Basic ctDNA Panel Promises Affordable Clinical Validity in Colon Cancer Patients but Not in Pancreas Cancer Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Samples Desciption
2.3. Somatic Analysis of Primary Tumor
2.4. The Development of the ctDNA Panel
2.5. Statistical Analysis
2.6. Naïve Detection of Somatic Mutations
3. Results
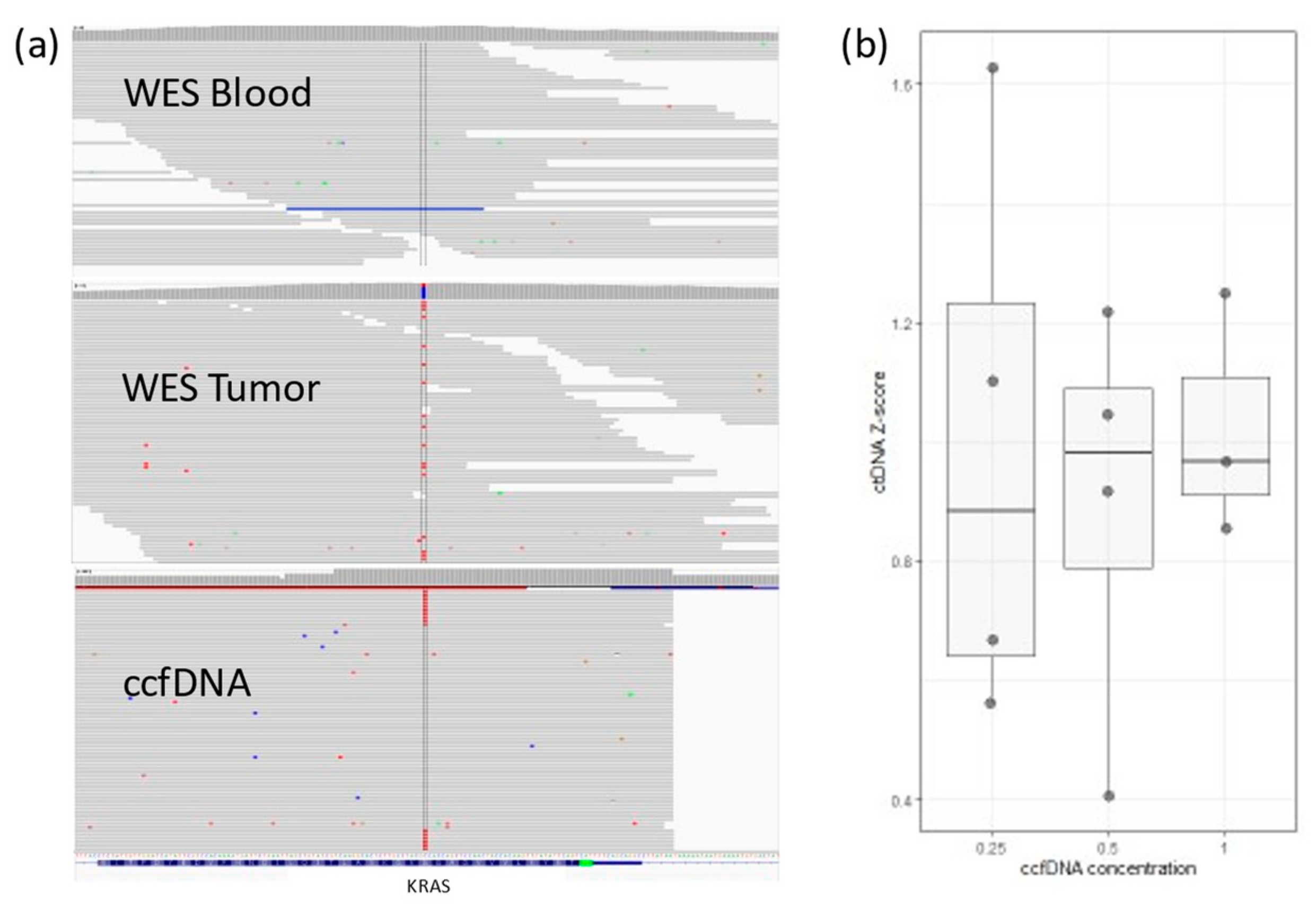
3.1. The Development of the ctDNA Panel
3.2. Quality Control Parameters and Limitations
3.3. ctDNA Presence in Colon and Pancreas Samples
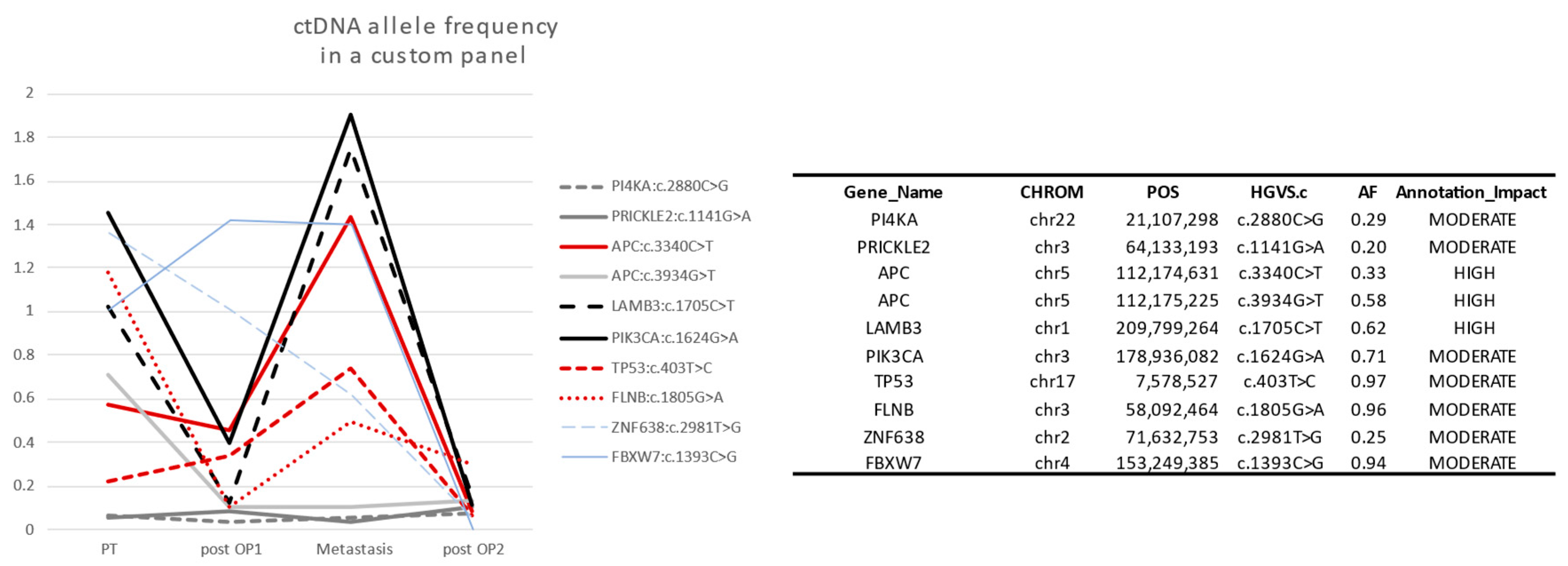
3.4. Case Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Figg, W.D., 2nd; Reid, J. Monitor tumor burden with circulating tumor DNA. Cancer Biol. Ther. 2013, 14, 697–698. [Google Scholar] [CrossRef]
- Brychta, N.; Krahn, T.; von Ahsen, O. Detection of KRAS Mutations in Circulating Tumor DNA by Digital PCR in Early Stages of Pancreatic Cancer. Clin. Chem. 2016, 62, 1482–1491. [Google Scholar] [CrossRef]
- McDonald, B.R.; Contente-Cuomo, T.; Sammut, S.J.; Odenheimer-Bergman, A.; Ernst, B.; Perdigones, N.; Chin, S.F.; Farooq, M.; Mejia, R.; Cronin, P.A.; et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 2019, 11, eaax7392. [Google Scholar] [CrossRef]
- Parikh, A.R.; Van Seventer, E.E.; Siravegna, G.; Hartwig, A.V.; Jaimovich, A.; He, Y.; Kanter, K.; Fish, M.G.; Fosbenner, K.D.; Miao, B.; et al. Minimal Residual Disease Detection using a Plasma-only Circulating Tumor DNA Assay in Patients with Colorectal Cancer. Clin. Cancer Res. 2021, 27, 5586–5594. [Google Scholar] [CrossRef]
- Panagopoulou, M.; Esteller, M.; Chatzaki, E. Circulating Cell-Free DNA in Breast Cancer: Searching for Hidden Information towards Precision Medicine. Cancers 2021, 13, 728. [Google Scholar] [CrossRef]
- Hou, W.; Yi, C.; Zhu, H. Predictive biomarkers of colon cancer immunotherapy: Present and future. Front. Immunol. 2022, 13, 1032314. [Google Scholar] [CrossRef]
- Lui, Y.Y.; Chik, K.W.; Chiu, R.W.; Ho, C.Y.; Lam, C.W.; Lo, Y.M. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin. Chem. 2002, 48, 421–427. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Murtaza, M.; Dawson, S.J.; Pogrebniak, K.; Rueda, O.M.; Provenzano, E.; Grant, J.; Chin, S.F.; Tsui, D.W.; Marass, F.; Gale, D.; et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun. 2015, 6, 8760. [Google Scholar] [CrossRef]
