A Noise-Tolerating Gene Association Network Uncovering an Oncogenic Regulatory Motif in Lymphoma Transcriptomics
Abstract
1. Introduction
Importance of Modeling Gene Association Networks with Biological Noises
2. Materials and Methods
2.1. A Framework for Regression-Based Modeling of GAN
2.2. Conventional Strategies for Estimating the Association Parameters
2.3. An Alternative Parameter Estimation Method
2.4. Modeling Biological Noises and Correlated Expressions
2.5. A Robust Distribution-Free Regression Method for Modeling GAN Using AWTE
- Step 1. Determine the common interaction component (i.e., the second source of the additive combination, xip) in Equation (6) among p predictor genes according to their observed expression levels; it can be made by
- Step 2. Estimate all the correlation parameters r1, r2, …, rp−1 in Equation (6), which can be achieved by
- Step 3. Obtain the association parameter βjk in Equation (7) under the constraint of Equation (6), by using
3. Results
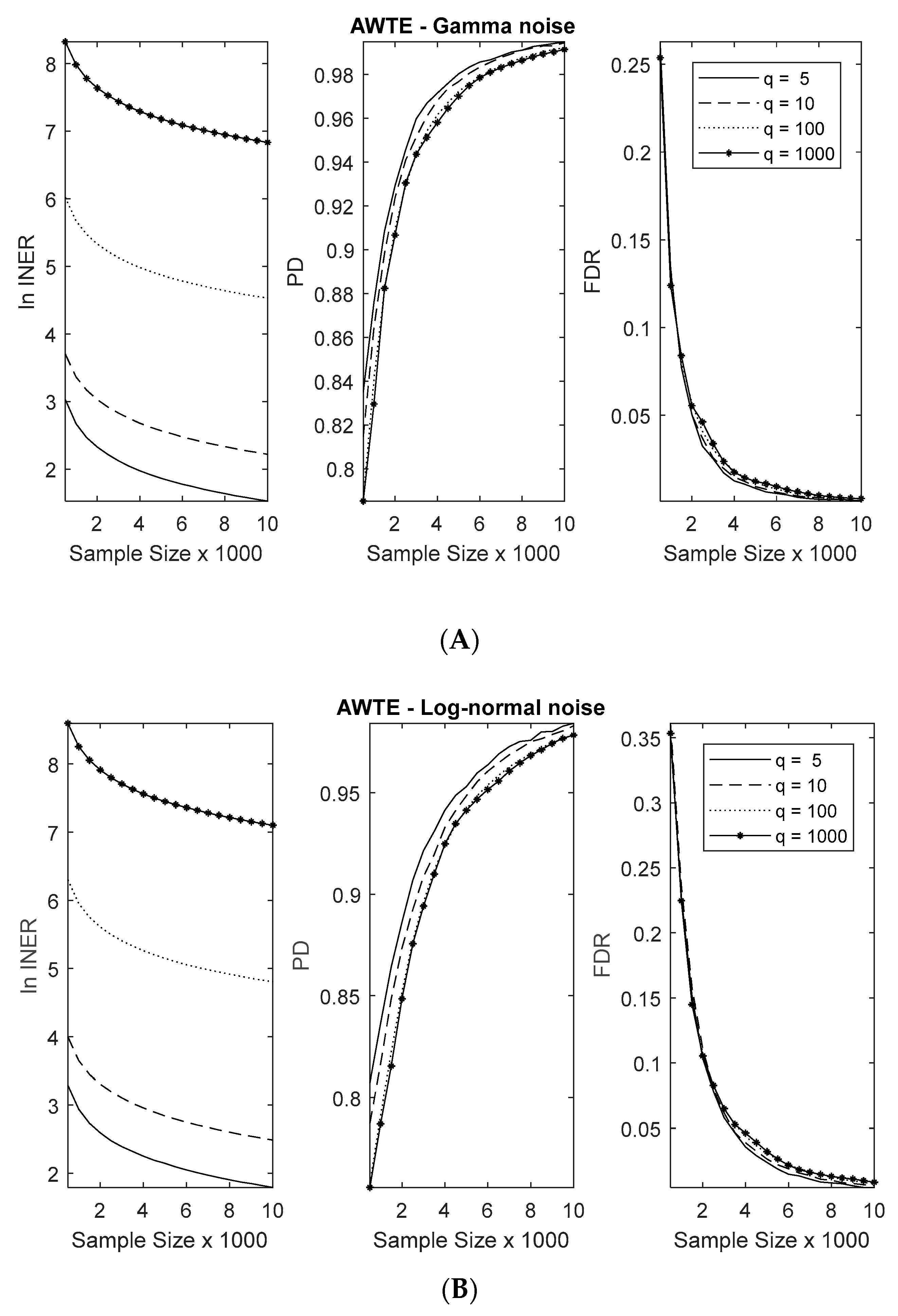
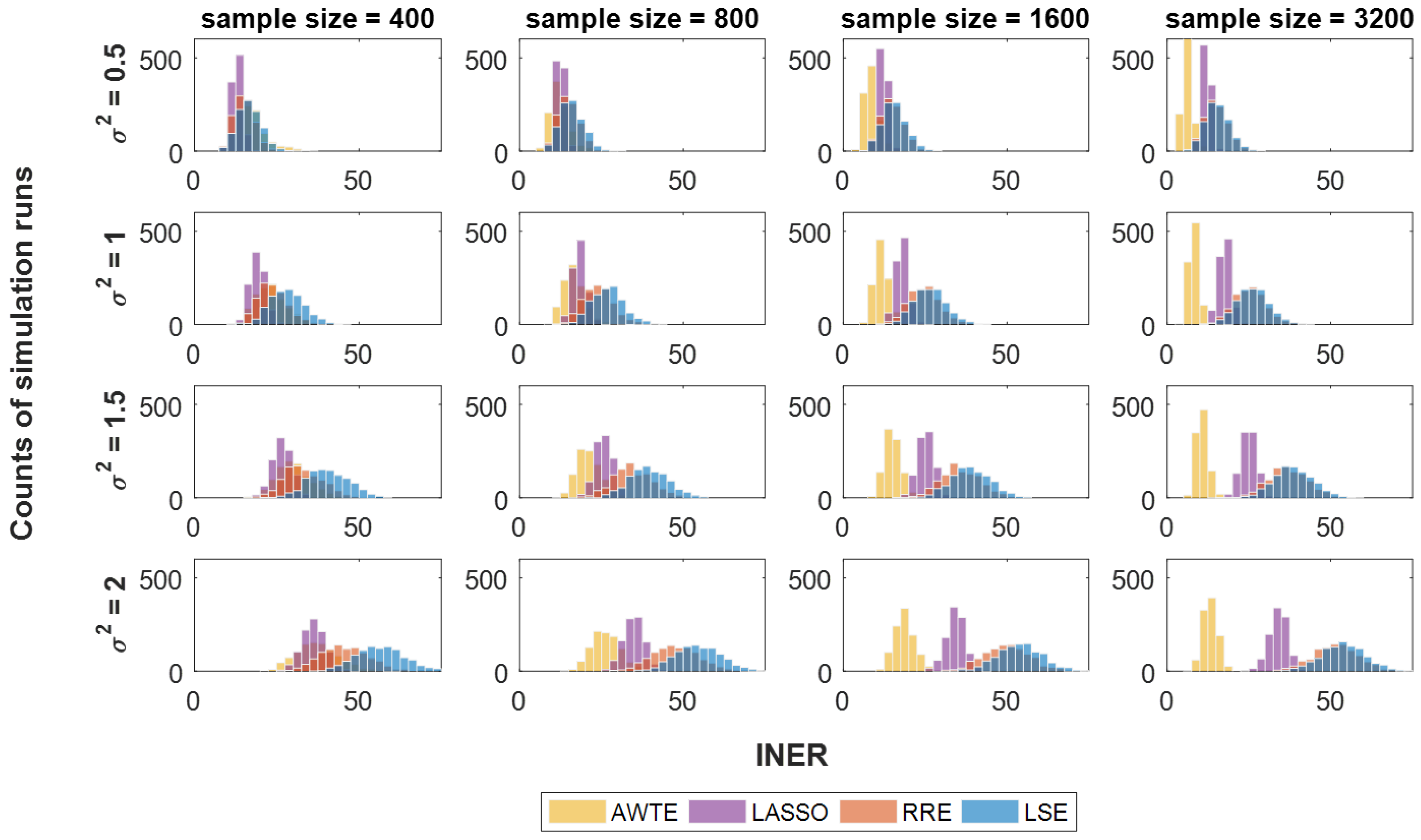
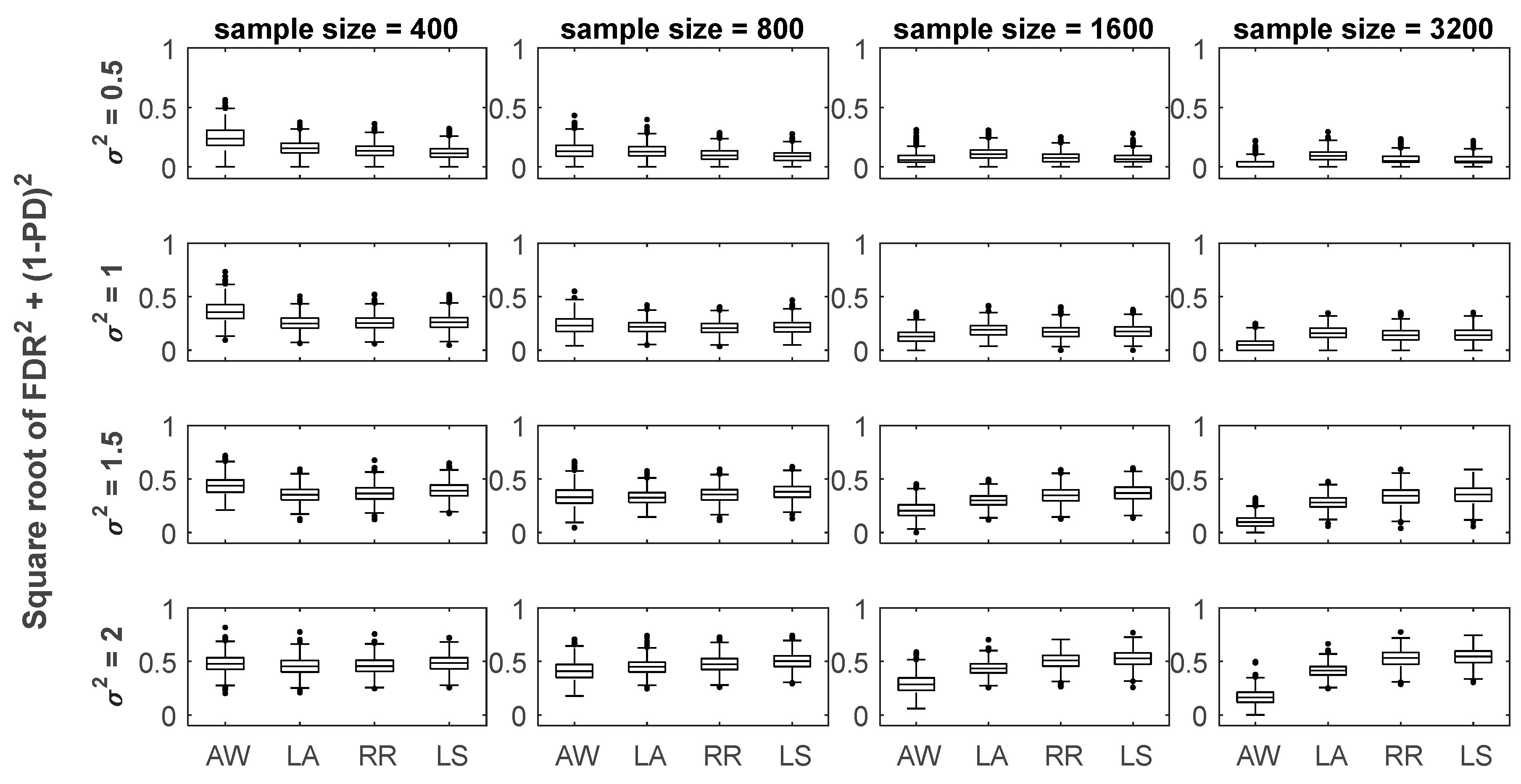
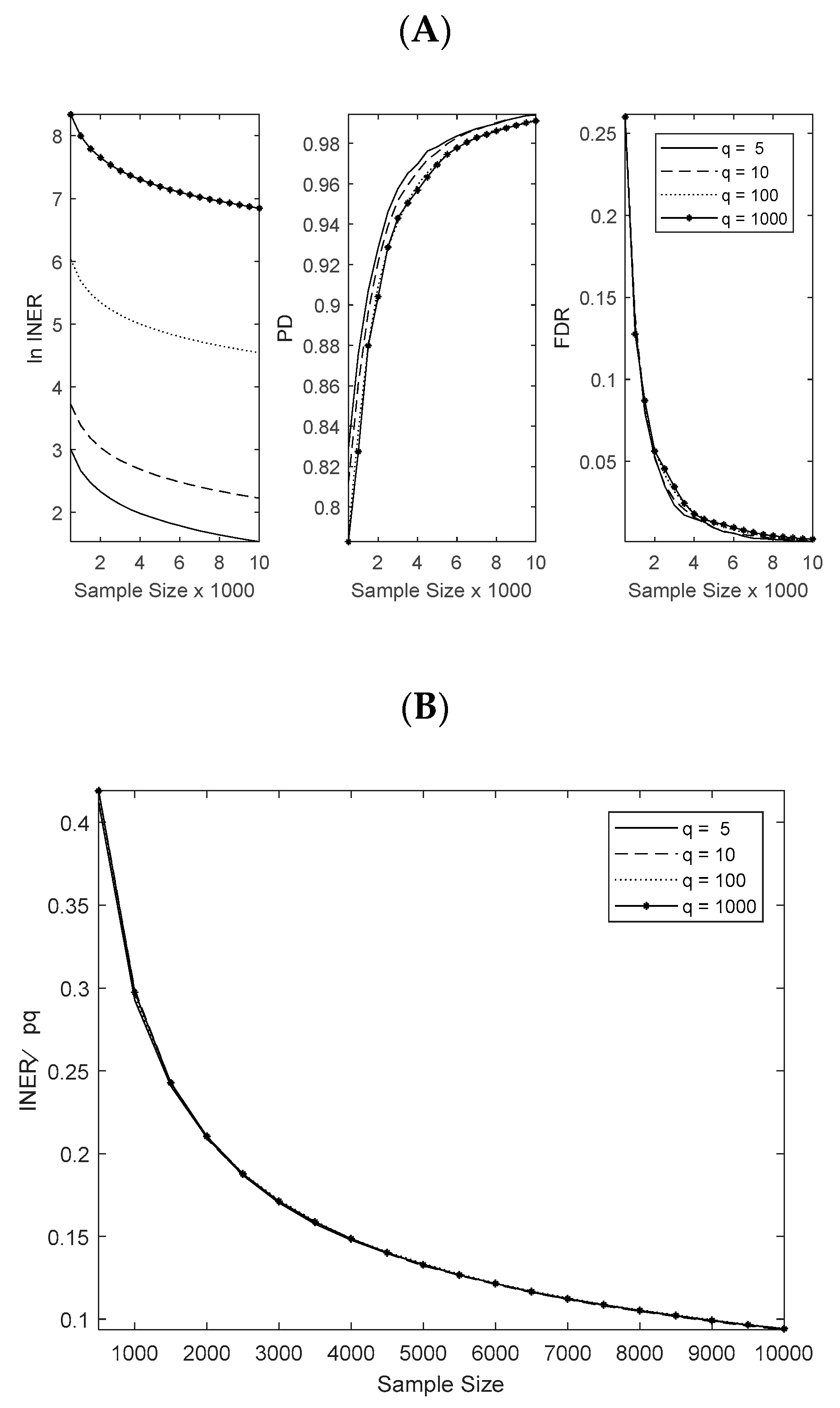
3.1. Numerical Simulation Settings
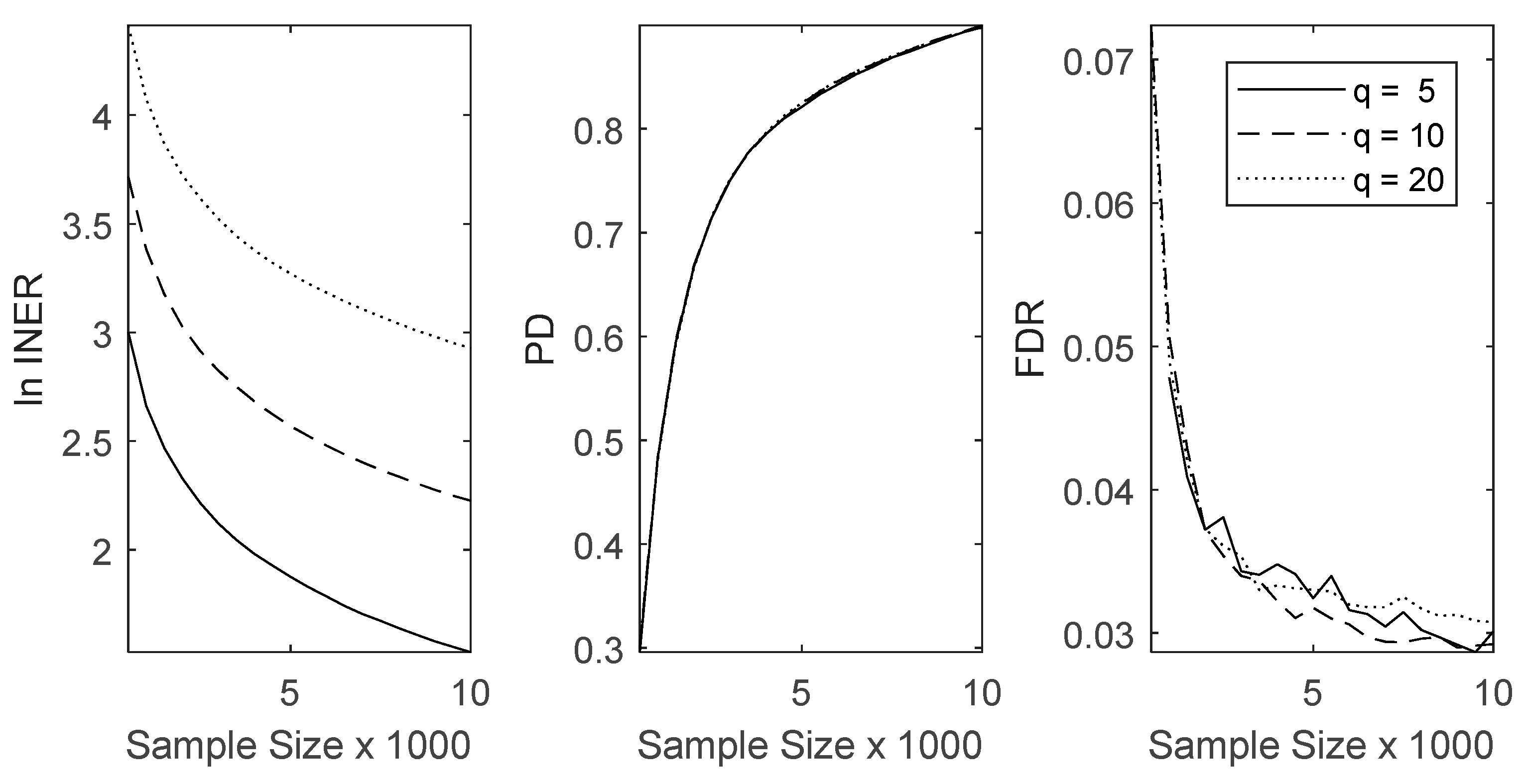
