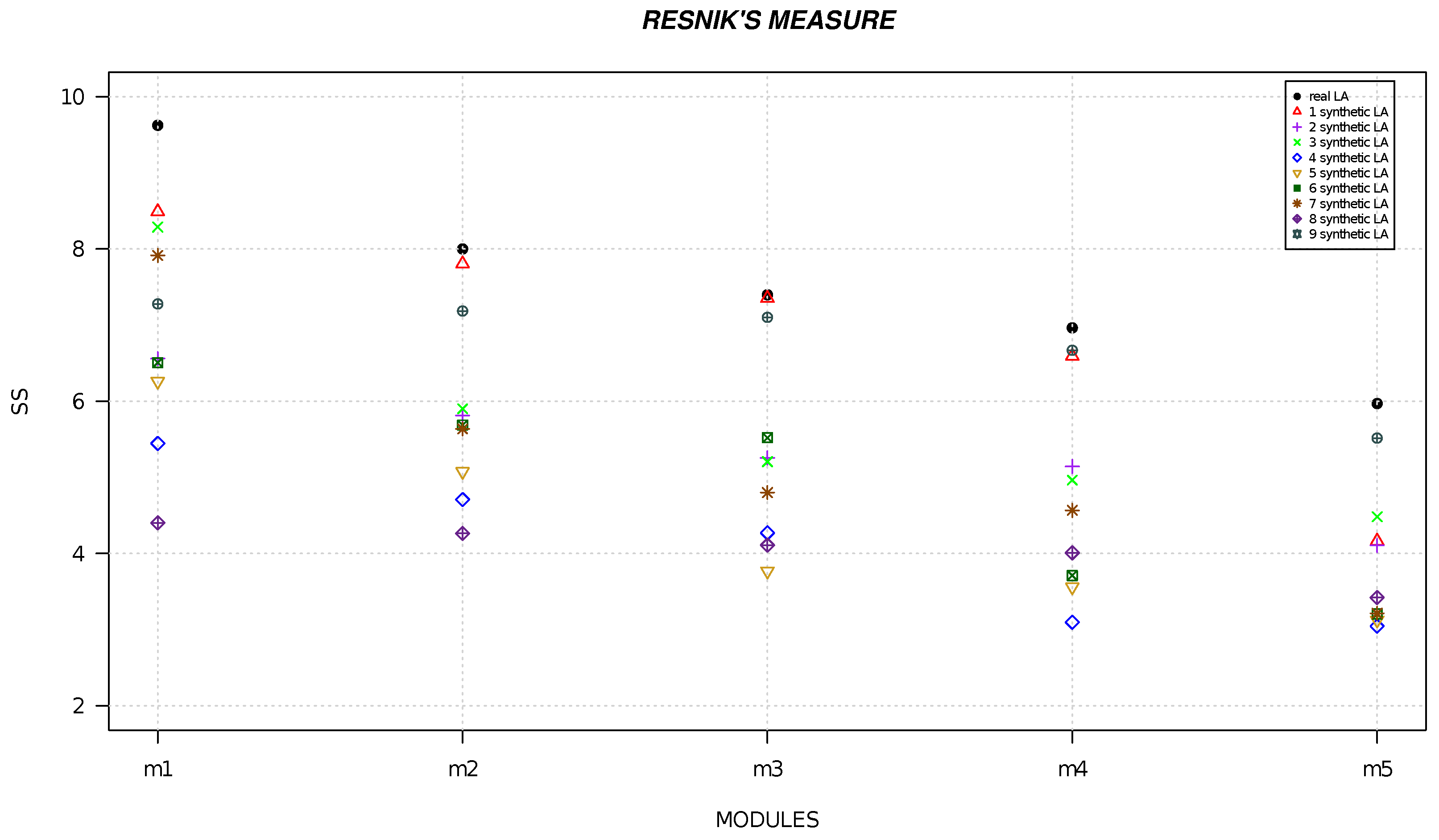
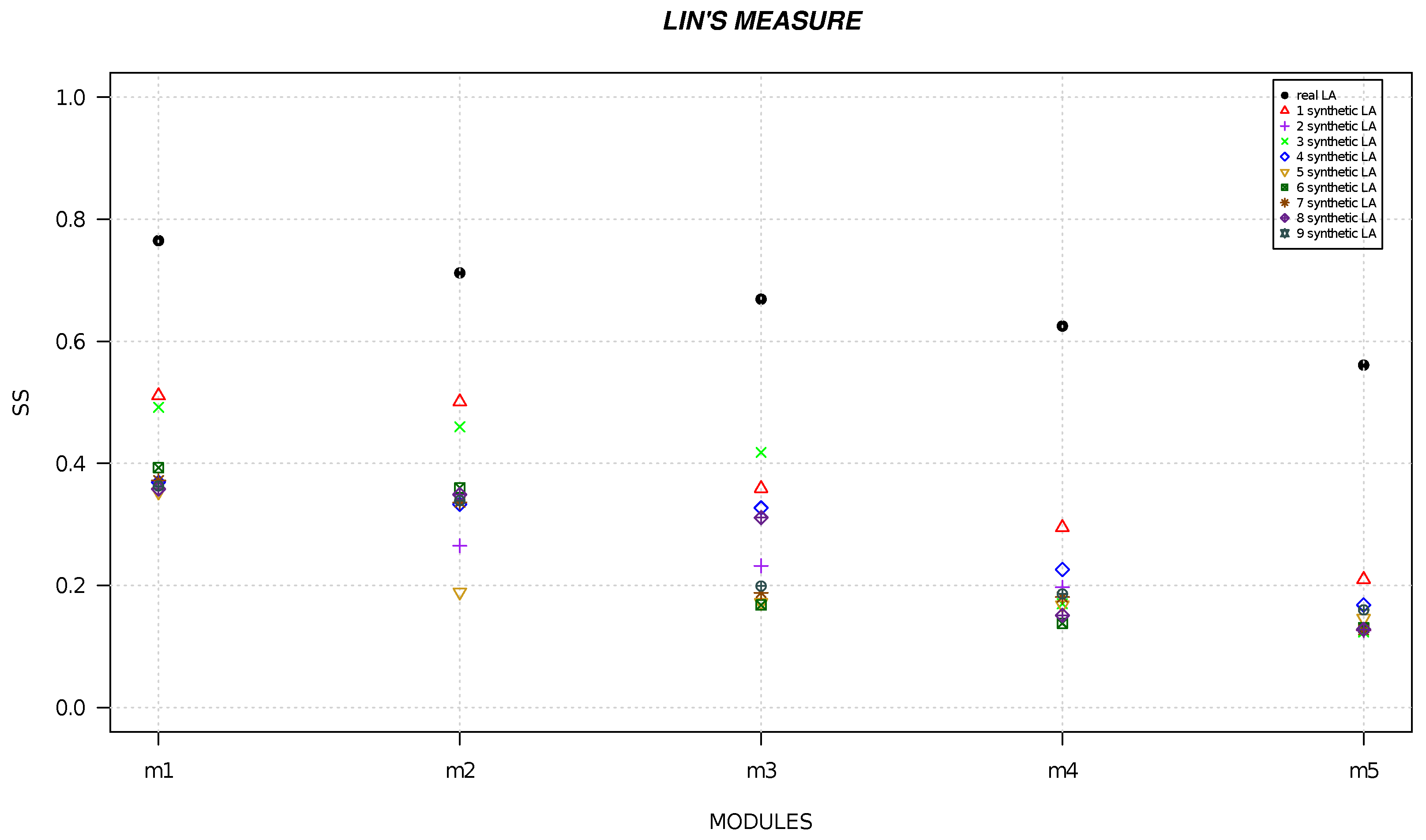
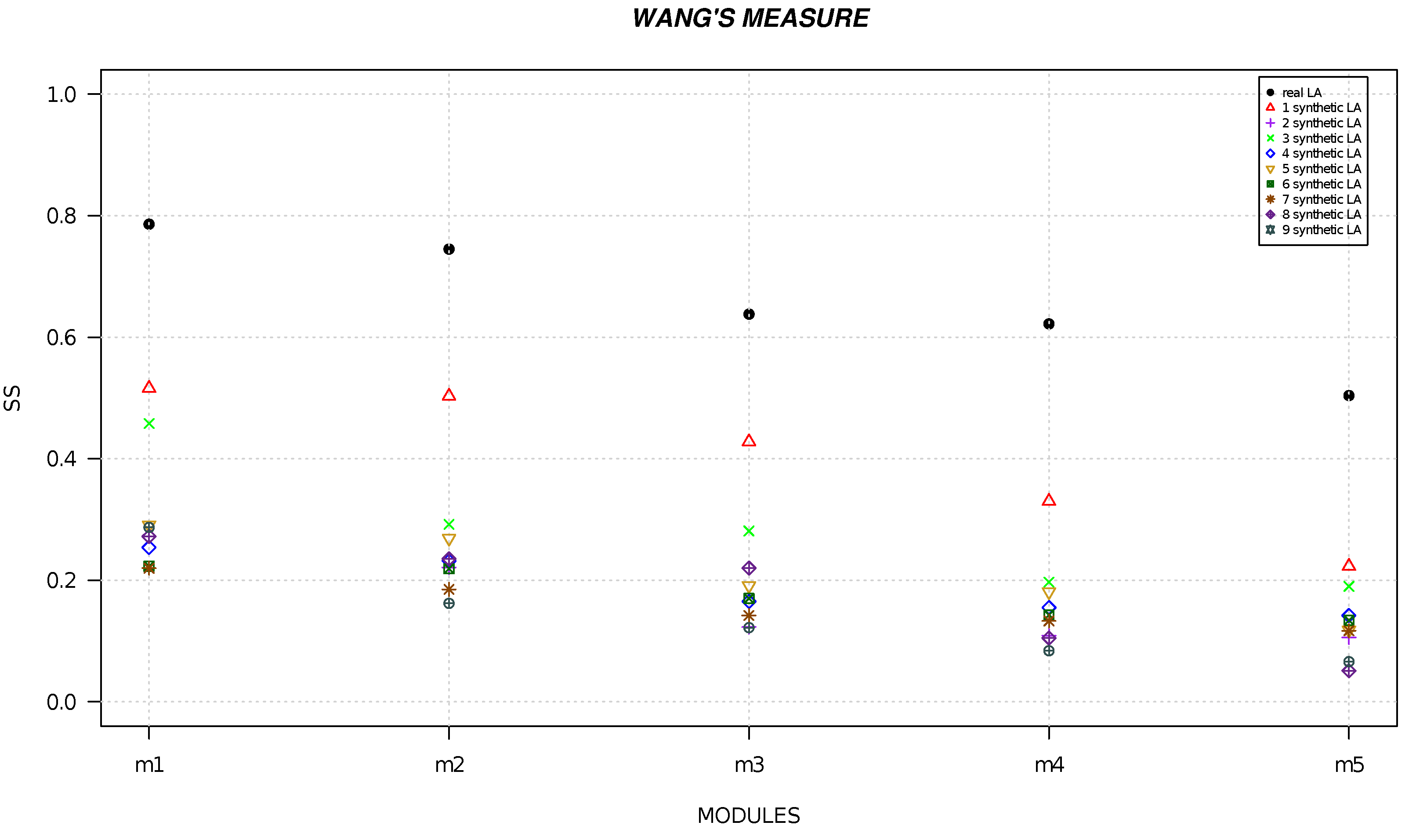
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| ND | Neurodegenerative diseases |
| AD | Alzheimer’s disease |
| PD | Parkinson’s disease |
| PPI | Protein-Protein Interaction |
| NA | Network Alignment |
| LNA | Local Network Alignment |
| GNA | Global Network Alignment |
| GO | Gene Ontology |
| SS | Semantic Similarity |
References
- Uwishema, O.; Mahmoud, A.; Sun, J.; Correia, I.F.S.; Bejjani, N.; Alwan, M.; Nicholas, A.; Oluyemisi, A.; Dost, B. Is Alzheimer’s disease an infectious neurological disease? A review of the literature. Brain Behav. 2022, 12, e2728. [Google Scholar] [CrossRef] [PubMed]
- Fragkiadaki, S.; Kontaxopoulou, D.; Stanitsa, E.; Angelopoulou, E.; Pavlou, D.; Šemrov, D.; Colnar, S.; Lustrek, M.; Blažica, B.; Vučica, I.; et al. How Well Did the Healthcare System Respond to the Healthcare Needs of Older People with and without Dementia during the COVID-19 Pandemic? The Perception of Healthcare Providers and Older People from the SI4CARE Project in the ADRION Region. Geriatrics 2023, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, F.; Zhang, S.; Xin, R.; Sun, Y. Genetic and environmental factors in Alzheimer’s and Parkinson’s diseases and promising therapeutic intervention via fecal microbiota transplantation. npj Park. Dis. 2021, 7, 70. [Google Scholar] [CrossRef]
- Smith, M.A.; Casadesus, G.; Joseph, J.A.; Perry, G. Amyloid-β and τ serve antioxidant functions in the aging and Alzheimer brain. Free Radic. Biol. Med. 2002, 33, 1194–1199. [Google Scholar] [CrossRef]
- Mandelkow, E.M.; Mandelkow, E. Tau in Alzheimer’s disease. Trends Cell Biol. 1998, 8, 425–427. [Google Scholar] [CrossRef]
- Eratne, D.; Loi, S.M.; Farrand, S.; Kelso, W.; Velakoulis, D.; Looi, J.C. Alzheimer’s disease: Clinical update on epidemiology, pathophysiology and diagnosis. Australas Psychiatry 2018, 26, 347–357. [Google Scholar] [CrossRef]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Emamzadeh, F.N.; Surguchov, A. Parkinson’s disease: Biomarkers, treatment, and risk factors. Front. Neurosci. 2018, 12, 612. [Google Scholar] [CrossRef] [PubMed]