- Lanman, R.B.; Mortimer, S.A.; Zill, O.A.; Sebisanovic, D.; Lopez, R.; Blau, S.; Collisson, E.A.; Divers, S.G.; Hoon, D.S.; Kopetz, E.S.; et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS ONE 2015, 10, e0140712. [Google Scholar] [CrossRef]
- Kowalchuk, R.O.; Kamdem Talom, B.C.; Van Abel, K.M.; Ma, D.M.; Waddle, M.R.; Routman, D.M. Estimated Cost of Circulating Tumor DNA for Posttreatment Surveillance of Human Papillomavirus-Associated Oropharyngeal Cancer. JAMA Netw. Open 2022, 5, e2144783. [Google Scholar] [CrossRef]
- Elazezy, M.; Joosse, S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 2018, 16, 370–378. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Källberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Ahlmann-Eltze, C.; Patil, I. ggsignif: R Package for Displaying Significance Brackets for ‘ggplot2’. PsyArxiv 2021. [Google Scholar] [CrossRef]
- Gerstung, M.; Beisel, C.; Rechsteiner, M.; Wild, P.; Schraml, P.; Moch, H.; Beerenwinkel, N. Reliable detection of subclonal single-nucleotide variants in tumour cell populations. Nat. Commun. 2012, 3, 811. [Google Scholar] [CrossRef]
- Henriksen, T.V.; Reinert, T.; Rasmussen, M.H.; Demuth, C.; Løve, U.S.; Madsen, A.H.; Gotschalck, K.A.; Iversen, L.H.; Andersen, C.L. Comparing single-target and multitarget approaches for postoperative circulating tumour DNA detection in stage II–III colorectal cancer patients. Mol. Oncol. 2022, 16, 3654–3665. [Google Scholar] [CrossRef]
- Rengifo, L.Y.; Smits, S.; Buedts, L.; Delforge, M.; Dehaspe, L.; Tousseyn, T.; Boeckx, N.; Lehnert, S.; Michaux, L.; Vermeesch, J.R.; et al. Ultra-low coverage whole genome sequencing of ccfDNA in multiple myeloma: A tool for laboratory routine? Cancer Treat. Res. Commun. 2021, 28, 100380. [Google Scholar] [CrossRef]
- Underhill, H.R. Leveraging the Fragment Length of Circulating Tumour DNA to Improve Molecular Profiling of Solid Tumour Malignancies with Next-Generation Sequencing: A Pathway to Advanced Non-invasive Diagnostics in Precision Oncology? Mol. Diagn. Ther. 2021, 25, 389–408. [Google Scholar] [CrossRef]
