Emergence of Rice Blast AVR-Pi9 Resistance Breaking Haplotypes in Yunnan Province, China
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Distribution of M. oryzae Isolates in Yunnan Province, China
3.2. Identification of AVR-Pi9 Haplotypes
3.3. Genetic Variation of the AVR-Pi9 Gene in Non-Rice Hosts
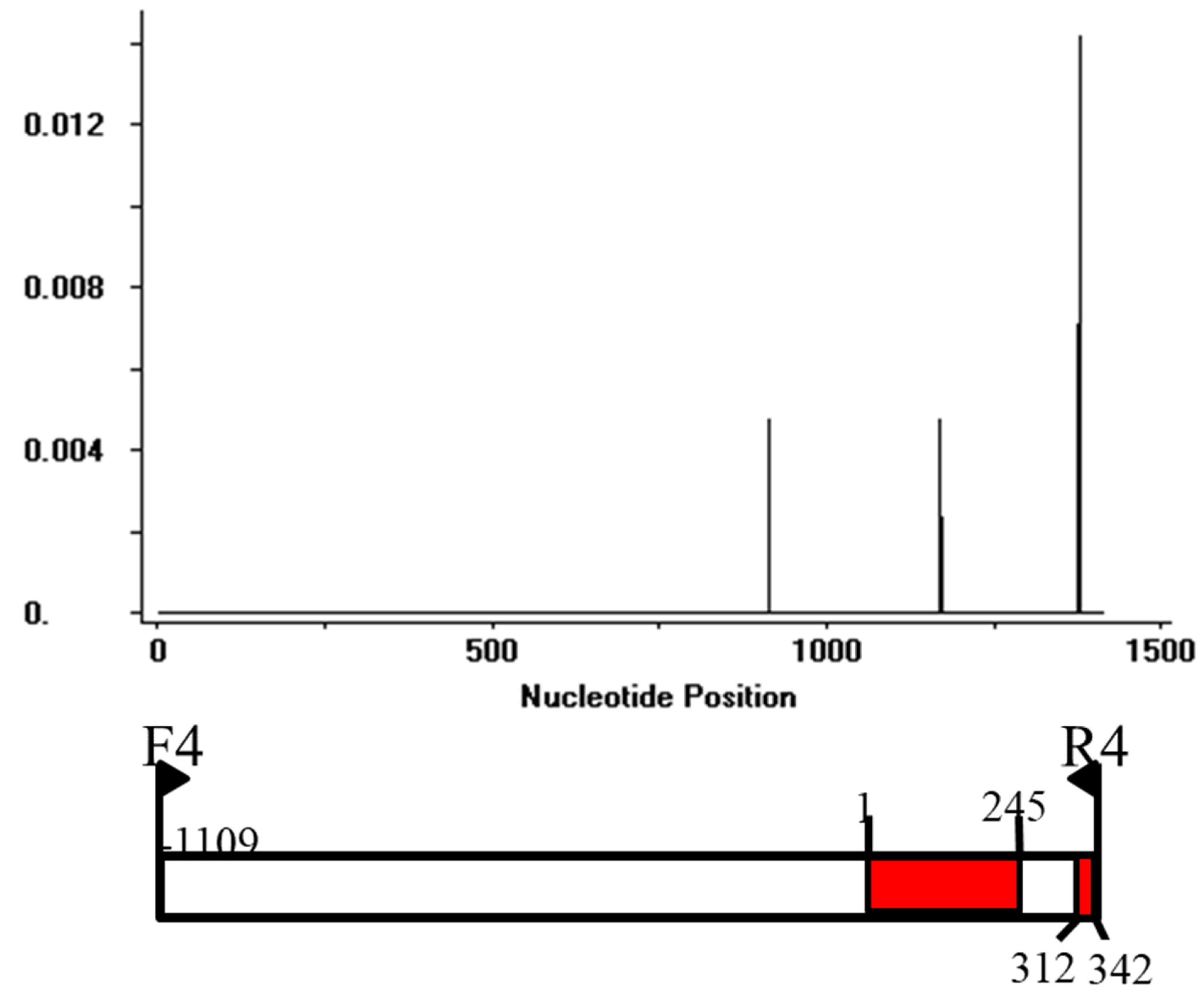
3.4. Selection Pressure of AVR-Pi9 in M. oryzae
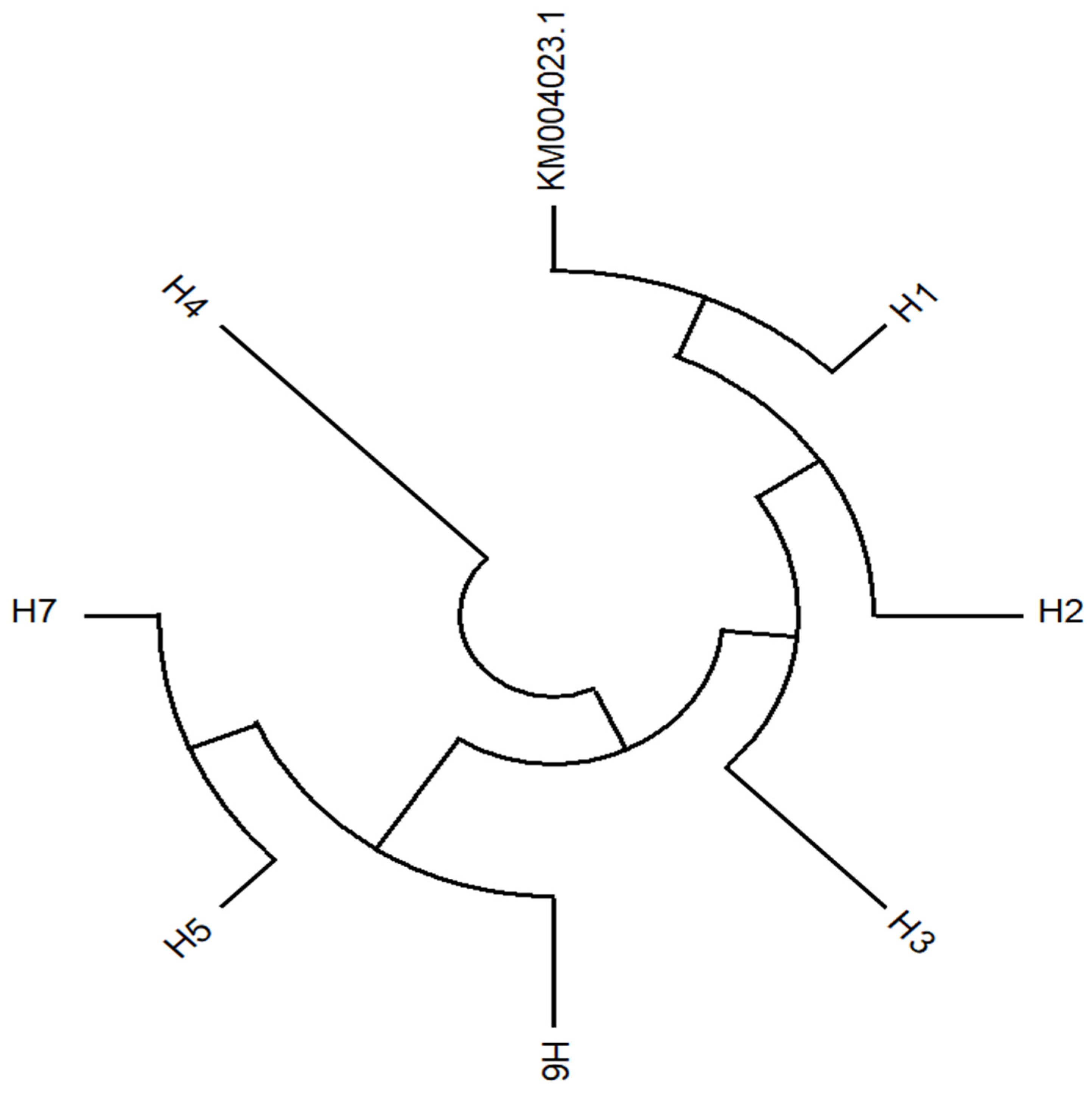
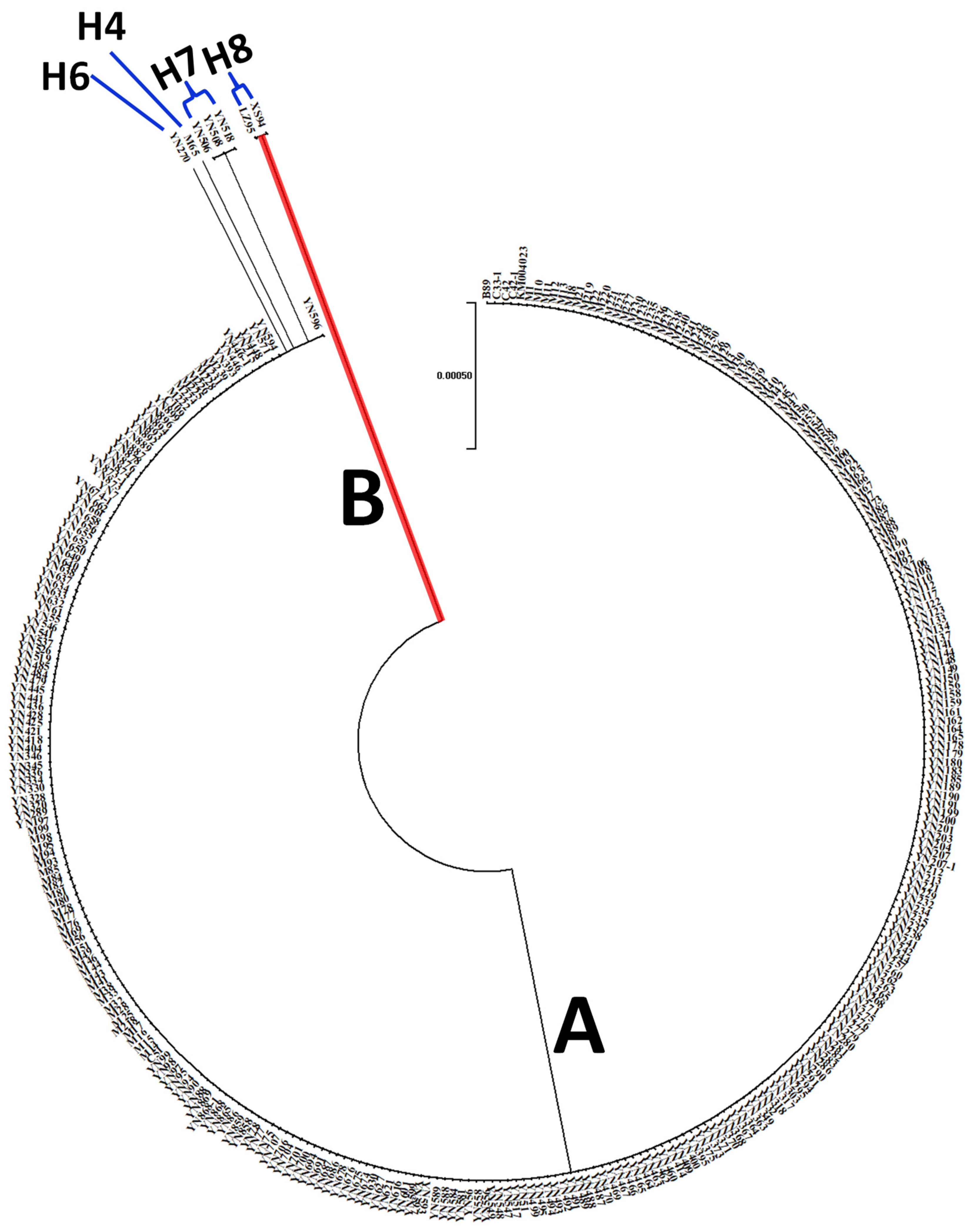
3.5. The Phylogenetic Relationship of AVR-Pi9 Haplotypes
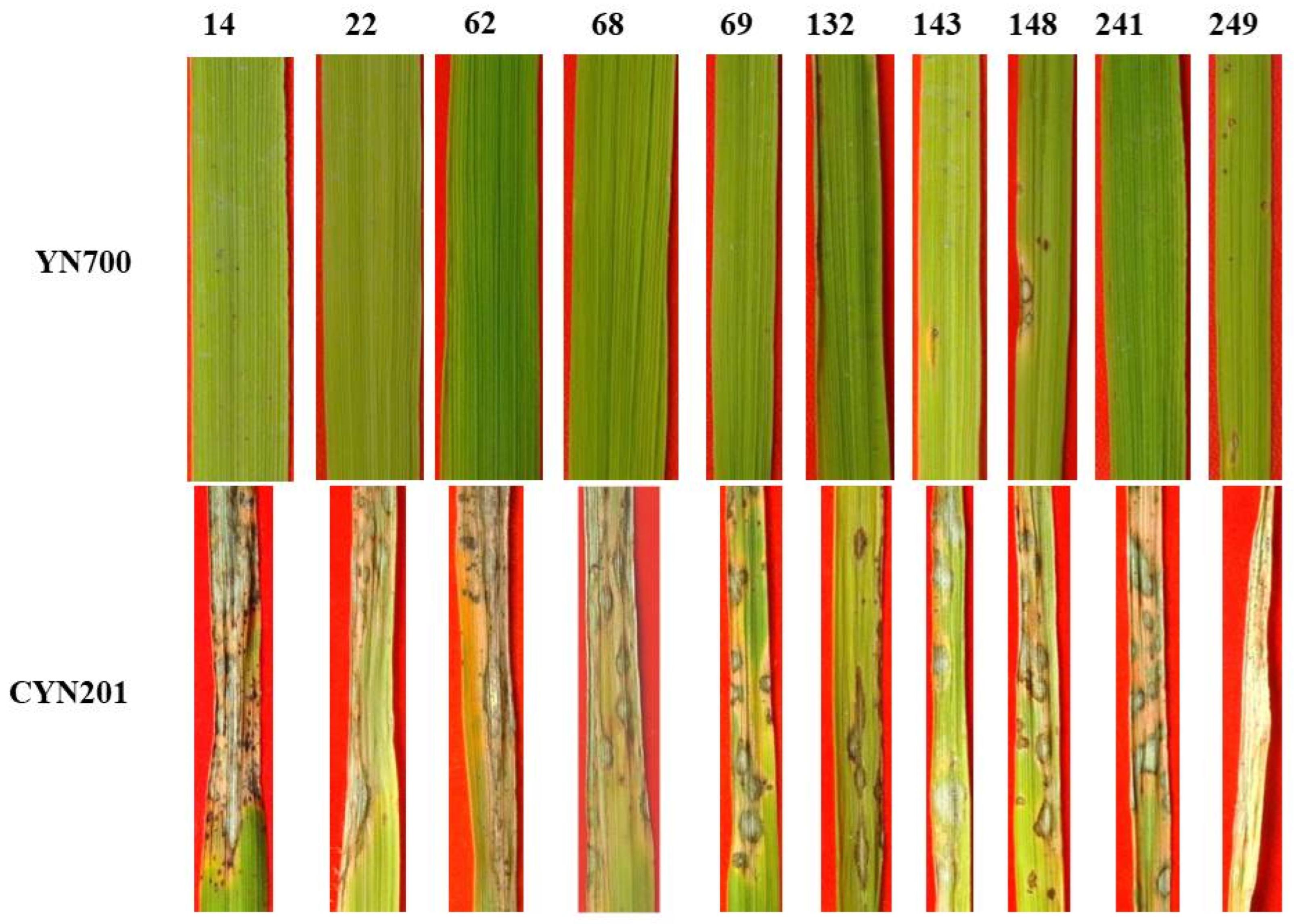
3.6. Pathogenicity Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coelho, M.A.D.O.; Torres, G.A.M.; Cecon, P.R.; Santana, F.M. Sowing Date Reduces the Incidence of Wheat Blast Disease. Pesqui. Agropecu. Bras. 2016, 51, 631–637. [Google Scholar] [CrossRef]
- Farman, M.L. Pyricularia Grisea Isolates Causing Gray Leaf Spot on Perennial Ryegrass (Lolium Perenne) in the United States: Relationship to P. Grisea Isolates from Other Host Plants. Phytopathology 2002, 92, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Skamnioti, P.; Gurr, S.J. Against the Grain: Safeguarding Rice from Rice Blast Disease. Trends Biotechnol. 2009, 27, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Heath, M.C.; Valent, B.; Howard, R.J.; Chumley, F.G. Interactions of Two Strains of Magnaporthe Grisea with Rice, Goosegrass, and Weeping Lovegrass. Can. J. Bot. 1990, 68, 1627–1637. [Google Scholar] [CrossRef]
- Singh, J.; Gupta, S.K.; Devanna, B.N.; Singh, S.; Upadhyay, A.; Sharma, T.R. Blast Resistance Gene Pi54 Over-Expressed in Rice to Understand Its Cellular and Sub-Cellular Localization and Response to Different Pathogens. Sci. Rep. 2020, 10, 5243. [Google Scholar] [CrossRef]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed Cell Death in the Plant Immune System. Cell Death Differ. 2011, 18, 1247–1256. [Google Scholar] [CrossRef]