3.2. Method Comparisons in Numerical Simulations
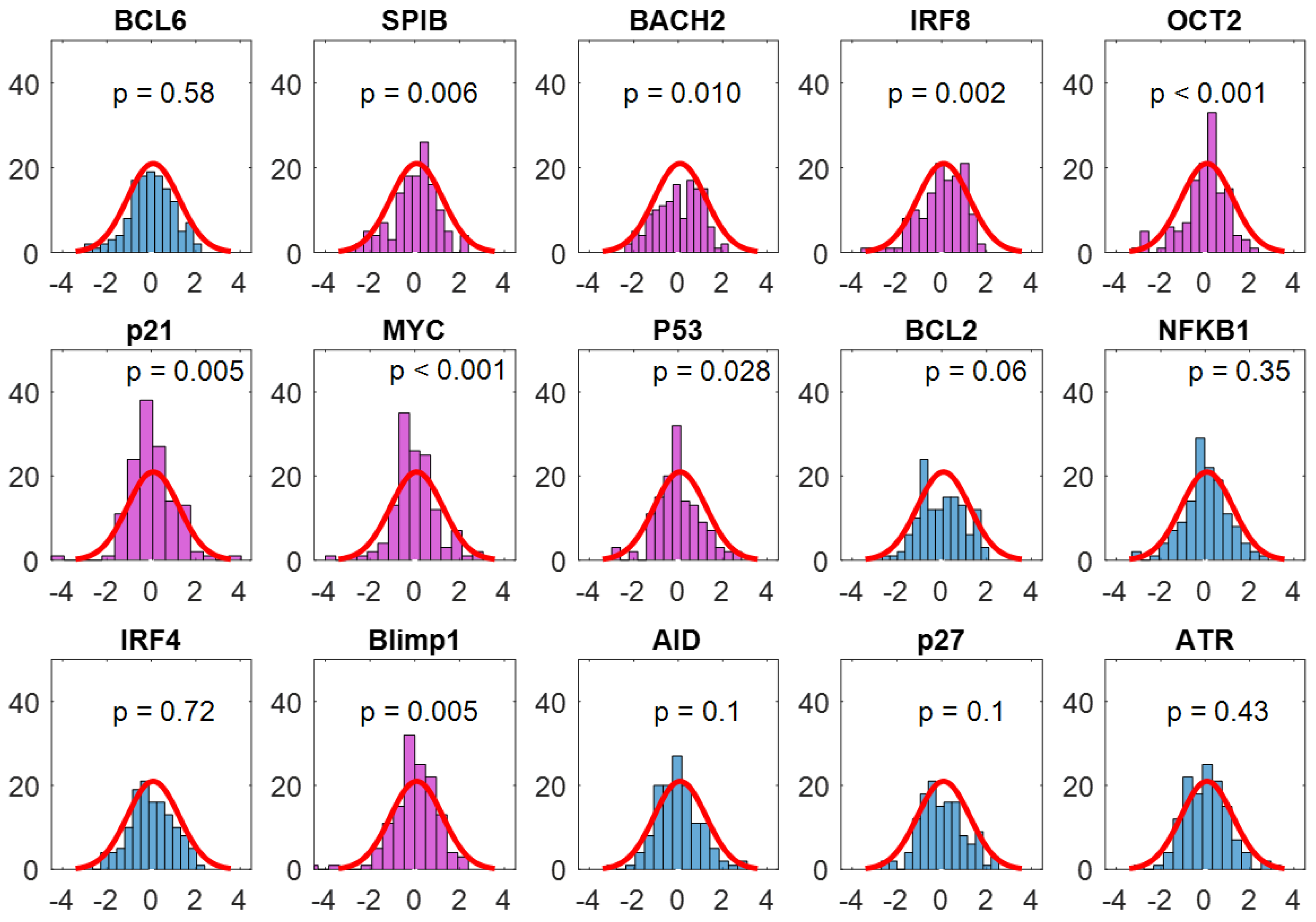
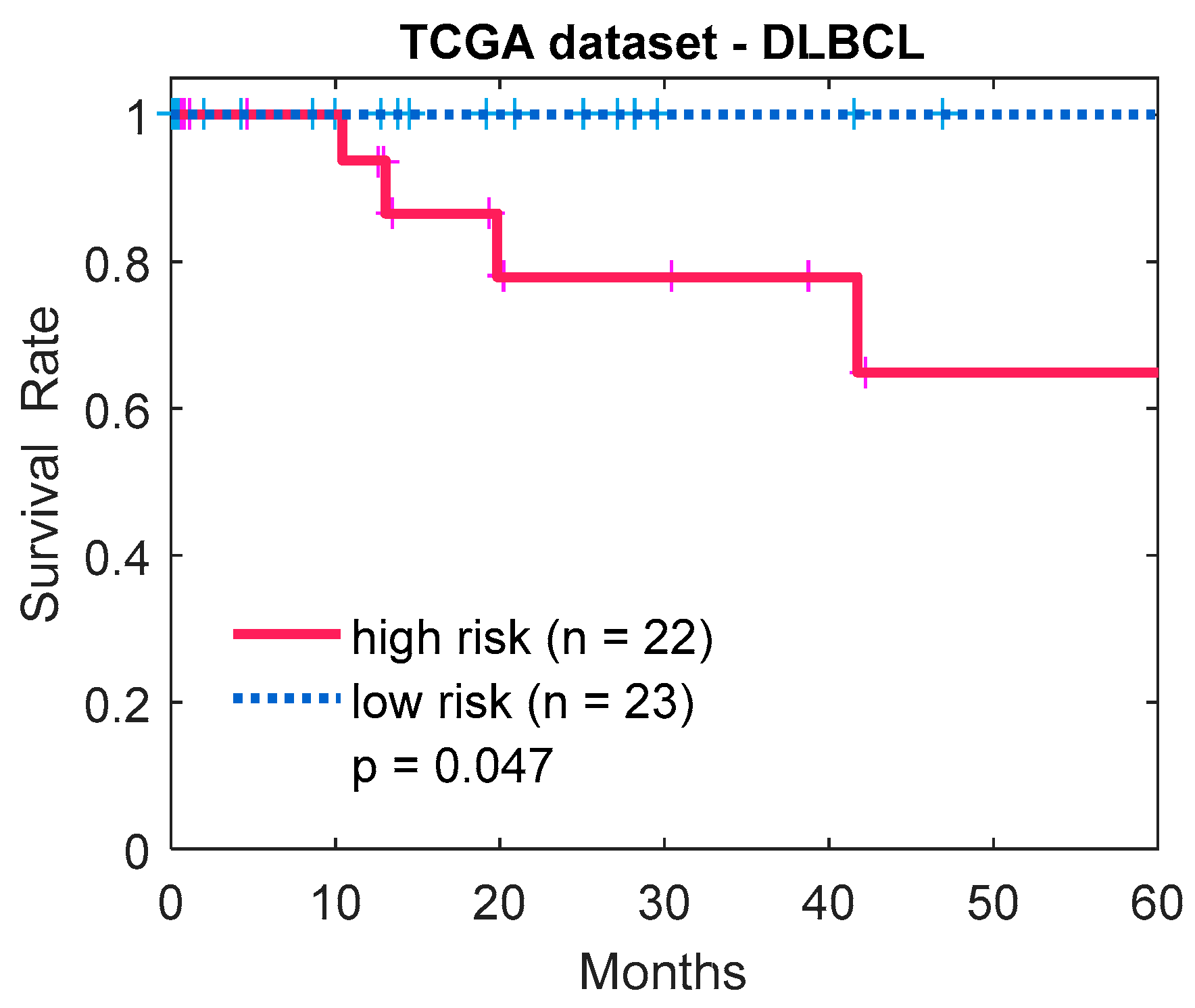
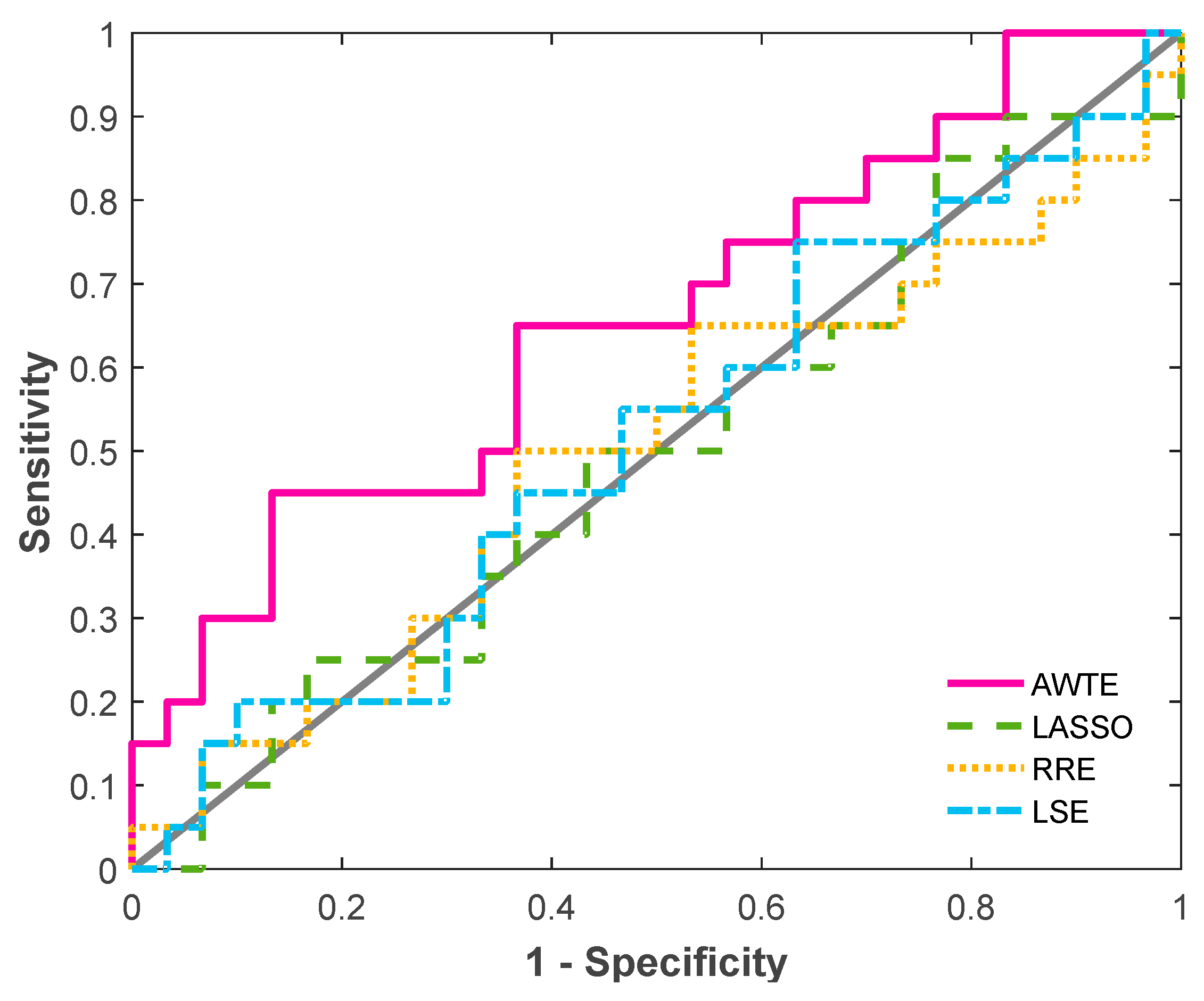
3.3. Method Comparisons Using an Actual Lymphoma Dataset
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
- (D1) The intrinsic noise of yj (i.e., εy1j, εy2j, …, εynj), the intrinsic noise of xk (i.e., εx1k, εx2k, …, εxnk), the extrinsic noise of yj (i.e., vy1j, vy2j, …, vynj), and the extrinsic noise of xk (i.e., vx1k, vx2k, …, vxnk), in regression model (7), for all positive integers j and k, are all independently and identically sampled random variables. Furthermore, their fourth moments are finite; that is,where gyj and gxk are unknown probability density functions of the intrinsic noises of yj and xk, respectively, and hyj and hxk are unknown probability density functions of the extrinsic noises of yj and xk, respectively.
- (D2) The expressions of unique component for predictor gene Xl (i.e., u1l, u2l, …, unl for l < p) and those of common interaction component (i.e., x1p, x2p, …, xnp) in Equation (6) are all independently and identically sampled random variables. Furthermore, their fourth moments are finite; that is,where ful and fxp are unknown probability density functions of uil and xip, respectively.
- (D3) The observed gene expressions x1p, x2p, …, xnp and the expressions of unique component u1l, u2l, …, unl for predictor gene Xl in Equation (6), for all positive integers l < p, are independent of intrinsic and extrinsic noises.
- (D4) The expressions of unique component u1t, u2t, …, unt for predictor gene Xt are independent of those of all the other unique component u1l, u2l, …, unl and common interaction component x1p, x2p, …, xnp in Equation (6), for all t not equal to l.
- (D5), for all real number c except for the case of c = 1 and k = p.
