Machine Learning Classification of Time since BNT162b2 COVID-19 Vaccination Based on Array-Measured Antibody Activity
Abstract
1. Introduction
2. Materials and Methods
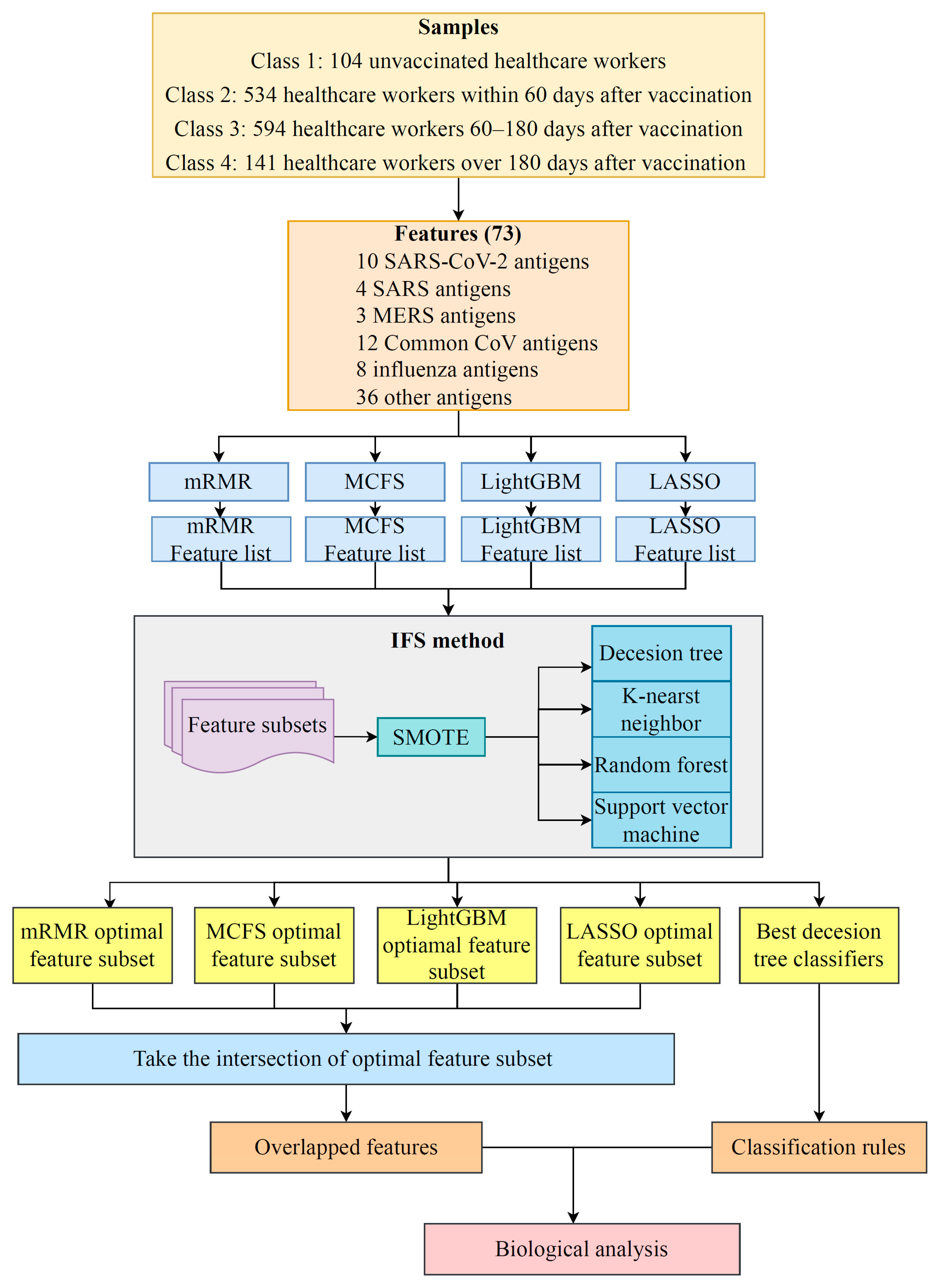
2.1. Data and Preprocessing
2.2. Feature Selection Methods
2.3. Incremental Feature Selection
2.4. Synthetic Minority Oversampling Technique
2.5. Classification Algorithms
2.6. Performance Assessment
2.7. Extraction of Essential Features for Each Class
3. Results
3.1. Results of Feature Selection Methods
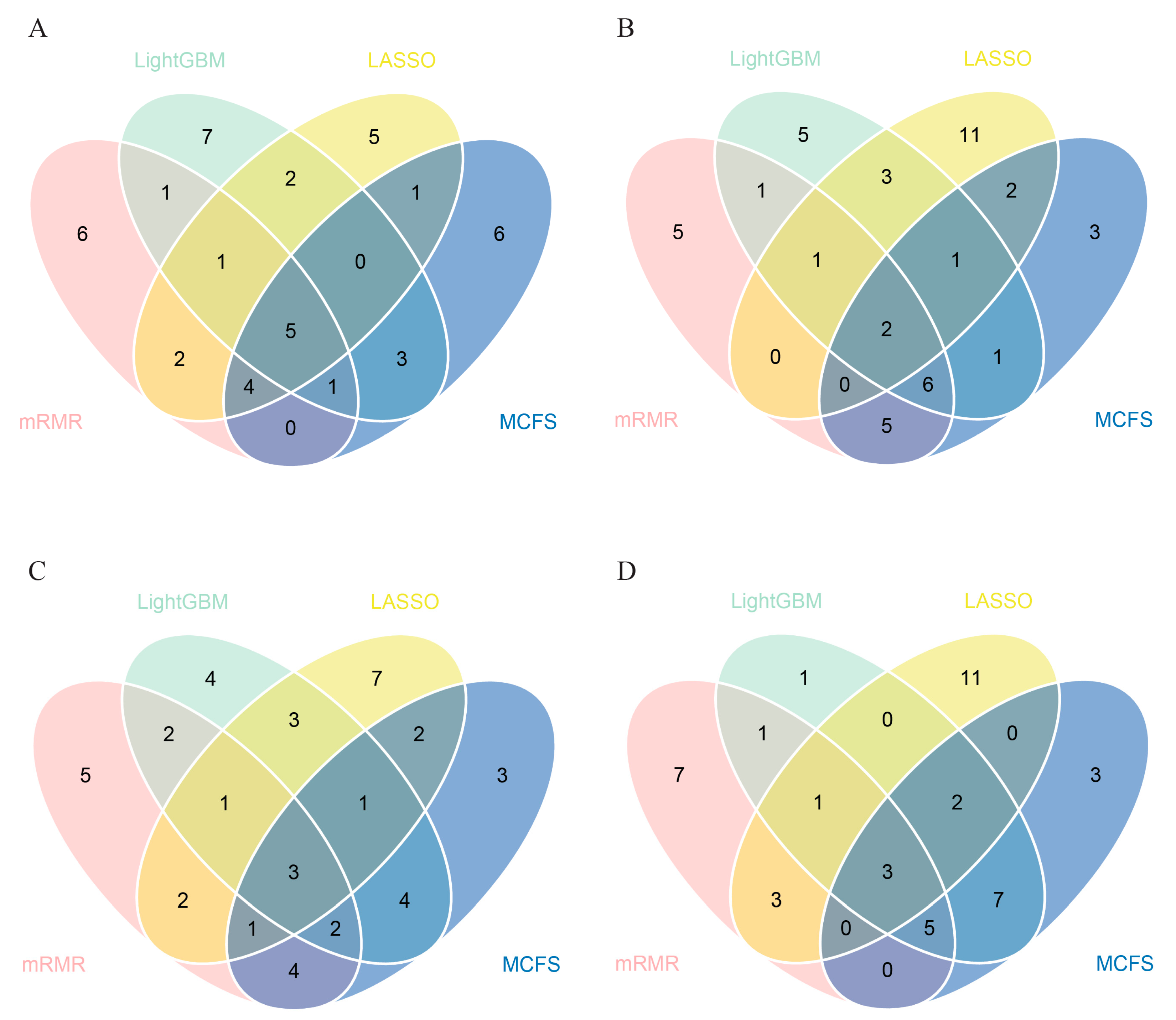
3.2. IFS Results and Feature Intersection
3.3. Essential Features for Each Class
3.4. Classification Rules
4. Discussion
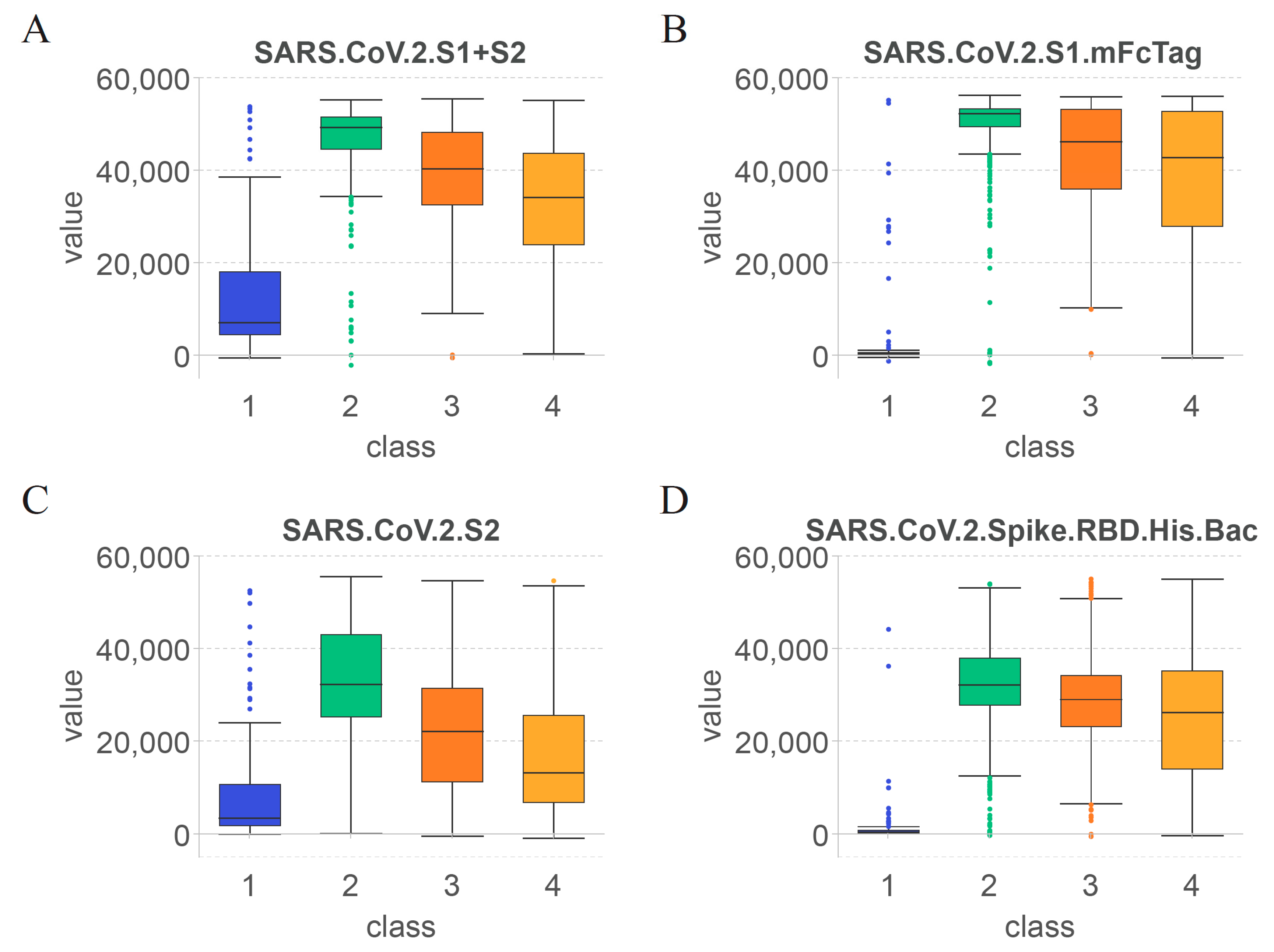
4.1. Key Features for Identifying the Effect of COVID-19 Vaccines on Antibody Production
4.2. Features Related to Time since Vaccination for Determining the Duration of Specific Antibodies after COVID-19 Vaccination
4.3. Rules for Quantitative Time after COVID-19 Vaccination and Antibody Levels
4.4. Limitations of this Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, S.; McNab, C.; Olson, R.M.; Bristol, N.; Nolan, C.; Bergstrøm, E.; Bartos, M.; Mabuchi, S.; Panjabi, R.; Karan, A.; et al. How an outbreak became a pandemic: A chronological analysis of crucial junctures and international obligations in the early months of the COVID-19 pandemic. Lancet 2021, 398, 2109–2124. [Google Scholar] [CrossRef] [PubMed]
- Adil, M.T.; Rahman, R.; Whitelaw, D.; Jain, V.; Al-Taan, O.; Rashid, F.; Munasinghe, A.; Jambulingam, P. SARS-CoV-2 and the pandemic of COVID-19. Postgrad. Med. J. 2021, 97, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Min, K.W.; Park, M.H.; Hong, S.R.; Lee, H.; Kwon, S.Y.; Hong, S.H.; Joo, H.J.; Park, I.A.; An, H.J.; Suh, K.S.; et al. Clear cell carcinomas of the ovary: A multi-institutional study of 129 cases in Korea with prognostic significance of Emi1 and Galectin-3. Int. J. Gynecol. Pathol. 2013, 32, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Parasher, A. COVID-19: Current understanding of its Pathophysiology, Clinical presentation and Treatment. Postgrad. Med. J. 2021, 97, 312–320. [Google Scholar] [CrossRef]
- Thakur, V.; Ratho, R.K. OMICRON (B.1.1.529): A new SARS-CoV-2 variant of concern mounting worldwide fear. J. Med. Virol. 2022, 94, 1821–1824. [Google Scholar] [CrossRef]
- Araf, Y.; Akter, F.; Tang, Y.-D.; Fatemi, R.; Parvez, M.S.A.; Zheng, C.; Hossain, M.G. Omicron variant of SARS-CoV-2: Genomics, transmissibility, and responses to current COVID-19 vaccines. J. Med. Virol. 2022, 94, 1825–1832. [Google Scholar] [CrossRef]
- Fiolet, T.; Kherabi, Y.; MacDonald, C.J.; Ghosn, J.; Peiffer-Smadja, N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: A narrative review. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. 2022, 28, 202–221. [Google Scholar] [CrossRef]
- Jin, Y.; Hou, C.; Li, Y.; Zheng, K.; Wang, C. mRNA Vaccine: How to Meet the Challenge of SARS-CoV-2. Front. Immunol. 2021, 12, 821538. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef]