- Xing, J.; Jia, Y.; Correll, J.C.; Lee, F.N.; Cartwright, R.; Cao, M.; Yuan, L. Analysis of Genetic and Molecular Identity among Field Isolates of the Rice Blast Fungus with an International Differential System, Rep-PCR, and DNA Sequencing. Plant Dis. 2013, 97, 491–495. [Google Scholar] [CrossRef]
- Flor, H.H. Current Status of the Gene-for-Gene Concept. Annu. Rev. Phytopathol. 1971, 9, 275. [Google Scholar] [CrossRef]
- Dodds, P.N.; Lawrence, G.J.; Catanzariti, A.-M.; Teh, T.; Wang, C.-I.; Ayliffe, M.A.; Kobe, B.; Ellis, J.G. Direct Protein Interaction Underlies Gene-for-Gene Specificity and Coevolution of the Flax Resistance Genes and Flax Rust Avirulence Genes. Proc. Natl. Acad. Sci. USA 2006, 103, 8888–8893. [Google Scholar] [CrossRef]
- Telebanco-Yanoria, M.J.; Koide, Y.; Fukuta, Y.; Imbe, T.; Tsunematsu, H.; Kato, H.; Ebron, L.A.; Nguyen, T.M.N.; Kobayashi, N. A Set of Near-Isogenic Lines of Indica-Type Rice Variety CO 39 as Differential Varieties for Blast Resistance. Mol. Breed. 2011, 27, 357–373. [Google Scholar] [CrossRef]
- Fang, W.W.; Liu, C.C.; Zhang, H.W.; Xu, H.; Zhou, S.; Fang, K.X.; Peng, Y.L.; Zhao, W.S. Selection of Differential Isolates of Magnaporthe Oryzae for Postulation of Blast Resistance Genes. Phytopathology 2018, 108, 878–884. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, M.; Sha, G.; Sun, Q.; Gong, Q.; Yang, Q.; Xie, K.; Yuan, M.; Mortimer, J.C.; Xie, W. The Genome of the Rice Variety LTH Provides Insight into Its Universal Susceptibility Mechanism to Worldwide Rice Blast Fungal Strains. Comput. Struct. Biotechnol. J. 2022, 20, 1012–1026. [Google Scholar] [CrossRef]
- Wang, B.; Ebbole, D.J.; Wang, Z. The Arms Race between Magnaporthe Oryzae and Rice: Diversity and Interaction of Avr and R Genes. J. Integr. Agric. 2017, 16, 2746–2760. [Google Scholar] [CrossRef]