- Konovalova, E.; Lopacheva, O.; Grivennikov, I.; Lebedeva, O.; Dashinimaev, E.; Khaspekov, L.; Fedotova, E.Y.; Illarioshkin, S. Mutations in the Parkinson’s disease-associated PARK2 gene are accompanied by imbalance in programmed cell death systems. Acta Nat. 2015, 7, 146–149. [Google Scholar] [CrossRef]
- Elkouzi, A.; Vedam-Mai, V.; Eisinger, R.S.; Okun, M.S. Emerging therapies in Parkinson disease—Repurposed drugs and new approaches. Nat. Rev. Neurol. 2019, 15, 204–223. [Google Scholar] [CrossRef]
- Armstrong, M.J.; Okun, M.S. Diagnosis and treatment of Parkinson disease: A review. JAMA 2020, 323, 548–560. [Google Scholar] [CrossRef]
- Calabrese, G.; Molzahn, C.; Mayor, T. Protein interaction networks in neurodegenerative diseases: From physiological function to aggregation. J. Biol. Chem. 2022, 298, 102062. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.S.; Xin, J.; Hu, Y.; Zhang, L.; Wang, J. Analyzing the genes related to Alzheimer’s disease via a network and pathway-based approach. Alzheimer’s Res. Ther. 2017, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Tomkins, J.E.; Manzoni, C. Advances in protein-protein interaction network analysis for Parkinson’s disease. Neurobiol. Dis. 2021, 155, 105395. [Google Scholar] [CrossRef] [PubMed]
- Cinaglia, P.; Cannataro, M. Network alignment and motif discovery in dynamic networks. Netw. Model. Anal. Health Inform. Bioinform. 2022, 11, 38. [Google Scholar] [CrossRef]
- Liu, Z.P.; Wang, Y.; Zhang, X.S.; Chen, L. Identifying dysfunctional crosstalk of pathways in various regions of Alzheimer’s disease brains. BMC Syst. Biol. 2010, 4, S11. [Google Scholar] [CrossRef]
- Krauthammer, M.; Kaufmann, C.A.; Gilliam, T.C.; Rzhetsky, A. Molecular triangulation: Bridging linkage and molecular-network information for identifying candidate genes in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2004, 101, 15148–15153. [Google Scholar] [CrossRef]