- Stephenson, K.E.; Le Gars, M.; Sadoff, J.; de Groot, A.M.; Heerwegh, D.; Truyers, C.; Atyeo, C.; Loos, C.; Chandrashekar, A.; McMahan, K.; et al. Immunogenicity of the Ad26.COV2.S Vaccine for COVID-19. JAMA 2021, 325, 1535–1544. [Google Scholar] [CrossRef]
- Thomas, S.J.; Moreira, E.D., Jr.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Polack, F.P.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine through 6 Months. N. Engl. J. Med. 2021, 385, 1761–1773. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, S.E.; Shurin, G.V.; Yost, M.; Anderson, A.; Pinto, L.; Wells, A.; Shurin, M.R. Differential Antibody Response to mRNA COVID-19 Vaccines in Healthy Subjects. Microbiol. Spectr. 2021, 9, e0034121. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A.; et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021, 592, 616–622. [Google Scholar] [CrossRef]
- Noori, M.; Nejadghaderi, S.A.; Arshi, S.; Carson-Chahhoud, K.; Ansarin, K.; Kolahi, A.-A.; Safiri, S. Potency of BNT162b2 and mRNA-1273 vaccine-induced neutralizing antibodies against severe acute respiratory syndrome-CoV-2 variants of concern: A systematic review of in vitro studies. Rev. Med. Virol. 2022, 32, e2277. [Google Scholar] [CrossRef]
- Shao, Y.; Wu, Y.; Feng, Y.; Xu, W.; Xiong, F.; Zhang, X. SARS-CoV-2 vaccine research and immunization strategies for improved control of the COVID-19 pandemic. Front. Med. 2022, 16, 185–195. [Google Scholar] [CrossRef]
- Meo, S.A.; Bukhari, I.A.; Akram, J.; Meo, A.S.; Klonoff, D.C. COVID-19 vaccines: Comparison of biological, pharmacological characteristics and adverse effects of Pfizer/BioNTech and Moderna Vaccines. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1663–1669. [Google Scholar] [CrossRef]
- Kim, W.; Zhou, J.Q.; Horvath, S.C.; Schmitz, A.J.; Sturtz, A.J.; Lei, T.; Liu, Z.; Kalaidina, E.; Thapa, M.; Alsoussi, W.B.; et al. Germinal centre-driven maturation of B cell response to mRNA vaccination. Nature 2022, 604, 141–145. [Google Scholar] [CrossRef]
- Feikin, D.R.; Higdon, M.M.; Abu-Raddad, L.J.; Andrews, N.; Araos, R.; Goldberg, Y.; Groome, M.J.; Huppert, A.; O’Brien, K.L.; Smith, P.G.; et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: Results of a systematic review and meta-regression. Lancet (Lond. Engl.) 2022, 399, 924–944. [Google Scholar] [CrossRef]
- Higdon, M.M.; Baidya, A.; Walter, K.K.; Patel, M.K.; Issa, H.; Espié, E.; Feikin, D.R.; Knoll, M.D. Duration of effectiveness of vaccination against COVID-19 caused by the omicron variant. Lancet Infect. Dis. 2022, 22, 1114–1116. [Google Scholar] [CrossRef]
- Mak, W.A.; Koeleman, J.G.M.; van der Vliet, M.; Keuren, F.; Ong, D.S.Y. SARS-CoV-2 antibody and T cell responses one year after COVID-19 and the booster effect of vaccination: A prospective cohort study. J. Infect. 2022, 84, 171–178. [Google Scholar] [CrossRef]
