Detection and Molecular Characterization of Canine Babesiosis Causative Agent Babesia canis in Naturally Infected Dogs in the Dobrogea Area (Southeastern Romania)
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Samples and Laboratory Investigations
2.3. DNA Isolation, PCR Amplification, and Sequencing
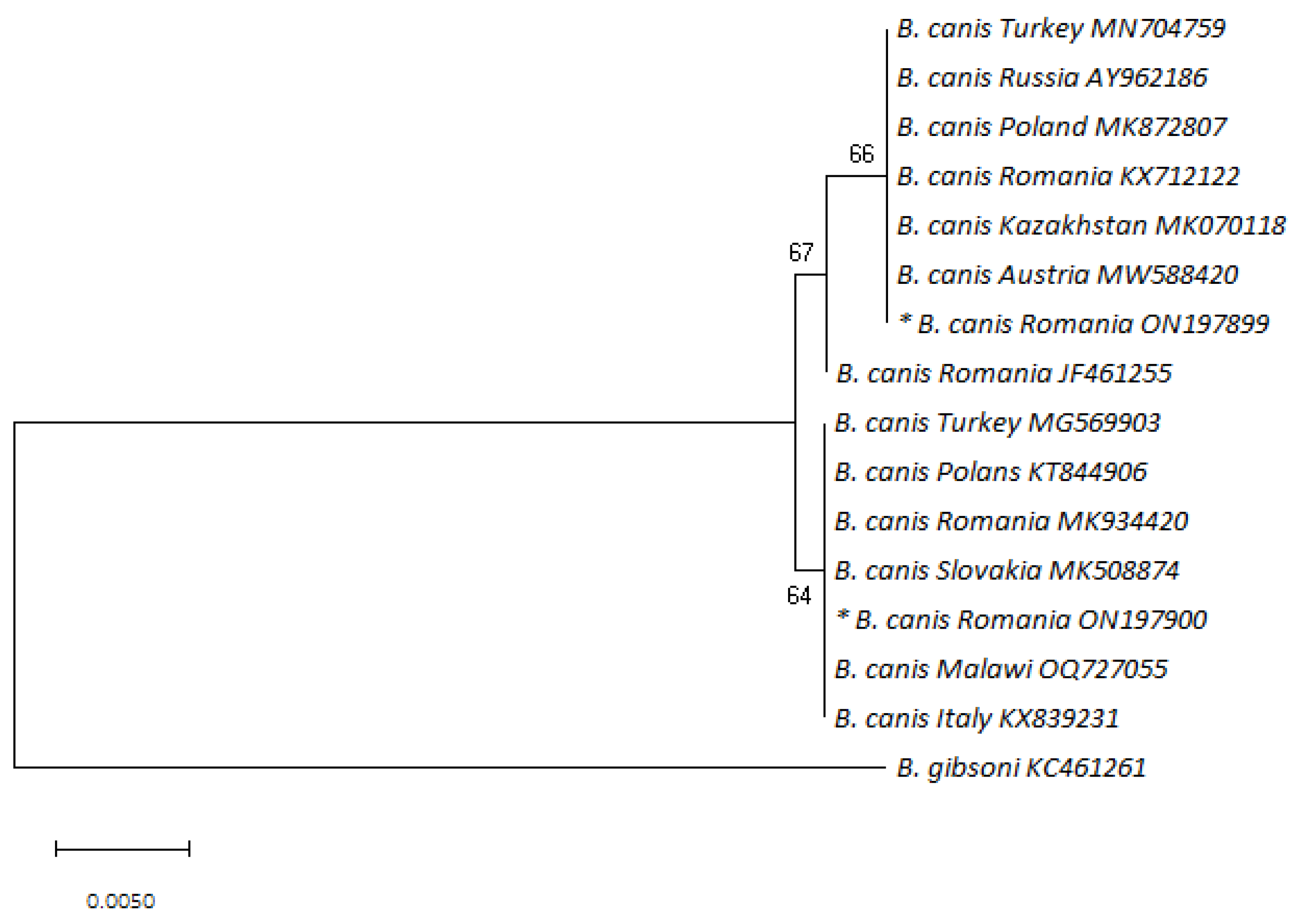
2.4. DNA Sequence and Phylogenetic Analysis
2.5. Statistical Analysis
3. Results
3.1. Clinical Cases
3.2. PCR and DNA Sequence Analysis
3.3. Clinicopathological Findings in B. canis-Infected Dogs
3.4. Clinicopathological Findings in the B. vogeli-Infected Dog
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boozer, A.L.; Macintire, D.K. Canine babesiosis. Vet. Clin. N. Am. Small Anim. Pract. 2003, 33, 885–904. [Google Scholar] [CrossRef] [PubMed]
- Solano-Gallego, L.; Sainz, Á.; Roura, X.; Estrada-Peña, A.; Miró, G. A review of canine babesiosis: The European perspective. Parasites Vectors 2016, 9, 336. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.P. Canine babesiosis: From molecular taxonomy to control. Parasites Vectors 2009, 2 (Suppl. S1), S4. [Google Scholar] [CrossRef] [PubMed]
- Solano-Gallego, L.; Baneth, G. Babesiosis in dogs and cats–expanding parasitological and clinical spectra. Vet. Parasitol. 2011, 181, 48–60. [Google Scholar] [CrossRef]
- Schnittger, L.; Rodriguez, A.E.; Florin-Christensen, M.; Morrison, D.A. Babesia: A world emerging. Infect. Genet. Evol. 2012, 12, 1788–1809. [Google Scholar] [CrossRef]
- Halos, L.; Lebert, I.; Abrial, D.; Danlois, F.; Garzik, K.; Rodes, D.; Schillmeier, M.; Ducrot, C.; Guillot, J. Questionnaire-based survey on the distribution and incidence of canine babesiosis in countries of Western Europe. Parasite 2014, 21, 13. [Google Scholar] [CrossRef]
- Matjila, T.P.; Nijhof, A.M.; Taoufik, A.; Houwers, D.; Teske, E.; Penzhorn, B.L.; Lange, T.; Jongejan, F. Autochthonous canine babesiosis in The Netherlands. Vet. Parasitol. 2005, 131, 23–29. [Google Scholar] [CrossRef]
- Øines, Ø.; Storli, K.; Brun-Hansen, H. First case of babesiosis caused by Babesia canis canis in a dog from Norway. Vet. Parasitol. 2010, 171, 350–353. [Google Scholar] [CrossRef]
- Rubel, F.; Brugger, K.; Monazahian, M.; Habedank, B.; Dautel, H.; Leverenz, S.; Kahl, O. The first German map of georeferenced ixodid tick locations. Parasites Vectors 2014, 7, 477. [Google Scholar] [CrossRef]
- Uilenberg, G.; Franssen, F.F.J.; Perie, M.; Spanger, A.A.M. Three groups of Babesia canis distinguished and a proposal for nomenclature. Vet. Q. 1989, 11, 33–40. [Google Scholar] [CrossRef]