- Goñi, J.; Esteban, F.J.; de Mendizábal, N.V.; Sepulcre, J.; Ardanza-Trevijano, S.; Agirrezabal, I.; Villoslada, P. A computational analysis of protein-protein interaction networks in neurodegenerative diseases. BMC Syst. Biol. 2008, 2, 52. [Google Scholar] [CrossRef]
- Lin, X.; Liu, M.; Chen, X.W. Assessing reliability of protein-protein interactions by integrative analysis of data in model organisms. BMC Bioinform. 2009, 10, S5. [Google Scholar] [CrossRef]
- Surguchov, A. Invertebrate models untangle the mechanism of neurodegeneration in Parkinson’s disease. Cells 2021, 10, 407. [Google Scholar] [CrossRef]
- Alexander, A.G.; Marfil, V.; Li, C. Use of Caenorhabditis elegans as a model to study Alzheimer’s disease and other neurodegenerative diseases. Front. Genet. 2014, 5, 279. [Google Scholar] [CrossRef]
- Milano, M.; Guzzi, P.H.; Tymofieva, O.; Xu, D.; Hess, C.; Veltri, P.; Cannataro, M. An extensive assessment of network alignment algorithms for comparison of brain connectomes. BMC Bioinform. 2017, 18, 235. [Google Scholar] [CrossRef] [PubMed]
- Chandler, R.J.; Cogo, S.; Lewis, P.A.; Kevei, E. Modelling the functional genomics of Parkinson’s disease in Caenorhabditis elegans: LRRK2 and beyond. Biosci. Rep. 2021, 41, BSR20203672. [Google Scholar] [CrossRef]
- Milano, M.; Agapito, G.; Cannataro, M. Challenges and limitations of biological network analysis. BioTech 2022, 11, 24. [Google Scholar] [CrossRef]
- Milano, M.; Guzzi, P.H.; Cannataro, M. Glalign: A novel algorithm for local network alignment. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 16, 1958–1969. [Google Scholar] [CrossRef] [PubMed]
- Milano, M.; Hayes, W.; Veltri, P.; Cannataro, M.; Guzzi, P.H. SL-GLAlign: Improving local alignment of biological networks through simulated annealing. Netw. Model. Anal. Health Inform. Bioinform. 2020, 9, 10. [Google Scholar] [CrossRef]