- Leon, J.; Merrill, A.E.; Rogers, K.; Kurt, J.; Dempewolf, S.; Ehlers, A.; Jackson, J.B.; Knudson, C.M. SARS-CoV-2 antibody changes in patients receiving COVID-19 convalescent plasma from normal and vaccinated donors. Transfus. Apher. Sci. Off. J. World Apher. Assoc. Off. J. Eur. Soc. Haemapheresis 2022, 61, 103326. [Google Scholar] [CrossRef]
- Plūme, J.; Galvanovskis, A.; Šmite, S.; Romanchikova, N.; Zayakin, P.; Linē, A. Early and strong antibody responses to SARS-CoV-2 predict disease severity in COVID-19 patients. J. Transl. Med. 2022, 20, 176. [Google Scholar] [CrossRef] [PubMed]
- Scheiblauer, H.; Nübling, C.M.; Wolf, T.; Khodamoradi, Y.; Bellinghausen, C.; Sonntagbauer, M.; Esser-Nobis, K.; Filomena, A.; Mahler, V.; Maier, T.J.; et al. Antibody response to SARS-CoV-2 for more than one year-kinetics and persistence of detection are predominantly determined by avidity progression and test design. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2022, 146, 105052. [Google Scholar] [CrossRef]
- Guo, L.; Wang, G.; Wang, Y.; Zhang, Q.; Ren, L.; Gu, X.; Huang, T.; Zhong, J.; Wang, Y.; Wang, X.; et al. SARS-CoV-2-specific antibody and T-cell responses 1 year after infection in people recovered from COVID-19: A longitudinal cohort study. Lancet Microbe 2022, 3, e348–e356. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Sun, Q. Antibodies and Vaccines Target RBD of SARS-CoV-2. Front. Mol. Biosci. 2021, 8, 671633. [Google Scholar] [CrossRef]
- Okyar Baş, A.; Hafizoğlu, M.; Akbiyik, F.; Güner Oytun, M.; Şahiner, Z.; Ceylan, S.; Ünsal, P.; Doğu, B.B.; Cankurtaran, M.; Çakir, B.; et al. Antibody response with SARS-CoV-2 inactivated vaccine (CoronaVac) in Turkish geriatric population. Age Ageing 2022, 51, afac088. [Google Scholar] [CrossRef]
- Sanz-Muñoz, I.; López-Mongil, R.; Sánchez-Martínez, J.; Sánchez-de Prada, L.; González, M.D.; Pérez-SanJose, D.; Rojo-Rello, S.; Hernán-García, C.; Fernández-Espinilla, V.; de Lejarazu-Leonardo, R.O.; et al. Evolution of antibody profiles against SARS-CoV-2 in experienced and naïve vaccinated elderly people. Front. Immunol. 2023, 14, 1128302. [Google Scholar] [CrossRef] [PubMed]
- Svetlova, J.; Gustin, D.; Manuvera, V.; Shirokov, D.; Shokina, V.; Prusakov, K.; Aldarov, K.; Kharlampieva, D.; Matyushkina, D.; Bespyatykh, J.; et al. Microarray Profiling of Vaccination-Induced Antibody Responses to SARS-CoV-2 Variants of Interest and Concern. Int. J. Mol. Sci. 2022, 23, 3220. [Google Scholar] [CrossRef]
- Khan, S.; Hosseinian, S.; Assis, R.; Khalil, G.; Luu, M.; Jain, A.; Horvath, P.; Nakajima, R.; Palma, A.; Hoang, A.; et al. Analysis and comparison of SARS-CoV-2 variant antibodies and neutralizing activity for 6 months after a booster mRNA vaccine in a healthcare worker population. Res. Sq. 2022. [Google Scholar] [CrossRef]
- Fisher, M.; Manor, A.; Abramovitch, H.; Fatelevich, E.; Afrimov, Y.; Bilinsky, G.; Lupu, E.; Ben-Shmuel, A.; Glinert, I.; Madar-Balakirski, N. A novel quantitative multi-component serological assay for SARS-CoV-2 vaccine evaluation. Anal. Chem. 2022, 94, 4380–4389. [Google Scholar] [CrossRef] [PubMed]