- Schetters, T.P.; Moubri, K.; Precigout, E.; Kleuskens, J.; Scholtes, N.C.; Gorenflot, A. Different Babesia canis isolates, different diseases. Parasitology 1997, 115, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Brandao, L.P.; Hagiwara, M.K.; Myiashiro, S.I. Humoral immunity and reinfection resistance in dogs experimentally inoculated with Babesia canis and either treated or untreated with imidocarb dipropionate. Vet. Parasitol. 2003, 114, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Penzhorn, B.L. Why is Southern African canine babesiosis so virulent? An evolutionary perspective. Parasites Vectors 2011, 4, 51. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, L.S.; Clark, A.I. The pathophysiology of canine babesiosis: New approaches to an old puzzle. J. S. Afr. Vet. Assoc. 1994, 65, 134–145. [Google Scholar]
- Reyers, F.; Leisewitz, A.L.; Lobetti, R.G.; Milner, R.J.; Jacobson, L.S.; van Zyl, M. Canine babesiosis in South Africa: More than one disease. Does this serve as a model for falciparum malaria? Ann. Trop. Med. Parasitol. 1998, 92, 503–511. [Google Scholar] [CrossRef]
- Köster, L.S.; Lobetti, R.G.; Kelly, P. Canine babesiosis: A perspective on clinical complications, biomarkers, and treatment. Vet. Med. Res. Rep. 2015, 6, 119–128. [Google Scholar]
- Shaw, E.S.; Michael, J.D.; Birtles, R.J.; Edward, B.B. Tick-borne infectious diseases of dogs. Trends Parasitol. 2001, 17, 74–80. [Google Scholar] [CrossRef]
- Hauschild, S.; Schein, E. The subspecies specificity of Babesia canis. Berl. Munch. Tierarztl. Wochenschr. 1996, 109, 216–219. [Google Scholar]
- Zahler, M.; Schein, E.; Rinder, H.; Gothe, R. Characteristic genotypes discriminate between Babesia canis isolates of differing vector specificity and pathogenicity to dogs. Parasitol. Res. 1998, 84, 544–548. [Google Scholar] [CrossRef]
- Ionita, M.; Mitrea, I.L.; Pfister, K.; Hamel, D.; Buzatu, C.M.; Silaghi, C. Canine babesiosis in Romania due to Babesia canis and Babesia vogeli: A molecular approach. Parasitol. Res. 2012, 110, 1659–1664. [Google Scholar] [CrossRef]
- Anghel, R.G.; Mitrea, I.L.; Ionita, M. Retrospective study on prevalence of canine vector borne diseases in Bucharest area. Rev. Rom. Med. Vet. 2016, 26, 27–32. [Google Scholar]
- Anghel, R.G.; Mitrea, I.L.; Ionita, M. Clinico-pathological findings in vector-borne pathogen co-infections in dogs, from Bucharest area. Sci. Works Ser. C Vet. Med. 2017, 63, 45–49. [Google Scholar]