- Apostolakou, A.E.; Sula, X.K.; Nastou, K.C.; Nasi, G.I.; Iconomidou, V.A. Exploring the conservation of Alzheimer-related pathways between H. sapiens and C. elegans: A network alignment approach. Sci. Rep. 2021, 11, 4572. [Google Scholar] [CrossRef]
- Cannataro, M.; Guzzi, P.H.; Veltri, P. Protein-to-protein interactions: Technologies, databases, and algorithms. ACM Comput. Surv. (CSUR) 2010, 43, 1–36. [Google Scholar] [CrossRef]
- Gao, X.; Xiao, B.; Tao, D.; Li, X. A survey of graph edit distance. Pattern Anal. Appl. 2010, 13, 113–129. [Google Scholar] [CrossRef]
- Guzzi, P.H.; Roy, S. Biological Network Analysis: Trends, Approaches, Graph Theory, and Algorithms; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Gu, S.; Jiang, M.; Guzzi, P.H.; Milenković, T. Modeling multi-scale data via a network of networks. Bioinformatics 2022, 38, 2544–2553. [Google Scholar] [CrossRef]
- Elhesha, R.; Sarkar, A.; Cinaglia, P.; Boucher, C.; Kahveci, T. Co-evolving Patterns in Temporal Networks of Varying Evolution. In Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Bcb’19, Niagara Falls, NY, USA, 7–10 September 2019; ACM: New York, NY, USA, 2019; pp. 494–503. [Google Scholar] [CrossRef]
- Tian, Y.; Mceachin, R.C.; Santos, C.; States, D.J.; Patel, J.M. SAGA: A subgraph matching tool for biological graphs. Bioinformatics 2006, 23, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Zager, L.A.; Verghese, G.C. Graph similarity scoring and matching. Appl. Math. Lett. 2008, 21, 86–94. [Google Scholar] [CrossRef]
- Raveaux, R.; Burie, J.C.; Ogier, J.M. A graph matching method and a graph matching distance based on subgraph assignments. Pattern Recognit. Lett. 2010, 31, 394–406. [Google Scholar] [CrossRef]
- Cinaglia, P.; Cannataro, M. A Method Based on Temporal Embedding for the Pairwise Alignment of Dynamic Networks. Entropy 2023, 25, 665. [Google Scholar] [CrossRef] [PubMed]
- Saraph, V.; Milenković, T. MAGNA: Maximizing accuracy in global network alignment. Bioinformatics 2014, 30, 2931–2940. [Google Scholar] [CrossRef] [PubMed]