- Hosseinian, S.; Powers, K.; Vasudev, M.; Palma, A.M.; de Assis, R.; Jain, A.; Horvath, P.; Birring, P.S.; Andary, R.; Au, C.; et al. Persistence of SARS-CoV-2 Antibodies in Vaccinated Health Care Workers Analyzed by Coronavirus Antigen Microarray. Front. Immunol. 2022, 13, 817345. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Better Subset Regression Using the Nonnegative Garrote. Technometrics 1995, 37, 373–384. [Google Scholar] [CrossRef]
- Tibshirani, R.J. Regression Shrinkage and Selection via the LASSO. J. R. Stat. Society. Ser. B Methodol. 1996, 73, 273–282. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finely, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. In Proceedings of the Advances in Neural Information Processing Systems 30 (NIP 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Micha, D.; Rada-Iglesias, A.; Enroth, S.; Wadelius, C.; Koronacki, J.; Komorowski, J. Monte Carlo feature selection for supervised classification. Bioinformatics 2008, 24, 110–117. [Google Scholar]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn Res. 2011, 12, 2825–2830. [Google Scholar]
- Chen, L.; Li, J.; Zhang, Y.H.; Feng, K.; Wang, S.; Zhang, Y.; Huang, T.; Kong, X.; Cai, Y.D. Identification of gene expression signatures across different types of neural stem cells with the Monte-Carlo feature selection method. J. Cell. Biochem. 2018, 119, 3394–3403. [Google Scholar] [CrossRef]
- Chen, X.; Jin, Y.; Feng, Y. Evaluation of Plasma Extracellular Vesicle MicroRNA Signatures for Lung Adenocarcinoma and Granuloma With Monte-Carlo Feature Selection Method. Front. Genet. 2019, 10, 367. [Google Scholar] [CrossRef]
- Huang, F.; Ma, Q.; Ren, J.; Li, J.; Wang, F.; Huang, T.; Cai, Y.-D. Identification of Smoking associated Transcriptome Aberration in Blood with Machine Learning Methods. BioMed Res. Int. 2023, 2023, 5333361. [Google Scholar] [CrossRef]
- Liu, H.A.; Setiono, R. Incremental feature selection. Appl. Intell. 1998, 9, 217–230. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Guo, W.; Zeng, T.; Zhang, S.; Chen, L.; Gamarra, M.; Mansour, R.F.; Escorcia-Gutierrez, J.; Huang, T.; Cai, Y.D. Identification of Microbiota Biomarkers With Orthologous Gene Annotation for Type 2 Diabetes. Front. Microbiol. 2021, 12, 711244. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Li, Z.; Zeng, T.; Pan, X.; Chen, L.; Liu, D.; Li, H.; Huang, T.; Cai, Y.D. Distinguishing Glioblastoma Subtypes by Methylation Signatures. Front. Genet. 2020, 11, 604336. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Fu, M.; Li, J.; Chen, L.; Feng, K.; Huang, T.; Cai, Y.-D. Analysis and Prediction of Protein Stability Based on Interaction Network, Gene Ontology, and KEGG Pathway Enrichment Scores. BBA Proteins Proteom. 2023, 1871, 140889. [Google Scholar] [CrossRef] [PubMed]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the International Joint Conference on Artificial Intelligence, Montréal, QC, Canada, 20–25 August 1995; pp. 1137–1145. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Pan, X.; Chen, L.; Liu, I.; Niu, Z.; Huang, T.; Cai, Y.D. Identifying protein subcellular locations with embeddings-based node2loc. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhou, X.; Guo, W.; Feng, K.; Huang, T.; VCai, Y.-D. Identification of Methylation Signatures and Rules for Sarcoma Subtypes by Machine Learning Methods. BioMed Res. Int. 2022, 2022, 5297235. [Google Scholar] [CrossRef]