- Jia, Y.; McAdams, S.A.; Bryan, G.T.; Hershey, H.P.; Valent, B. Direct Interaction of Resistance Gene and Avirulence Gene Products Confers Rice Blast Resistance. EMBO J. 2000, 19, 4004–4014. [Google Scholar] [CrossRef]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y. The Rice Resistance Protein Pair RGA4/RGA5 Recognizes the Magnaporthe Oryzae Effectors AVR-Pia and AVR1-CO39 by Direct Binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Correll, J.C.; Jia, Y. Characterization of Rice Blast Resistance Genes in Rice Germplasm with Monogenic Lines and Pathogenicity Assays. Crop Prot. 2015, 72, 132–138. [Google Scholar] [CrossRef]
- Liu, G.; Lu, G.; Zeng, L.; Wang, G.-L. Two Broad-Spectrum Blast Resistance Genes, Pi9 (t) and Pi2 (t), Are Physically Linked on Rice Chromosome 6. Mol. Genet. Genom. 2002, 267, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.; Liu, G.; Zhou, B. The Broad-Spectrum Blast Resistance Gene Pi9 Encodes a Nucleotide-Binding Site Leucine-Rich Repeat Protein and Isa Member of a Multigene Family in Rice. Genetics 2006, 172, 1901–1914. [Google Scholar]
- Wu, J.; Kou, Y.; Bao, J.; Li, Y.; Tang, M.; Zhu, X.; Ponaya, A.; Xiao, G.; Li, J.; Li, C. Comparative Genomics Identifies the Magnaporthe Oryzae Avirulence Effector AvrPi9 That Triggers Pi9-Mediated Blast Resistance in Rice. New Phytol. 2015, 206, 1463–1475. [Google Scholar] [CrossRef]
- Sutthiphai, T.; Damchuay, K.; Neupane, R.C.; Longya, A.; Sriwongchai, T.; Songkumarn, P.; Parinthawong, N.; Darwell, K.; Jantasuriyarat, C. Genetic Variation of Avirulence Genes (AVR-Pi9, AVR-Pik, AVR-Pita1) and Genetic Diversity of Rice Blast Fungus, Pyricularia Oryzae, in Thailand. Plant Pathol. 2022, 71, 322–333. [Google Scholar] [CrossRef]
- Perfect, S.E.; Green, J.R. Infection Structures of Biotrophic and Hemibiotrophic Fungal Plant Pathogens. Mol. Plant Pathol. 2001, 2, 101–108. [Google Scholar] [CrossRef]
- Zhang, Q. Strategies for Developing Green Super Rice. Proc. Natl. Acad. Sci. USA 2007, 104, 16402–16409. [Google Scholar] [CrossRef]