- Milano, M.; Milenković, T.; Cannataro, M.; Guzzi, P.H. L-hetnetaligner: A novel algorithm for local alignment of heterogeneous biological networks. Sci. Rep. 2020, 10, 3901. [Google Scholar] [CrossRef]
- Satuluri, V.; Parthasarathy, S.; Ucar, D. Markov clustering of protein interaction networks with improved balance and scalability. In Proceedings of the First ACM International Conference on Bioinformatics and Computational Biology, Niagara Falls, NY, USA, 2–4 August 2010; pp. 247–256. [Google Scholar]
- Cannataro, M.; Guzzi, P.H.; Mazza, T.; Tradigo, G.; Veltri, P. Using ontologies for preprocessing and mining spectra data on the Grid. Future Gener. Comput. Syst. 2007, 23, 55–60. [Google Scholar] [CrossRef]
- Resnik, P. Semantic Similarity in a Taxonomy: An Information-Based Measure and its Application to Problems of Ambiguity in Natural Language. arXiv 2011, arXiv:1105.5444. [Google Scholar] [CrossRef]
- Lin, D. An information-theoretic definition of similarity. In Proceedings of the ICML ’98: Proceedings of the Fifteenth International Conference on Machine Learning, Madison, WI, USA, 24–27 July 1998; Volume 98, pp. 296–304. [Google Scholar]
- Jiang, J.J.; Conrath, D.W. Semantic Similarity Based on Corpus Statistics and Lexical Taxonomy. arXiv 1997, arXiv:cmp-lg/9709008. [Google Scholar]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Kim, W.; Underwood, R.S.; Greenwald, I.; Shaye, D.D. OrthoList 2: A new comparative genomic analysis of human and Caenorhabditis elegans genes. Genetics 2018, 210, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.O. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef] [PubMed]
- Resnik, P. Using information content to evaluate semantic similarity in a taxonomy. arXiv 1995, arXiv:cmp-lg/9511007. [Google Scholar]
- Wang, J.Z.; Du, Z.; Payattakool, R.; Yu, P.S.; Chen, C.F. A new method to measure the semantic similarity of GO terms. Bioinformatics 2007, 23, 1274–1281. [Google Scholar] [CrossRef] [PubMed]