- Leica, L.; Mitrea, I.L.; Ionita, M. Clinical occurrence of canine babesiosis in the coastal area of the Black Sea (Dobrogea) in southeastern Romania and associated epidemiological implications. J. Parasitol. 2019, 105, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Lobetti, R.G. Canine babesiosis. In Manual of Canine and Feline Haematology and Transfusion Medicine; Day, M., Mackin, A., Littlewood, J., Eds.; British Small Animal Veterinary Association: Gloucester, UK, 2000; pp. 85–91. [Google Scholar]
- Jacobson, L.S. The south african form of severe and complicated canine babesiosis: Clinical advances 1994–2004. Vet. Parasitol. 2006, 138, 126–139. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Rozsa, L.; Reiczigel, J.; Majoros, G. Quantifying parasites in samples of hosts. J. Parasitol. 2000, 86, 228–232. [Google Scholar] [CrossRef]
- Casati, S.; Sager, H.; Gern, L.; Piffaretti, J.C. Presence of potentially pathogenic Babesia sp. for human in Ixodes ricinus in Switzerland. Ann. Agric. Environ. Med. 2006, 13, 65–70. [Google Scholar]
- Caccio, S.M.; Antunovic, B.; Moretti, A.; Mangili, V.; Marinculic, A.; Baric, R.R.; Slemenda, S.B.; Pieniazek, N.J. Molecular characterisation of Babesia canis canis and Babesia canis vogeli from naturally infected European dogs. Vet. Parasitol. 2002, 106, 285–292. [Google Scholar] [CrossRef]
- Duh, D.; Tozon, N.; Pertovec, M.; Strasek, K.; Avsic-Zupanc, T. Canine babesiosis in Slovenia: Molecular evidence of Babesia canis canis and Babesia canis vogeli. Vet. Res. 2004, 35, 363–368. [Google Scholar] [CrossRef]
- Földvari, G.; Hell, E.; Farkas, R. Babesia canis canis in dogs from Hungary: Detection by PCR and sequencing. Vet. Parasitol. 2005, 127, 221–226. [Google Scholar] [CrossRef]
- Rar, V.A.; Maksimova, T.G.; Zakharenko, L.P.; Bolykhina, S.A.; Dobrotvorsky, A.K.; Morozova, O.V. Babesia DNA detection in canine blood and Dermacentor reticulatus ticks in southwestern Siberia, Russia. Vector Borne Zoonotic Dis. 2005, 5, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Adaszek, L.; Winiarczyk, S. Molecular characterization of Babesia canis canis isolates from naturally infected dogs in Poland. Vet. Parasitol. 2008, 152, 235–241. [Google Scholar] [CrossRef]
- Beck, R.; Vojta, L.; Mrljak, V.; Marinculic, A.; Beck, A.; Zivicnjak, T.; Caccio, S.M. Diversity of Babesia and Theileria species in symptomatic and asymptomatic dogs in Croatia. Int. J. Parasitol. 2009, 39, 843–848. [Google Scholar] [CrossRef]