- Scheuermann, K.K.; Jia, Y. Identification of a Pi9-Containing Rice Germplasm with a Newly Developed Robust Marker. Phytopathology 2016, 106, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Berruyer, R.; Adreit, H.; Milazzo, J.; Gaillard, S.; Berger, A.; Dioh, W.; Lebrun, M.-H.; Tharreau, D. Identification and Fine Mapping of Pi33, the Rice Resistance Gene Corresponding to the Magnaporthe Grisea Avirulence Gene ACE1. Theor. Appl. Genet. 2003, 107, 1139–1147. [Google Scholar] [CrossRef] [PubMed]
- Sirisathaworn, T.; Srirat, T.; Longya, A.; Jantasuriyarat, C. Evaluation of Mating Type Distribution and Genetic Diversity of Three Magnaporthe Oryzae Avirulence Genes, PWL-2, AVR-Pii and Avr-Piz-t, in Thailand Rice Blast Isolates. Agric. Nat. Resour. 2017, 51, 7–14. [Google Scholar] [CrossRef]
- Belyavskaya, L.G.; Belyavsky, Y.V. Interaction of Modern Soybean Varieties with Biological Preparations of Complex Action and Their Impact on the Yield. Mikrobiolohichnyi Zhurnal 2016, 78, 61–68. [Google Scholar] [CrossRef]
- Tai, T.H.; Tanksley, S.D. A Rapid and Inexpensive Method for Isolation of Total DNA from Dehydrated Plant Tissue. Plant Mol. Biol. Rep. 1990, 8, 297–303. [Google Scholar] [CrossRef]
- Williams, S.T.; Goodfellow, M.; Alderson, G.; Wellington, E.M.H.; Sneath, P.H.A.; Sackin, M.J. Numerical Classification of Streptomyces and Related Genera. Microbiology 1983, 129, 1743–1813. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Roff, D.A.; Bentzen, P. The Statistical Analysis of Mitochondrial DNA Polymorphisms: Chi 2 and the Problem of Small Samples. Mol. Biol. Evol. 1989, 6, 539–545. [Google Scholar]
- Rozas, J.; Ferrer-Matta, A.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 32, 3299–3302. [Google Scholar] [CrossRef]
- Nei, M.; Miller, J.C. A Simple Method for Estimating Average Number of Nucleotide Substitutions within and between Populations from Restriction Data. Genetics 1990, 125, 873–879. [Google Scholar] [CrossRef]
- Korneliussen, T.S.; Albrechtsen, A.; Nielsen, R. ANGSD: Analysis of next Generation Sequencing Data. BMC Bioinform. 2014, 15, 356. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. The Effect of Change in Population Size on DNA Polymorphism. Genetics 1989, 123, 597–601. [Google Scholar] [CrossRef]