- Safavian, S.R.; Landgrebe, D. A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern. 1991, 21, 660–674. [Google Scholar] [CrossRef]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Zhou, X.; Ding, S.; Wang, D.; Chen, L.; Feng, K.; Huang, T.; Li, Z.; Cai, Y.-D. Identification of cell markers and their expression patterns in skin based on single-cell RNA-sequencing profiles. Life 2022, 12, 550. [Google Scholar] [CrossRef]
- Wu, C.; Chen, L. A model with deep analysis on a large drug network for drug classification. Math. Biosci. Eng. 2023, 20, 383–401. [Google Scholar] [CrossRef] [PubMed]
- Ran, B.; Chen, L.; Li, M.; Han, Y.; Dai, Q. Drug-Drug interactions prediction using fingerprint only. Comput. Math. Methods Med. 2022, 2022, 7818480. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, L. Identification of human protein subcellular location with multiple networks. Curr. Proteom. 2022, 19, 344–356. [Google Scholar]
- Tang, S.; Chen, L. iATC-NFMLP: Identifying classes of anatomical therapeutic chemicals based on drug networks, fingerprints and multilayer perceptron. Curr. Bioinform. 2022, 17, 814–824. [Google Scholar]
- Ren, J.; Zhang, Y.; Guo, W.; Feng, K.; Yuan, Y.; Huang, T.; Cai, Y.-D. Identification of Genes Associated with the Impairment of Olfactory and Gustatory Functions in COVID-19 via Machine-Learning Methods. Life 2023, 13, 798. [Google Scholar] [CrossRef]
- Wang, H.; Chen, L. PMPTCE-HNEA: Predicting metabolic pathway types of chemicals and enzymes with a heterogeneous network embedding algorithm. Curr. Bioinform. 2023. [Google Scholar] [CrossRef]
- Gorodkin, J. Comparing two K-category assignments by a K-category correlation coefficient. Comput. Biol. Chem. 2004, 28, 367–374. [Google Scholar] [CrossRef]
- De Assis, R.R.; Jain, A.; Nakajima, R.; Jasinskas, A.; Felgner, J.; Obiero, J.M.; Norris, P.J.; Stone, M.; Simmons, G.; Bagri, A. Analysis of SARS-CoV-2 antibodies in COVID-19 convalescent blood using a coronavirus antigen microarray. Nat. Commun. 2021, 12, 6. [Google Scholar] [CrossRef]
- Bruckner, T.A.; Parker, D.M.; Bartell, S.M.; Vieira, V.M.; Khan, S.; Noymer, A.; Drum, E.; Albala, B.; Zahn, M.; Boden-Albala, B. Estimated seroprevalence of SARS-CoV-2 antibodies among adults in Orange County, California. Sci. Rep. 2021, 11, 3081. [Google Scholar] [CrossRef]
- Assis, R.; Jain, A.; Nakajima, R.; Jasinskas, A.; Khan, S.; Davies, H.; Corash, L.; Dumont, L.J.; Kelly, K.; Simmons, G.; et al. Distinct SARS-CoV-2 antibody reactivity patterns in coronavirus convalescent plasma revealed by a coronavirus antigen microarray. Sci. Rep. 2021, 11, 7554. [Google Scholar] [CrossRef] [PubMed]