- Tiskina, V.; Capligina, V.; Must, K.; Berzina, I.; Ranka, R.; Jokelainen, P. Fatal Babesia canis canis infection in a splenectomized Estonian dog. Acta Vet. Scand. 2016, 58, 7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hodzic, A.; Mrowietz, N.; Cezanne, R.; Bruckschwaiger, P.; Punz, S.; Habler, V.E.; Tomsik, V.; Lazar, J.; Duscher, G.G.; Glawischnig, W.; et al. Occurrence and diversity of arthropod-transmitted pathogens in red foxes (Vulpes vulpes) in western Austria, and possible vertical (transplacental) transmission of Hepatozoon canis. Parasitology 2018, 145, 335–344. [Google Scholar] [CrossRef]
- D’Amico, G.; Ionică, A.M.; Györke, A.; Dumitrache, M.O. Epidemiological Survey of the Main Tick-Borne Pathogens Infecting Dogs from the Republic of Moldova. Pathogens 2022, 11, 1267. [Google Scholar] [CrossRef]
- Carret, C.; Walas, F.; Carey, B.; Grande, N.; Precigout, E.; Moubri, K.; Schetters, T.P.; Gorenflot, A. Babesia canis canis, Babesia canis vogeli, Babesia canis rossi: Differentiation of the three subspecies by a restriction fragment length polymorphism analysis on amplified small subunit ribosomal RNA genes. J. Eukaryot. Microbiol. 1999, 46, 298–303. [Google Scholar] [CrossRef]
- Paulauskas, A.; Radzijevskaja, J.; Karveliene, B.; Grigonis, A.; Aleksandraviciene, A.; Zamokas, G.; Babickaitė, L.; Sabūnas, V.; Petkevičius, S. Detection and molecular characterization of canine babesiosis causative agent Babesia canis in the naturally infected dog in Lithuania. Vet. Parasitol. 2014, 205, 702–706. [Google Scholar] [CrossRef] [PubMed]
- Mierzejewska, E.J.; Pawelczyk, A.; Radkowski, M.; Welc-Faleciak, R.; Bajer, A. Pathogens vectored by the tick, Dermacentor reticulatus, in endemic regions and zones of expansion in Poland. Parasites Vectors 2015, 8, 490. [Google Scholar] [CrossRef]
- Seleznova, M.; Kivrane, A.; Namina, A.; Krumins, R.; Aleinikova, D.; Lazovska, M.; Akopjana, S.; Capligina, V.; Ranka, R. Babesiosis in Latvian domestic dogs, 2016–2019. Ticks Tick. Borne Dis. 2020, 11, 101459. [Google Scholar] [CrossRef]
- Passos, L.M.F.; Geiger, S.M.; Ribeiro, M.F.B.; Pfister, K.; Zahler-Rinder, M. First molecular detection of Babesia vogeli in dogs from Brazil. Vet. Parasitol. 2005, 127, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Mumcuoglu, K.Y.; Arslan-Akveran, G.; Aydogdu, S.; Karasartova, D.; Koşar, A.; Savci, U.; Keskin, A.; Taylan-Ozkan, A. Pathogens in ticks collected in Israel: II. Bacteria and protozoa found in Rhipicephalus sanguineus sensu lato and Rhipicephalus turanicus. Ticks Tick. Borne Dis. 2022, 13, 101986. [Google Scholar] [CrossRef] [PubMed]
- Imre, M.; Farkas, R.; Ilie, M.S.; Imre, K.; Darabus, G. Survey of babesiosis in symptomatic dogs from Romania: Occurrence of Babesia gibsoni associated with breed. Ticks Tick. Dis. 2013, 4, 500–502. [Google Scholar] [CrossRef]