Appendix B

| p21 | MYC | p53 | BCL2 | NFKB1 | IRF4 | Blimp1 | AID | p27 | ATR | |
|---|---|---|---|---|---|---|---|---|---|---|
| BCL6 | 0.059 | −0.267 | 0.158 | −0.686 | 0.108 | −0.667 | −0.022 | 0.053 | −0.263 | −0.049 |
| SPIB | −0.110 | 0.235 | −0.236 | 0.093 | 0.018 | 0.677 | 0.026 | 0.650 * | −0.042 | −0.122 |
| BACH2 | −0.108 | −0.052 | 0.048 | 0.168 | −0.303 | −0.255 | −0.296 | −0.563 * | 0.132 | 0.158 |
| IRF8 | −0.234 | −0.233 | −0.050 | 0.101 | −0.249 | −0.329 * | −0.189 | −0.226 | 0.003 | 0.068 |
| OCT2 | −0.134 | 0.055 | 0.001 | 0.361 | −0.113 | 0.554 | 0.163 | 0.532 | −0.017 | −0.005 |
| p21 | MYC | p53 | BCL2 | NFKB1 | IRF4 | Blimp1 | AID | p27 | ATR | |
|---|---|---|---|---|---|---|---|---|---|---|
| BCL6 | 0.733 | 0.227 | 0.242 | 1.0 × 10−6 | 0.261 | 1.4 × 10−4 | 0.820 | 0.736 | 0.003 | 0.521 |
| SPIB | 0.467 | 0.222 | 0.047 | 0.427 | 0.828 | 1.2 × 10−5 | 0.759 | 6.0 × 10−6 | 0.575 | 0.069 |
| BACH2 | 0.355 | 0.723 | 0.595 | 0.062 | 5.9 × 10−6 | 0.027 | 8.9 × 10−6 | 4.4 × 10−7 | 0.024 | 0.002 |
| IRF8 | 0.096 | 0.192 | 0.644 | 0.351 | 0.002 | 0.018 | 0.015 | 0.078 | 0.971 | 0.268 |
| OCT2 | 0.424 | 0.795 | 0.997 | 0.006 | 0.224 | 0.001 | 0.079 | 6.7 × 10−4 | 0.834 | 0.944 |
| p21 | MYC | p53 | BCL2 | NFKB1 | IRF4 | Blimp1 | AID | p27 | ATR | |
|---|---|---|---|---|---|---|---|---|---|---|
| BCL6 | 0.041 | −0.181 | 0.060 | −0.586 | 0.099 | −0.372 | −0.017 | 0.299 | −0.286 | −0.048 |
| SPIB | 0.221 | 0.711 | −0.311 | 0.039 | −0.116 | 0.509 | −0.107 | 0.615 | 0.211 | 0.099 |
| BACH2 | −0.171 | −0.040 | 0.055 | 0.113 | −0.245 | −0.078 | −0.298 | −0.358 | 0.116 | 0.134 |
| IRF8 | −0.079 | −0.364 | 0.057 | 0.163 | −0.094 | −0.371 | −0.045 | −0.225 | 0.002 | 0.047 |
| OCT2 | −0.443 | −0.689 | 0.323 | 0.259 | 0.025 | 0.205 | 0.250 | 0.119 | −0.260 | −0.206 |
| p21 | MYC | p53 | BCL2 | NFKB1 | IRF4 | Blimp1 | AID | p27 | ATR | |
|---|---|---|---|---|---|---|---|---|---|---|
| BCL6 | 0.029 | −0.170 | 0.051 | −0.536 | 0.071 | −0.321 | −0.018 | 0.285 | −0.267 | −0.039 |
| SPIB | 0.172 | 0.646 | −0.248 | 0.034 | −0.077 | 0.431 | −0.076 | 0.581 | 0.186 | 0.069 |
| BACH2 | −0.187 | −0.072 | 0.081 | 0.110 | −0.207 | −0.104 | −0.266 | −0.359 | 0.100 | 0.108 |
| IRF8 | −0.064 | −0.330 | 0.036 | 0.152 | −0.094 | −0.310 | −0.053 | −0.199 | 0.008 | 0.049 |
| OCT2 | −0.390 | −0.654 | 0.282 | 0.243 | 0.006 | 0.194 | 0.218 | 0.120 | −0.245 | −0.174 |
| p21 | MYC | p53 | BCL2 | NFKB1 | IRF4 | Blimp1 | AID | p27 | ATR | |
|---|---|---|---|---|---|---|---|---|---|---|
| BCL6 | 0.022 | −0.122 | 0.039 | −0.514 | 0.067 | −0.307 | −0.010 | 0.228 | −0.236 | −0.032 |
| SPIB | 0.156 | 0.541 | −0.242 | 0.024 | −0.079 | 0.440 | −0.074 | 0.544 | 0.157 | 0.073 |
| BACH2 | −0.166 | −0.070 | 0.062 | 0.094 | −0.208 | −0.073 | −0.254 | −0.316 | 0.083 | 0.109 |
| IRF8 | −0.047 | −0.266 | 0.035 | 0.137 | −0.077 | −0.306 | −0.035 | −0.155 | 0.001 | 0.038 |
| OCT2 | −0.381 | −0.600 | 0.281 | 0.236 | 0.014 | 0.163 | 0.197 | 0.084 | −0.218 | −0.176 |
| GAN Method Comparison * | Close Form for Estimating Equation | Collinear Impact Adjustment | Noise/Heterogeneity Tolerating | Large Scale Predictor Gene Selection |
|---|---|---|---|---|
| LSE | Yes | No | No | No |
| RRE | Yes | Yes | No | No |
| LASSO | No | Yes | No | Yes |
| AWTE | Yes | Yes | Yes | No |
References
- Alon, U. Biological networks: The tinkerer as an engineer. Science 2003, 301, 1866–1867. [Google Scholar] [CrossRef] [PubMed]
- Prill, R.J.; Marbach, D.; Saez-Rodriguez, J.; Sorger, P.K.; Alexopoulos, L.G.; Xue, X.; Clarke, N.D.; Altan-Bonnet, G.; Stolovitzky, G. Towards a rigorous assessment of systems biology models: The DREAM3 challenges. PLoS ONE 2010, 5, e9202. [Google Scholar] [CrossRef]
- Barabási, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Chiang, A.W.T.; Liu, W.C.; Charusanti, P.; Hwang, M.J. Understanding system dynamics of an adaptive enzyme network from globally profiled kinetic parameters. BMC Syst. Biol. 2014, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, J.; Strimmer, K. An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics 2005, 21, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Song, T.; Yuan, C. Inference of gene regulatory networks from genetic perturbations with linear regression model. PLoS ONE 2013, 8, e83263. [Google Scholar] [CrossRef]
- Huang, X.; Zi, Z. Inferring cellular regulatory networks with Bayesian model averaging for linear regression (BMALR). Mol. Biosyst. 2014, 10, 2023–2030. [Google Scholar] [CrossRef]
- Bansal, M.; Belcastro, V.; Ambesi-Impiombato, A.; di Bernardo, D. How to infer gene networks from expression profiles. Mol. Syst. Biol. 2007, 3, 78. [Google Scholar] [CrossRef]
- Althubaiti, A.; Donev, A. Non-Gaussian Berkson errors in bioassay. Stat. Methods Med. Res. 2016, 25, 430–445. [Google Scholar] [CrossRef]
- Emmert-Streib, F.; Altay, G. Local network-based measures to assess the inferability of different regulatory networks. IET Syst. Biol. 2010, 4, 277–288. [Google Scholar] [CrossRef]
- Pedraza, J.M.; van Oudenaarden, A. Noise propagation in gene networks. Science 2005, 307, 1965–1969. [Google Scholar] [CrossRef] [PubMed]
- Swain, P.S.; Elowitz, M.B.; Siggia, E.D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl. Acad. Sci. USA 2002, 99, 12795–12800. [Google Scholar] [CrossRef] [PubMed]
- Thattai, M.; van Oudenaarden, A. Intrinsic noise in gene regulatory networks. Proc. Natl. Acad. Sci. USA 2001, 98, 8614–8619. [Google Scholar] [CrossRef] [PubMed]