- Assis, R.; Jain, A.; Nakajima, R.; Jasinskas, A.; Khan, S.; Palma, A.; Parker, D.M.; Chau, A.; Obiero, J.M.; Tifrea, D.; et al. Distinct SARS-CoV-2 antibody reactivity patterns elicited by natural infection and mRNA vaccination. npj Vaccines 2021, 6, 132. [Google Scholar] [CrossRef] [PubMed]
- Altmann, D.M.; Boyton, R.J. COVID-19 vaccination: The road ahead. Science 2022, 375, 1127–1132. [Google Scholar] [CrossRef]
- Begum, J.; Mir, N.A.; Dev, K.; Buyamayum, B.; Wani, M.Y.; Raza, M. Challenges and prospects of COVID-19 vaccine development based on the progress made in SARS and MERS vaccine development. Transbound. Emerg. Dis. 2021, 68, 1111–1124. [Google Scholar] [CrossRef]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef]
- Dong, Y.; Dai, T.; Wei, Y.; Zhang, L.; Zheng, M.; Zhou, F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target. Ther. 2020, 5, 237. [Google Scholar] [CrossRef] [PubMed]
- Suthar, M.S.; Zimmerman, M.G.; Kauffman, R.C.; Mantus, G.; Linderman, S.L.; Hudson, W.H.; Vanderheiden, A.; Nyhoff, L.; Davis, C.W.; Adekunle, O.; et al. Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Rep. Med. 2020, 1, 100040. [Google Scholar] [CrossRef]
- Kirtipal, N.; Bharadwaj, S.; Kang, S.G. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2020, 85, 104502. [Google Scholar] [CrossRef]
- Morse, J.S.; Lalonde, T.; Xu, S.; Liu, W.R. Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV. Chembiochem A Eur. J. Chem. Biol. 2020, 21, 730–738. [Google Scholar] [CrossRef]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Wec, A.Z.; Wrapp, D.; Herbert, A.S.; Maurer, D.P.; Haslwanter, D.; Sakharkar, M.; Jangra, R.K.; Dieterle, M.E.; Lilov, A.; Huang, D.; et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 2020, 369, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Mateus, J.; Coelho, C.H.; Dan, J.M.; Moderbacher, C.R.; Gálvez, R.I.; Cortes, F.H.; Grifoni, A.; Tarke, A.; Chang, J.; et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell 2022, 185, 2434–2451. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Casner, R.G.; Nair, M.S.; Yu, J.; Guo, Y.; Wang, M.; Chan, J.F.W.; Cerutti, G.; Iketani, S.; Liu, L.; et al. A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses. Emerg. Microbes Infect. 2022, 11, 147–157. [Google Scholar] [CrossRef]
- Dolgin, E. Pan-coronavirus vaccine pipeline takes form. Nat. Rev. Drug Discov. 2022, 21, 324–326. [Google Scholar] [CrossRef]
- Rosenberg, E.S.; Dorabawila, V.; Easton, D.; Bauer, U.E.; Kumar, J.; Hoen, R.; Hoefer, D.; Wu, M.; Lutterloh, E.; Conroy, M.B.; et al. COVID-19 Vaccine Effectiveness in New York State. N. Engl. J. Med. 2022, 386, 116–127. [Google Scholar] [CrossRef]
- Takeda, M. Proteolytic activation of SARS-CoV-2 spike protein. Microbiol. Immunol. 2022, 66, 15–23. [Google Scholar] [CrossRef]