- Cimpan, A.A.; Nachum-Biala, Y.; Ben-Shitrit, B.; Miron, L.; Baneth, G. Epidemiological study of canine babesiosis and hepatozoonosis in the South of Romania. Acta Parasitol. 2020, 65, 669–678. [Google Scholar] [CrossRef]
- Ciucă, L.; Martinescu, G.; Miron, L.D.; Roman, C.; Acatrinei, D.; Cringoli, G.; Rinaldi, L.; Maurelli, M.P. Occurrence of Babesia species and co-infection with Hepatozoon canis in symptomatic dogs and in their ticks in Eastern Romania. Pathogens 2021, 10, 1339. [Google Scholar] [CrossRef]
- Łyp, P.; Bartnicki, M.; Staniec, M.; Winiarczyk, S.; Adaszek, Ł. Occurrence of different strains of Babesia canis in dogs in eastern Poland. J. Vet. Res. 2016, 60, 423–427. [Google Scholar] [CrossRef][Green Version]
- Schaarschmidt, D.; Gilli, U.; Gottstein, B.; Marreros, N.; Kuhnert, P.; Daeppen, J.A.; Rosenberg, G.; Hirt, D.; Frey, C.F. Questing Dermacentor reticulatus harbouring Babesia canis DNA associated with outbreaks of canine babesiosis in the Swiss Midlands. Ticks Tick. Borne Dis. 2013, 4, 334–340. [Google Scholar] [CrossRef][Green Version]
- Radzijevskaja, J.; Mardosaitė-Busaitienė, D.; Aleksandravičienė, A.; Karvelienė, B.; Razgūnaitė, M.; Stadalienė, I.; Paulauskas, A. Genetic Diversity of Babesia canis Strains in Dogs in Lithuania. Microorganisms 2022, 10, 1446. [Google Scholar] [CrossRef]
- Reddy, G.; Chakrabarti, D.; Yowell, C.A.; Dame, J.B. Sequence microheterogeneity of the three small subunit ribosomal RNA genes of Babesia bigemina: Expression in erythrocyte culture. Nucleic Acids Res. 1991, 19, 3641–3645. [Google Scholar] [CrossRef] [PubMed]
- Dalrymple, B.P.; Dimmock, C.M.; Parrodi, F.; Wright, I.G. Babesia bovis, Babesia bigemina, Babesia canis, Babesia microti and Babesia rodhaini: Comparison of ribosomal RNA gene organization. Int. J. Parasitol. 1992, 22, 851–855. [Google Scholar] [CrossRef]
- Birkenheuer, A.J.; Levy, M.G.; Breitschwerdt, E.B. Development and evaluation of a seminested PCR for detection and differentiation of Babesia gibsoni (Asian genotype) and B. canis DNA in canine blood samples. J. Clin. Microbiol. 2003, 41, 4172–4177. [Google Scholar] [CrossRef]
- Schoeman, J.P. Canine babesiosis. Onderstepoort J. Vet. Res. 2009, 76, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Solano-Gallego, L.; Trotta, M.; Carli, E.; Carcy, B.; Caldin, M.; Furlanello, T. Babesia canis canis and Babesia canis vogeli clinicopathological findings and DNA detection by means of PCR-RFLP in blood from Italian dogs suspected of tick-borne disease. Vet. Parasitol. 2008, 157, 211–221. [Google Scholar] [CrossRef]