- Wang, J.C.; Jia, Y.; Wen, J.W.; Liu, W.P.; Liu, X.M.; Li, L.; Jiang, Z.Y.; Zhang, J.H.; Guo, X.L.; Ren, J.P. Identification of Rice Blast Resistance Genes Using International Monogenic Differentials. Crop Prot. 2013, 45, 109–116. [Google Scholar] [CrossRef]
- Li, W.; Wang, B.; Wu, J.; Lu, G.; Hu, Y.; Zhang, X.; Zhang, Z.; Zhao, Q.; Feng, Q.; Zhang, H. The Magnaporthe Oryzae Avirulence Gene AvrPiz-t Encodes a Predicted Secreted Protein That Triggers the Immunity in Rice Mediated by the Blast Resistance Gene Piz-T. Mol. Plant-Microbe Interact. 2009, 22, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Wang, L.; Pan, Q. Identification and Characterization of a New Blast Resistance Gene Located on Rice Chromosome 1 through Linkage and Differential Analyses. Phytopathology 2004, 94, 515–519. [Google Scholar] [CrossRef]
- Jia, Y.; Valent, B.; Lee, F.N. Determination of Host Responses to Magnaporthe Grisea on Detached Rice Leaves Using a Spot Inoculation Method. Plant Dis. 2003, 87, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Telebanco-Yanoria, M.J.; Koide, Y.; Fukuta, Y.; Imbe, T.; Kato, H.; Tsunematsu, H.; Kobayashi, N. Development of Near-Isogenic Lines of Japonica-Type Rice Variety Lijiangxintuanheigu as Differentials for Blast Resistance. Breed. Sci. 2010, 60, 629–638. [Google Scholar] [CrossRef]
- Tian, D.; Guo, X.; Zhang, Z.; Wang, M.; Wang, F. Improving Blast Resistance of the Rice Restorer Line, Hui 316, by Introducing Pi9 or Pi2 with Marker-Assisted Selection. Biotechnol. Biotechnol. Equip. 2019, 33, 1195–1203. [Google Scholar] [CrossRef]
- Ling, Z.; Mew, T.; Wang, J.; Lei, C.; Huang, N. Development of Chinese Near-Isogenic Lines of Rice and Their Differentiating Ability to Pathogenic Races of Pyricularia Grisea. Sci. Agric. Sin. 2000, 33, 1–8. [Google Scholar]