- Cucunawangsih, C.; Wijaya, R.S.; Lugito, N.P.H.; Suriapranata, I. Antibody response to the inactivated SARS-CoV-2 vaccine among healthcare workers, Indonesia. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2021, 113, 15–17. [Google Scholar] [CrossRef]
- Widge, A.T.; Rouphael, N.G.; Jackson, L.A.; Anderson, E.J.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med. 2021, 384, 80–82. [Google Scholar] [CrossRef]
- Levi, R.; Azzolini, E.; Pozzi, C.; Ubaldi, L.; Lagioia, M.; Mantovani, A.; Rescigno, M. One dose of SARS-CoV-2 vaccine exponentially increases antibodies in individuals who have recovered from symptomatic COVID-19. J. Clin. Investig. 2021, 131, 149154. [Google Scholar] [CrossRef] [PubMed]
- Pieri, M.; Nicolai, E.; Ciotti, M.; Nuccetelli, M.; Sarubbi, S.; Pelagalli, M.; Bernardini, S. Antibody response to COVID-19 vaccine: A point of view that can help to optimize dose distribution. Int. Immunopharmacol. 2022, 102, 108406. [Google Scholar] [CrossRef] [PubMed]
- Bates, T.A.; Weinstein, J.B.; Farley, S.; Leier, H.C.; Messer, W.B.; Tafesse, F.G. Cross-reactivity of SARS-CoV structural protein antibodies against SARS-CoV-2. Cell Rep. 2021, 34, 108737. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Minn, D.; Chang, S.-H.; Suh, J.-S. Comparing SARS-CoV-2 Antibody Responses after Various COVID-19 Vaccinations in Healthcare Workers. Vaccines 2022, 10, 193. [Google Scholar] [CrossRef] [PubMed]
- Lange, A.; Borowik, A.; Bocheńska, J.; Rossowska, J.; Jaskuła, E. Immune Response to COVID-19 mRNA Vaccine-A Pilot Study. Vaccines 2021, 9, 488. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Y.; Liang, T.; Yang, N.; Li, T.; Zheng, C.; Ning, N.; Luo, D.; Yang, X.; He, Z.; et al. Longitudinal and proteome-wide analyses of antibodies in COVID-19 patients reveal features of the humoral immune response to SARS-CoV-2. J. Adv. Res. 2022, 37, 209–219. [Google Scholar] [CrossRef]
- Lv, H.; Wu, N.C.; Tsang, O.T.-Y.; Yuan, M.; Perera, R.A.P.M.; Leung, W.S.; So, R.T.Y.; Chan, J.M.C.; Yip, G.K.; Chik, T.S.H.; et al. Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep. 2020, 31, 107725. [Google Scholar] [CrossRef]
- Levin, E.G.; Lustig, Y.; Cohen, C.; Fluss, R.; Indenbaum, V.; Amit, S.; Doolman, R.; Asraf, K.; Mendelson, E.; Ziv, A.; et al. Waning Immune Humoral Response to BNT162b2 COVID-19 Vaccine over 6 Months. N. Engl. J. Med. 2021, 385, e84. [Google Scholar] [CrossRef]
- Reynolds, C.J.; Pade, C.; Gibbons, J.M.; Butler, D.K.; Otter, A.D.; Menacho, K.; Fontana, M.; Smit, A.; Sackville-West, J.E.; Cutino-Moguel, T.; et al. Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science 2021, 372, 1418–1423. [Google Scholar] [CrossRef]
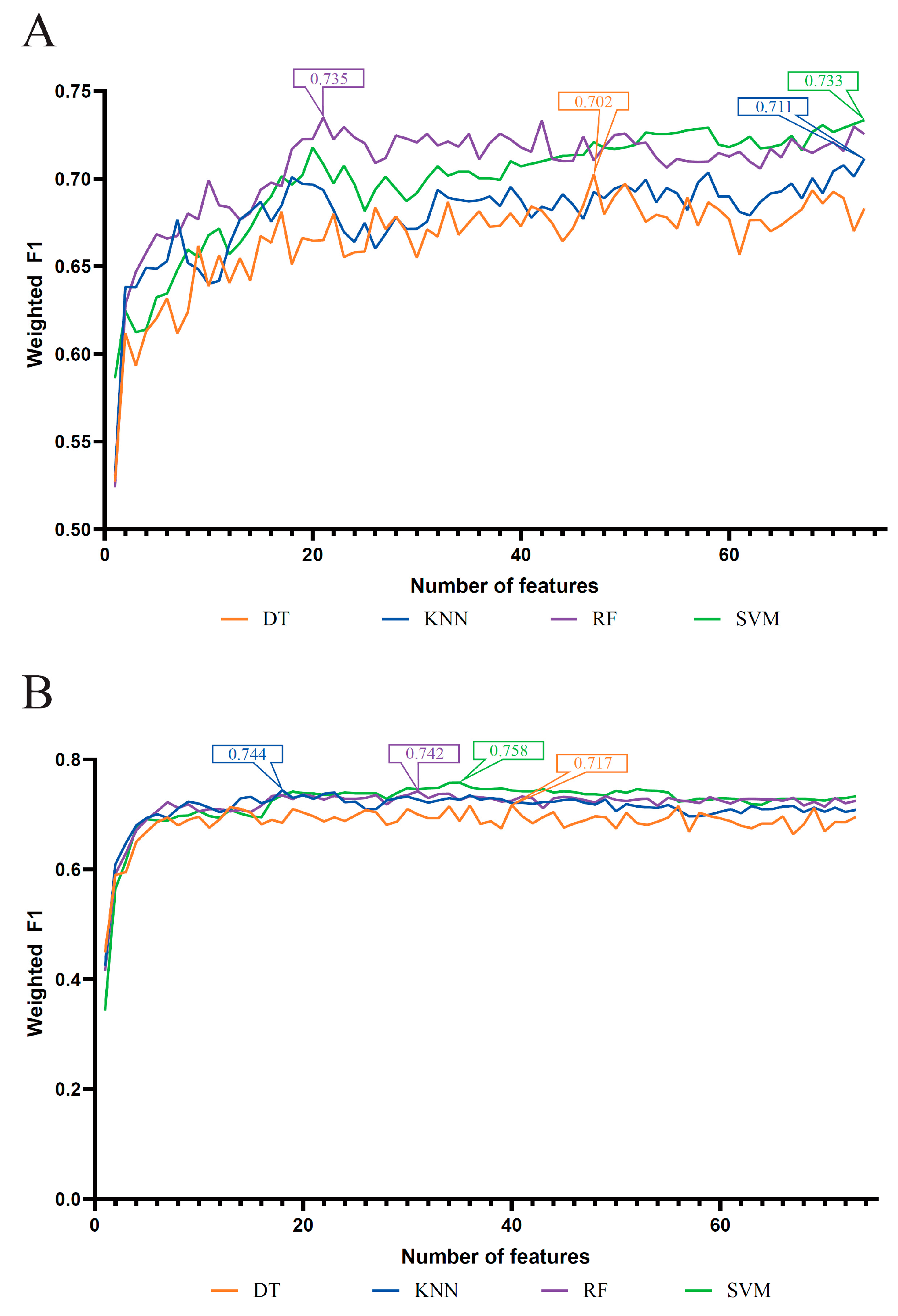


| Feature List | Classification Algorithm | Number of Features | ACC | MCC | Macro F1 | Weighted F1 |
|---|---|---|---|---|---|---|
| LASSO feature list | DT | 47 | 0.704 | 0.554 | 0.744 | 0.702 |
| KNN | 73 | 0.716 | 0.574 | 0.776 | 0.711 | |
| RF | 21 | 0.741 | 0.622 | 0.787 | 0.735 | |
| SVM | 73 | 0.737 | 0.603 | 0.796 | 0.733 | |
| LightGBM feature list | DT | 40 | 0.720 | 0.573 | 0.762 | 0.717 |