- Rawangchue, T.; Sungpradit, S. Clinicopathological and molecular profiles of Babesia vogeli infection and Ehrlichia canis coinfection. Vet. World 2020, 13, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- De Tommasi, A.S.; Otranto, D.; Dantas-Torres, F.; Capelli, G.; Breitschwerdt, E.B.; De Caprariis, D. Are vector-borne pathogen co-infections complicating the clinical presentation in dogs? Parasites Vectors 2013, 6, 97. [Google Scholar] [CrossRef] [PubMed]
- Ionita, M.; Silaghi, C.; Mitrea, I.L.; Edouard, S.; Parola, P.; Pfister, K. Molecular detection of Rickettsia conorii and other zoonotic spotted fever group rickettsiae in ticks, Romania. Ticks Tick. Borne Dis. 2016, 7, 150–153. [Google Scholar] [CrossRef]
- Ionita, M.; Mitrea, I.L.; Buzatu, M.C.; Dascalu, L. Seasonal dynamics of tick (Acari: Ixodidae) populations in different areas of Romania and the associated risks of tick-borne diseases. In Proceedings of the XIIth International Congress of Parasitology (ICOPA), Melbourne, VIC, Australia, 15–20 August 2010; Medimond International Proceedings: Bologna, Italy, 2010; pp. 91–95. [Google Scholar]
| Variable | No. (%) | Clinical Presentation | Uncomplicated and Complicated (C-) Babesiosis | ||||
|---|---|---|---|---|---|---|---|
| Mild | Moderate | Severe | Uncomplicated | * C-SOD | ** C-MOD | ||
| Total dogs (n = 23) | 6 (26.1) | 7 (30.4) | 10 (43.5) | 10 (43.5) | 9 (39.1) | 4 (17.4) | |
| Age (years) | |||||||
| <1 | 6 (26.1) | 3 (50.0) | 2 (33.3) | 1 (16.7) | 4 (66.7) | 2 (33.3) | 0 |
| 1–3 | 8 (34.8) | 2 (25.0) | 3 (37.5) | 3 (37.5) | 3 (37.5) | 4 (50.0) | 1 (12.5) |
| 4–6 | 3 (8.7) | 0 | 0 | 3 (100) | 0 | 1 (33.3) | 2 (66.7) |
| 7–9 | 2 (8.7) | 1 (50.0) | 0 | 1 (50.0) | 1 (50.0) | 1 (50.0) | 0 |
| ≥10 | 4 (17.4) | 0 | 2 (50.0) | 2 (50.0) | 2 (50.0) | 1 (25.0) | 1 (25.0) |
| Sex | |||||||
| male | 17 (73.9) | 5 (29.4) | 2 (11.8) | 10 (58.8) | 8 (47.1) | 5 (29.4) | 4 (23.5) |
| female | 6 (26.1) | 1 (16.7) | 5 (83.3) | 0 | 2 (33.3) | 4 (66.7) | 0 |
| Breed | |||||||
| pure breed | 17 (73.9) | 4 (23.5) | 6 (35.3) | 7 (41.2) | 7 (41.2) | 6 (35.3) | 4 (23.5) |
| mixed breed | 6 (26.1) | 2 (33.3) | 1 (16.7) | 3 (50.0) | 3 (50.0) | 3 (50.0) | 0 |
| Babesia species | |||||||
| Babesia vogeli | 1 (4.4) | 0 | 0 | 1 (100) | 1 (100) | 0 | 0 |
| Babesia canis | 22 (95.6) | 6 (27.3) | 7 (31.8) | 9 (40.9) | 9 (40.9) | 9 (40.9) | 4 (18.2) |
| B. canis genotypes | |||||||
| AG | 12 (54.5) | 4 (33.3) | 7 (58.3) | 1 (8.4) | 6 (50) | 6 (50) | 0 |
| GA | 2 (9.1) | 0 | 0 | 2 (100) | 0 | 1 (50.0) | 1 (50.0) |
| RR | 8 (36.4) | 2 (25.0) | 0 | 6 (75.0%) | 3 (37.5) | 2 (25.0) | 3 (37.5) |
| p-value | 1.000 | 0.012 | 0.002 | 0.571 | 0.571 | 0.051 | |