- Hu, Z.-J.; Huang, Y.-Y.; Lin, X.-Y.; Feng, H.; Zhou, S.-X.; Xie, Y.; Liu, X.-X.; Liu, C.; Zhao, R.-M.; Zhao, W.-S. Loss and Natural Variations of Blast Fungal Avirulence Genes Breakdown Rice Resistance Genes in the Sichuan Basin of China. Front. Plant Sci. 2022, 13, 788876. [Google Scholar] [CrossRef]
- Chuma, I.; Isobe, C.; Hotta, Y.; Ibaragi, K.; Futamata, N.; Kusaba, M.; Yoshida, K.; Terauchi, R.; Fujita, Y.; Nakayashiki, H. Multiple Translocation of the AVR-Pita Effector Gene among Chromosomes of the Rice Blast Fungus Magnaporthe Oryzae and Related Species. PLoS Pathog. 2011, 7, e1002147. [Google Scholar] [CrossRef]
- Yoshida, K.; Saunders, D.G.O.; Mitsuoka, C.; Natsume, S.; Kosugi, S.; Saitoh, H.; Inoue, Y.; Chuma, I.; Tosa, Y.; Cano, L.M. Host Specialization of the Blast Fungus Magnaporthe Oryzae Is Associated with Dynamic Gain and Loss of Genes Linked to Transposable Elements. BMC Genom. 2016, 17, 370. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, J.; Lu, L.; He, C.; Li, C. Novel Variation and Evolution of AvrPiz-t of Magnaporthe Oryzae in Field Isolates. Front. Genet. 2020, 11, 746. [Google Scholar] [CrossRef]
- Jeuken, M.J.W.; Zhang, N.W.; McHale, L.K.; Pelgrom, K.; Den Boer, E.; Lindhout, P.; Michelmore, R.W.; Visser, R.G.F.; Niks, R.E. Rin4 Causes Hybrid Necrosis and Race-Specific Resistance in an Interspecific Lettuce Hybrid. Plant Cell 2009, 21, 3368–3378. [Google Scholar] [CrossRef]
- Chen, X.; Shang, J.; Chen, D.; Lei, C.; Zou, Y.; Zhai, W.; Liu, G.; Xu, J.; Ling, Z.; Cao, G. AB-lectin Receptor Kinase Gene Conferring Rice Blast Resistance. Plant J. 2006, 46, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, S.; Saka, N.; Koga, H.; Ono, K.; Shimizu, T.; Ebana, K.; Hayashi, N.; Takahashi, A.; Hirochika, H.; Okuno, K. Loss of Function of a Proline-Containing Protein Confers Durable Disease Resistance in Rice. Science 2009, 325, 998–1001. [Google Scholar] [CrossRef] [PubMed]
| Host | Locations/Years | No. of Isolates | PCR Detection | Pathogenicity Assay a | ||
|---|---|---|---|---|---|---|