- Katzav, A.; Kivity, S.; Blank, M.; Shoenfeld, Y.; Chapman, J. Adjuvant immunization induces high levels of pathogenic antiphospholipid antibodies in genetically prone mice: Another facet of the ASIA syndrome. Lupus 2012, 21, 210–216. [Google Scholar] [CrossRef]
- Hilfinger, A.; Paulsson, J. Separating intrinsic from extrinsic fluctuations in dynamic biological systems. Proc. Natl. Acad. Sci. USA 2011, 108, 12167–12172. [Google Scholar] [CrossRef]
- Wei, G.; Wang, Z.; Lam, J.; Fraser, K.; Rao, G.P.; Liu, X. Robust filtering for stochastic genetic regulatory networks with time-varying delay. Math. Biosci. 2009, 220, 73–80. [Google Scholar] [CrossRef]
- Wu, Y.J.; Fang, W.Q. Consistent estimation approach to tackling collinearity and Berkson-type measurement error in linear regression using adjusted Wald-type estimator. Commun. Stat. Theory Methods 2017, 46, 5501–5516. [Google Scholar] [CrossRef]
- Batt, G.; Yordanov, B.; Weiss, R.; Belta, C. Robustness analysis and tuning of synthetic gene networks. Bioinformatics 2007, 23, 2415–2422. [Google Scholar] [CrossRef]
- Chiang, A.W.T.; Hwang, M.J. A computational pipeline for identifying kinetic motifs to aid in the design and improvement of synthetic gene circuits. BMC Bioinform. 2013, 14 (Suppl. 16), S5. [Google Scholar] [CrossRef]
- Chen, B.S.; Wu, W.S.; Wang, Y.C.; Li, W.H. On the robust circuit design schemes of biochemical networks: Steady-state approach. IEEE Trans. Biomed. Circuits Syst. 2007, 1, 91–104. [Google Scholar] [CrossRef]
- Chkrebtii, O.A.; Campbell, D.A.; Calderhead, B.; Girolami, M.A. Bayesian solution uncertainty quantification for differential equations. Bayesian Anal. 2016, 11, 1239–1267. [Google Scholar] [CrossRef]
- Cai, L.; Li, Q.; Du, Y.; Yun, J.; Xie, Y.; DeBerardinis, R.J.; Xiao, G. Genomic regression analysis of coordinated expression. Nat. Commun. 2017, 8, 2187. [Google Scholar] [CrossRef] [PubMed]
- Fujita, A.; Patriota, A.G.; Sato, J.R.; Miyano, S. The impact of measurement errors in the identification of regulatory networks. BMC Bioinform. 2009, 10, 412. [Google Scholar] [CrossRef]
- Göbl, C.S.; Bozkurt, L.; Tura, A.; Pacini, G.; Kautzky-Willer, A.; Mittlböck, M. Application of penalized regression techniques in modelling insulin sensitivity by correlated metabolic parameters. PLoS ONE 2015, 10, e0141524. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Z.; Wu, F.X.; Zhang, W.J. A group LASSO-based method for robustly inferring gene regulatory networks from multiple time-course datasets. BMC Syst. Biol. 2014, 8, S1. [Google Scholar] [CrossRef] [PubMed]
- Rencher, A.C.; Schaalje, G.B. Linear Models in Statistics; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Tibshirani, R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. Ser. B Stat. Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Hoerl, A.E.; Kennard, R.W. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics 1970, 12, 55–67. [Google Scholar] [CrossRef]
- Neto, E.C.; Bare, J.C.; Margolin, A.A. Simulation studies as designed experiments: The comparison of penalized regression models in the “large p, small n” setting. PLoS ONE 2014, 9, e107957. [Google Scholar]
- Wald, A. Fitting of straight lines if both variables are subject to error. Ann. Math. Stat. 1940, 11, 284–300. [Google Scholar] [CrossRef]
- Wansbeek, T.; Meijer, E. Measurement Error and Latent Variables in Econometrics; North-Holland: Amsterdam, The Netherlands, 2000. [Google Scholar]
- Pakes, A. On the asymptotic bias of the Wald-type estimators of a straight line when both variables are subject to error. Int. Econ. Rev. 1982, 23, 491–497. [Google Scholar] [CrossRef]
- Theil, H.; van Yzeren, J. On the efficiency of Wald’s method of fitting straight lines. Rev. Inst. Int. Stat. 1956, 24, 17–26. [Google Scholar] [CrossRef]
- Zidek, J.V.; Wong, H.; Le, N.D.; Burnett, R. Causality, measurement error and multicollinearity in epidemiology. Environmentrics 1996, 7, 441–451. [Google Scholar] [CrossRef]
- Hedjazi, L.; Le Lann, M.V.; Kempowsky, T.; Dalenc, F.; Aguilar-Martin, J.; Favre, G. Symbolic data analysis to defy low signal-to-noise ratio in microarray data for breast cancer prognosis. J. Comput. Biol. 2013, 20, 610–620. [Google Scholar] [CrossRef]
- Lo, K.; Raftery, A.E.; Dombek, K.M.; Zhu, J.; Schadt, E.E.; Bumgarner, R.E.; Yeung, K.Y. Integrating external biological knowledge in the construction of regulatory networks from time-series expression data. BMC Syst. Biol. 2012, 6, 1. [Google Scholar] [CrossRef]
- Singh, A. Transient changes in intercellular protein variability identify sources of noise in gene expression. Biophys. J. 2014, 107, 2214–2220. [Google Scholar] [CrossRef] [PubMed]
- Sima, C.; Hua, J.; Jung, S. Inference of gene regulatory networks using time-series data: A survey. Curr. Genom. 2009, 10, 416–429. [Google Scholar] [CrossRef]
- Kibria, B.M.G.; Månsson, K.; Shukur, G. A simulation study of some biasing parameters for the ridge type estimation of Poisson regression. Commun. Stat. Simul. Comput. 2015, 44, 943–957. [Google Scholar] [CrossRef]
- Logsdon, B.A.; Mezey, J. Gene expression network reconstruction by convex feature selection when incorporating genetic perturbations. PLoS Comput. Biol. 2010, 6, e1001014. [Google Scholar] [CrossRef]
- Lenz, G.; Staudt, L.M. Aggressive lymphomas. N. Engl. J. Med. 2010, 362, 1417–1429. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef]
- Gene Expression Omnibus. Available online: http://www.ncbi.nlm.nih.gov/geo/ (accessed on 25 May 2023).