- Milano, M. Computing Languages for Bioinformatics: R. In Encyclopedia of Bioinformatics and Computational Biology—Volume 1; Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 199–205. [Google Scholar] [CrossRef]
- Guzzi, P.H.; Cortese, F.; Mannino, G.C.; Pedace, E.; Succurro, E.; Andreozzi, F.; Veltri, P. Analysis of age-dependent gene-expression in human tissues for studying diabetes comorbidities. Sci. Rep. 2023, 13, 10372. [Google Scholar] [CrossRef]

| Module | # Protein |
|---|---|
| 1 | 28 |
| 2 | 28 |
| 3 | 12 |
| 4 | 12 |
| 5 | 2 |
| 6 | 2 |
| Module | # Protein |
|---|---|
| 1 | 50 |
| 2 | 30 |
| 3 | 35 |
| 4 | 21 |
| 5 | 15 |
| Local Alignment | Resnik | Lin | Wang |
|---|---|---|---|
| module 1 | 22 | 0.913 | 0.870 |
| module 2 | 21.678 | 0.819 | 0.810 |
| module 3 | 16.900 | 0.766 | 0.567 |
| module 4 | 15.478 | 0.664 | 0.552 |
| module 5 | 14.000 | 0.444 | 0.400 |
| module 6 | 12.030 | 0.405 | 0.387 |
| Local Alignment | Resnik | Lin | Wang |
|---|---|---|---|
| module 1 | 9.624 | 0.765 | 0.786 |
| module 2 | 8 | 0.712 | 0.745 |
| module 3 | 7.398 | 0.669 | 0.638 |
| module 4 | 6.592 | 0.625 | 0.622 |
| module 5 | 5.97 | 0.561 | 0.504 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Milano, M.; Cinaglia, P.; Guzzi, P.H.; Cannataro, M. Aligning Cross-Species Interactomes for Studying Complex and Chronic Diseases. Life 2023, 13, 1520. https://doi.org/10.3390/life13071520
Milano M, Cinaglia P, Guzzi PH, Cannataro M. Aligning Cross-Species Interactomes for Studying Complex and Chronic Diseases. Life. 2023; 13(7):1520. https://doi.org/10.3390/life13071520
Chicago/Turabian StyleMilano, Marianna, Pietro Cinaglia, Pietro Hiram Guzzi, and Mario Cannataro. 2023. "Aligning Cross-Species Interactomes for Studying Complex and Chronic Diseases" Life 13, no. 7: 1520. https://doi.org/10.3390/life13071520
APA StyleMilano, M., Cinaglia, P., Guzzi, P. H., & Cannataro, M. (2023). Aligning Cross-Species Interactomes for Studying Complex and Chronic Diseases. Life, 13(7), 1520. https://doi.org/10.3390/life13071520