| No. of Isolates with AVR-Pi9 | Frequency (%) | No. of Avirulence Isolates | Frequency (%) | |||
| Rice | Northeastern/2012 | 44 | 44 | 100.0 | 44 | 100.0 |
| Central/2013 | 88 | 88 | 100.0 | 88 | 100.0 | |
| Southeastern/2013 | 21 | 21 | 100.0 | 21 | 100.0 | |
| Western/2013 | 127 | 127 | 100.0 | 122 | 96.1 | |
| Northwestern/2014 | 21 | 21 | 100.0 | 21 | 100.0 | |
| Southwestern/2014 | 25 | 25 | 100.0 | 25 | 100.0 | |
| Total | 326 | 326 | 100.0 | 321 | 98.5 | |
| Non-rice | Western/2014 | 1 | 1 | 100.0 | 1 | 100.0 |
| Central/2014 | 1 | 1 | 100.0 | 1 | 100.0 | |
| Total | 2 | 2 | 100.0 | 2 | 100.0 | |
| Haplotype | No. of Isolates | Frequency (%) | No. of Isolates and Frequency (%) in Each Region | Production a | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Northeastern | Central | Southeastern | Western | Northwestern | Southwestern | XI | GJ | |||
| H01 | 218 | 66.9 | 42 (95.5) | 48 (54.5) | 12 (57.1) | 95 (74.8) | 8 (38.1) | 13 (52.0) | 120 (69.4) | 98 (64.1) |
| H02 | 87 | 26.7 | 1 (2.3) | 30 (34.1) | 9 (42.9) | 25 (19.7) | 13 (61.9) | 9 (36.0) | 43 (24.9) | 44 (28.8) |
| H03 | 12 | 3.7 | 0 | 7 (8.0) | 0 | 5 (3.9) | 0 | 0 | 5 (2.9) | 7 (4.6) |
| H04 | 2 | 0.6 | 0 | 0 | 0 | 0 | 0 | 2 (8.0) | 2 (1.2) | 0 |
| H05 | 3 | 0.9 | 0 | 0 | 0 | 2 (1.6) | 0 | 1 (4.0) | 3 (1.7) | 0 |
| H06 | 1 | 0.3 | 1 (2.3) | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.7) |
| H07 | 3 | 0.9 | 0 | 3 (3.4) | 0 | 0 | 0 | 0 | 0 | 3 (2.0) |
| Total | 326 | 100 | 44 | 88 | 21 | 127 | 21 | 25 | 173 | 153 |
| No. of haplotypes | 3 | 4 | 2 | 4 | 2 | 4 | 5 | 5 | ||
| Index of diversity b | 0.088 | 0.579 | 0.490 | 0.400 | 0.472 | 0.592 | 0.456 | 0.505 | ||
| Haplotype | No. of Isolates | Frequency (%) | Variant Locus a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Promoter Region | CDS | Intron | ||||||||||
| Between 168–169 | Between 812–827 | 912 | 1139 | 1169 | 1310 | Between 1342–1343 | 1345 | 1374 | 1376 | |||
| KM004023.1 | - | CTCCTACACTGGGGCT | T | C | C | C | - | G | G | C | ||
| H1 | 218 | 66.5 | . | . | . | . | . | . | - | . | . | . |