- Yamane, A.; Resch, W.; Kuo, N.; Kuchen, S.; Li, Z.; Sun, H.-W.; Robbiani, D.F.; McBride, K.; Nussenzweig, M.C.; Casellas, R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat. Immunol. 2011, 12, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Tarlinton, D.M. Bcl6: Where too much complexity is barely enough. Eur. J. Immunol. 2011, 41, 2148–2151. [Google Scholar] [CrossRef] [PubMed]
- Carotta, S.; Willis, S.N.; Hasbold, J.; Inouye, M.; Pang, S.H.M.; Emslie, D.; Light, A.; Chopin, M.; Shi, W.; Wang, H.; et al. The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. J. Exp. Med. 2014, 211, 2169–2181. [Google Scholar] [CrossRef] [PubMed]
- Green, M.; Gandhi, M.K.; Camilleri, E.; Marlton, P.; Lea, R.; Griffiths, L. High levels of BACH2 associated with lower levels of BCL2 transcript abundance in t(14; 18)(q21; q34) translocation positive non-Hodgkin’s lymphoma. Leuk. Res. 2009, 33, 731–734. [Google Scholar] [CrossRef]
- Chen, B.S.; Lin, Y.P. A unifying mathematical framework for genetic robustness, environmental robustness, network robustness and their trade-offs on phenotype robustness in biological networks. part III: Synthetic gene networks in synthetic biology. Evol. Bioinform. 2013, 9, 87–109. [Google Scholar] [CrossRef]
- Zhang, P.W.; Chen, L.; Huang, T.; Zhang, N.; Kong, X.-Y.; Cai, Y.-D. Classifying ten types of major cancers based on reverse phase protein array profiles. PLoS ONE 2015, 10, e0123147. [Google Scholar] [CrossRef]
- Necela, B.M.; Crozier, J.A.; Andorfer, C.A.; Lewis-Tuffin, L.; Kachergus, J.M.; Geiger, X.J.; Kalari, K.; Serie, D.J.; Sun, Z.; Moreno-Aspitia, A.; et al. Folate receptor-α (FOLR1) expression and function in triple negative tumors. PLoS ONE 2015, 10, e0122209. [Google Scholar]
- UK Biobank. Available online: https://www.ukbiobank.ac.uk/ (accessed on 25 May 2023).
- Fujita, A.; Sato, J.R.; Garay-Malpartida, H.M.; Yamaguchi, R.; Miyano, S.; Sogayar, M.C.; Ferreira, C.E. Modeling gene expression regulatory networks with the sparse vector autoregressive model. BMC Syst. Biol. 2007, 1, 39. [Google Scholar] [CrossRef]
- Park, R.E.; Mitchell, B.M. Estimating the autocorrelated error model with trended data. J. Econom. 1980, 13, 185–201. [Google Scholar] [CrossRef]
- Bierens, H.J. Introduction to the Mathematical and Statistical Foundations of Econometrics; Cambridge University Press: New York, NY, USA, 2004. [Google Scholar]
- Care, M.A.; Cocco, M.; Laye, J.P.; Barnes, N.; Huang, Y.; Wang, M.; Barrans, S.; Du, M.; Jack, A.; Westhead, D.; et al. SPIB and BATF provide alternate determinants of IRF4 occupancy in diffuse large B-cell lymphoma linked to disease heterogeneity. Nucleic Acids Res. 2014, 42, 7591–7610. [Google Scholar] [CrossRef]
- Hodson, D.J.; Shaffer, A.L.; Xiao, W.; Wright, G.W.; Schmitz, R.; Phelan, J.D.; Yang, Y.; Webster, D.E.; Rui, L.; Kohlhammer, H.; et al. Regulation of normal B-cell differentiation and malignant B-cell survival by OCT2. Proc. Natl. Acad. Sci. USA 2016, 113, E2039–E2046. [Google Scholar] [CrossRef] [PubMed]
- Heckman, C.A.; Duan, H.; Garcia, P.B.; Boxer, L.M. Oct transcription factors mediate t(14; 18) lymphoma cell survival by directly regulating bcl-2 expression. Oncogene 2006, 25, 888–898. [Google Scholar] [CrossRef] [PubMed]
- Takatsu, K.; Nakajima, H. IL-5 and eosinophilia. Curr. Opin. Immunol. 2008, 20, 288–294. [Google Scholar] [CrossRef]
- Park, S.R.; Zan, H.; Pal, Z.; Zhang, J.; Al-Qahtani, A.; Pone, E.J.; Xu, Z.; Mai, T.; Casali, P. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA re-combination and somatic hypermutation. Nat. Immunol. 2009, 10, 540–550. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, W.-Q.; Wu, Y.-L.; Hwang, M.-J. A Noise-Tolerating Gene Association Network Uncovering an Oncogenic Regulatory Motif in Lymphoma Transcriptomics. Life 2023, 13, 1331. https://doi.org/10.3390/life13061331
Fang W-Q, Wu Y-L, Hwang M-J. A Noise-Tolerating Gene Association Network Uncovering an Oncogenic Regulatory Motif in Lymphoma Transcriptomics. Life. 2023; 13(6):1331. https://doi.org/10.3390/life13061331
Chicago/Turabian StyleFang, Wei-Quan, Yu-Le Wu, and Ming-Jing Hwang. 2023. "A Noise-Tolerating Gene Association Network Uncovering an Oncogenic Regulatory Motif in Lymphoma Transcriptomics" Life 13, no. 6: 1331. https://doi.org/10.3390/life13061331
APA StyleFang, W.-Q., Wu, Y.-L., & Hwang, M.-J. (2023). A Noise-Tolerating Gene Association Network Uncovering an Oncogenic Regulatory Motif in Lymphoma Transcriptomics. Life, 13(6), 1331. https://doi.org/10.3390/life13061331