| KNN | 18 | 0.747 | 0.618 | 0.802 | 0.744 | |
| RF | 31 | 0.752 | 0.649 | 0.796 | 0.742 | |
| SVM | 35 | 0.761 | 0.640 | 0.806 | 0.758 | |
| MCFS feature list | DT | 17 | 0.729 | 0.589 | 0.771 | 0.727 |
| KNN | 20 | 0.742 | 0.611 | 0.799 | 0.739 | |
| RF | 23 | 0.756 | 0.649 | 0.801 | 0.747 | |
| SVM | 41 | 0.768 | 0.652 | 0.811 | 0.765 | |
| mRMR feature list | DT | 14 | 0.730 | 0.594 | 0.763 | 0.728 |
| KNN | 24 | 0.741 | 0.612 | 0.797 | 0.737 | |
| RF | 26 | 0.754 | 0.646 | 0.797 | 0.745 | |
| SVM | 30 | 0.762 | 0.643 | 0.805 | 0.758 |
| Target Antigens | LASSO | LightGBM | MCFS | mRMR |
|---|---|---|---|---|
| SARS.CoV.2.S1.mFcTag | ✔ | ✔ | ✔ | ✔ |
| MERS.CoV.S1.RBD.367.606.rFcTag | ✔ | ✔ | ✔ | ✔ |
| SARS.CoV.2.Spike.RBD.His.Bac | ✔ | ✔ | ✔ | ✔ |
| SARS.CoV.S1.HisTag | ✔ | ✔ | ✔ | ✔ |
| SARS.CoV.2.S1.RBD.mFc | ✔ | ✔ | ✔ | ✔ |
| SARS.CoV.2.S1 + S2 | ✔ | ✔ | ✔ | ✔ |
| SARS.CoV.2.S2 | ✔ | ✔ | ✔ | |
| hCoV.HKU1.NP | ✔ | ✔ | ✔ | |
| SARS.CoV.2.Spike.RBD.rFc | ✔ | ✔ | ✔ | |
| SARS.CoV.2.S1 | ✔ | ✔ | ✔ | |
| SARS.CoV.2.S1.HisTag | ✔ | ✔ | ✔ | |
| SARS.CoV.S1.RBD.HisTag | ✔ | ✔ | ✔ | |
| hCoV.229E.S1 | ✔ | ✔ | ✔ |
| Rules | Criteria | Predicted Class (Days after Vaccination) |
|---|---|---|
| Rule 0 | SARS.CoV.2.S1.mFcTag ≤ 5354.39 | Unvaccinated healthcare workers |
| −383.87 < SARS.CoV.2.S1.HisTag | ||
| −414.30 < SARS.CoV.2.S1.RBD.mFc ≤ 3773.83 | ||
| 414.54 < hCoV.OC43.HE | ||
| Rule 1 | SARS.CoV.2.S1.mFcTag ≤ 54,010.17 | Healthcare workers within 60 days after vaccination |
| 37,653.75 < SARS.CoV.2.S2 | ||
| 48,882.58 < SARS.CoV.2.S1 + S2 | ||
| Rule 2 | 5354.39 < SARS.CoV.2.S1.mFcTag | Healthcare workers between 60 and 180 days after vaccination |
| 3773.83 < SARS.CoV.2.S1.RBD.mFc ≤ 33,656.48 | ||
| 400.30 < SARS.CoV.S1.HisTag ≤ 15,087.42 | ||
| Rule 3 | 5354.39 < SARS.CoV.2.S1.mFcTag ≤ 34,194.92 | Healthcare workers over 180 days after vaccination |
| 3773.83 < SARS.CoV.2.S1.RBD.mFc |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Q.-L.; Huang, F.-M.; Guo, W.; Feng, K.-Y.; Huang, T.; Cai, Y.-D. Machine Learning Classification of Time since BNT162b2 COVID-19 Vaccination Based on Array-Measured Antibody Activity. Life 2023, 13, 1304. https://doi.org/10.3390/life13061304
Ma Q-L, Huang F-M, Guo W, Feng K-Y, Huang T, Cai Y-D. Machine Learning Classification of Time since BNT162b2 COVID-19 Vaccination Based on Array-Measured Antibody Activity. Life. 2023; 13(6):1304. https://doi.org/10.3390/life13061304
Chicago/Turabian StyleMa, Qing-Lan, Fei-Ming Huang, Wei Guo, Kai-Yan Feng, Tao Huang, and Yu-Dong Cai. 2023. "Machine Learning Classification of Time since BNT162b2 COVID-19 Vaccination Based on Array-Measured Antibody Activity" Life 13, no. 6: 1304. https://doi.org/10.3390/life13061304
APA StyleMa, Q.-L., Huang, F.-M., Guo, W., Feng, K.-Y., Huang, T., & Cai, Y.-D. (2023). Machine Learning Classification of Time since BNT162b2 COVID-19 Vaccination Based on Array-Measured Antibody Activity. Life, 13(6), 1304. https://doi.org/10.3390/life13061304