| H2 | 87 | 26.5 | . | - | . | . | . | . | - | . | . | . |
| H3 | 12 | 3.7 | T | - | . | . | . | . | - | . | . | . |
| H4 | 1 | 0.3 | . | - | C | . | . | . | - | . | . | . |
| H5 | 4 | 1.2 | T | . | . | . | . | . | - | . | . | . |
| H6 | 1 | 0.3 | . | . | . | . | T | . | - | . | . | . |
| H7 | 3 | 0.9 | . | . | . | . | . | . | - | . | . | T |
| H8 * | 2 | 0.6 | . | . | . | T | . | T | GCCCTGTACAATGCTTTTT | T | T | . |
| 328 | 100 | |||||||||||
| Host and Isolate | ||||||||
|---|---|---|---|---|---|---|---|---|
| Rice | Rice | Perennial Ryegrass | Perennial Ryegrass | Wheat | Wheat | Setaria viridis (L.) Beauv | ||
| Query | Size (bp) | P131 | PY34 | PgKY | PGPA | PY5033 | PY6045 | US71 |
| AVR-Pi9_KM004023.1 | 3496 letters | 2301/2302 (99%) | 3490/3502 (99%), Gaps = 9/3502 (0%) | 1242/1258 (98%), Gaps = 10/1258 (0%) | 1242/1258 (98%), Gaps = 10/1258 (0%) | 1237/1274 (97%), Gaps = 27/1274 (2%) | 1237/1274 (97%), Gaps = 27/1274 (2%) | 1372/1393 (98%), Gaps = 19/1393 (1%) |
| m | S | π | D |
|---|---|---|---|
| 328 | 7 | 0.00006 | −1.85719 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, L.; Wang, Q.; Shi, Z.; Li, C.; Guo, Z.; Li, J. Emergence of Rice Blast AVR-Pi9 Resistance Breaking Haplotypes in Yunnan Province, China. Life 2023, 13, 1320. https://doi.org/10.3390/life13061320
Lu L, Wang Q, Shi Z, Li C, Guo Z, Li J. Emergence of Rice Blast AVR-Pi9 Resistance Breaking Haplotypes in Yunnan Province, China. Life. 2023; 13(6):1320. https://doi.org/10.3390/life13061320
Chicago/Turabian StyleLu, Lin, Qun Wang, Zhufeng Shi, Chengyun Li, Zhixiang Guo, and Jinbin Li. 2023. "Emergence of Rice Blast AVR-Pi9 Resistance Breaking Haplotypes in Yunnan Province, China" Life 13, no. 6: 1320. https://doi.org/10.3390/life13061320
APA StyleLu, L., Wang, Q., Shi, Z., Li, C., Guo, Z., & Li, J. (2023). Emergence of Rice Blast AVR-Pi9 Resistance Breaking Haplotypes in Yunnan Province, China. Life, 13(6), 1320. https://doi.org/10.3390/life13061320