| GenBank Accession No. | Nucleotide Position | |
|---|---|---|
| 609 | 610 | |
| AY072926.1 | G | A |
| #ON197899.1 | A | G |
| #ON197900.1 | G | A |
| #Mixed RR | G/A | G/A |
| Pathological Changes in Babesia canis-Positive Dogs, Stratified by B. canis 18S rRNA Genotypes: Number of Dogs; Percentage (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Anemia | Thrombocytopenia | * T-bil↑ | Azotemia | ** H↑ | *** P↑ | MODs | |||||||
| Total | Mild | Moderate | Severe | Total | Mild | Moderate | Severe | ||||||
| Total (n = 22) | 14 (63.6) | 2 (14.2) | 8 (57.1) | 4 (28.6) | 13 (59.1) | 3 (23.1) | 3 (23.1) | 7 (53.8) | 16 (72.7) | 13 (59.1) | 10 (45.4) | 1 (4.5) | 4 (18.2) |
| B. canis genotypes | |||||||||||||
| AG (n = 12) | 5 (41.7) | 1 (8.33) | 4 (33.3) | 0 | 4 (33.3) | 2 (16.7) | 1 (8.3) | 1 (8.3) | 8 (66.7) | 5 (41.7) | 5 (41.7) | 0 | 0 |
| GA (n = 2) | 2 (100) | 0 | 0 | 2 (100) | 2 (100) | 0 | 0 | 2 (100) | 2 (100) | 2 (100) | 1 (50.0) | 0 | 1 (50) |
| RR (mixed) (n = 8) | 7 (87.5) | 1 (12.5) | 4 (50.0) | 2 (25.0) | 7 (87.5) | 1 (12.5) | 2 (25.0) | 4 (50.0) | 6 (75.0) | 6 (75.0) | 4 (50.0) | 1 (12.5) | 3 (37.5) |
| p-value | 0.074 | 1.000 | 0.549 | 0.004 | 0.009 | 1.000 | 0.657 | 0.017 | 1.000 | 0.202 | 1.000 | 1.000 | 0.051 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ionita, M.; Leica, L.; Wassermann, M.; Mitrea, E.; Nicorescu, I.M.; Mitrea, I.L. Detection and Molecular Characterization of Canine Babesiosis Causative Agent Babesia canis in Naturally Infected Dogs in the Dobrogea Area (Southeastern Romania). Life 2023, 13, 1354. https://doi.org/10.3390/life13061354
Ionita M, Leica L, Wassermann M, Mitrea E, Nicorescu IM, Mitrea IL. Detection and Molecular Characterization of Canine Babesiosis Causative Agent Babesia canis in Naturally Infected Dogs in the Dobrogea Area (Southeastern Romania). Life. 2023; 13(6):1354. https://doi.org/10.3390/life13061354
Chicago/Turabian StyleIonita, Mariana, Laurentiu Leica, Marion Wassermann, Emanuel Mitrea, Isabela Madalina Nicorescu, and Ioan Liviu Mitrea. 2023. "Detection and Molecular Characterization of Canine Babesiosis Causative Agent Babesia canis in Naturally Infected Dogs in the Dobrogea Area (Southeastern Romania)" Life 13, no. 6: 1354. https://doi.org/10.3390/life13061354
APA StyleIonita, M., Leica, L., Wassermann, M., Mitrea, E., Nicorescu, I. M., & Mitrea, I. L. (2023). Detection and Molecular Characterization of Canine Babesiosis Causative Agent Babesia canis in Naturally Infected Dogs in the Dobrogea Area (Southeastern Romania). Life, 13(6), 1354. https://doi.org/10.3390/life13061354