- Loupakis, F.; Sharma, S.; Derouazi, M.; Murgioni, S.; Biason, P.; Rizzato, M.D.; Rasola, C.; Renner, D.; Shchegrova, S.; Koyen Malashevich, A.; et al. Detection of Molecular Residual Disease Using Personalized Circulating Tumor DNA Assay in Patients with Colorectal Cancer Undergoing Resection of Metastases. JCO Precis. Oncol. 2021, 5, 1166–1177. [Google Scholar] [CrossRef]
- Sahin, I.H.; Lin, Y.; Yothers, G.; Lucas, P.C.; Deming, D.; George, T.J.; Kopetz, S.; Lieu, C.H.; Dasari, A. Minimal Residual Disease-Directed Adjuvant Therapy for Patients with Early-Stage Colon Cancer: CIRCULATE-US. Oncology 2022, 36, 604–608. [Google Scholar] [CrossRef]
- Maron, S.B.; Chase, L.M.; Lomnicki, S.; Kochanny, S.; Moore, K.L.; Joshi, S.S.; Landron, S.; Johnson, J.; Kiedrowski, L.A.; Nagy, R.J.; et al. Circulating Tumor DNA Sequencing Analysis of Gastroesophageal Adenocarcinoma. Clin. Cancer Res. 2019, 25, 7098–7112. [Google Scholar] [CrossRef]
- Postel, M.; Roosen, A.; Laurent-Puig, P.; Taly, V.; Wang-Renault, S.F. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: A cancer diagnostic perspective. Expert. Rev. Mol. Diagn. 2018, 18, 7–17. [Google Scholar] [CrossRef]
- Lin, C.; Liu, X.; Zheng, B.; Ke, R.; Tzeng, C.M. Liquid Biopsy, ctDNA Diagnosis through NGS. Life 2021, 11, 890. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Lahouel, K.; Lo, S.N.; Wang, Y.; Kosmider, S.; Wong, R.; Shapiro, J.; Lee, M.; Harris, S.; et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N. Engl. J. Med. 2022, 386, 2261–2272. [Google Scholar] [CrossRef]
- Stoffel, E.M.; Brand, R.E.; Goggins, M. Pancreatic Cancer: Changing Epidemiology and New Approaches to Risk Assessment, Early Detection, and Prevention. Gastroenterology 2023, 164, 752–765. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, H.; Chen, N.; Hao, J.; Jin, H.; Ma, X. Diagnostic value of various liquid biopsy methods for pancreatic cancer: A systematic review and meta-analysis. Medicine 2020, 99, e18581. [Google Scholar] [CrossRef]


| ID | Cancer | Detected ctDNA | ctDNA Concordant with Clinical Data |
|---|---|---|---|
| sample1 | Colon | ND | - |
| sample2 | Colon | Detected | Concordant with clinical data |
| sample3 | Colon | Detected | Concordant with clinical data |
| sample5 | Colon | Detected | Concordant with clinical data |
| sample7 | Colon | Detected | Concordant with clinical data |
| sample8 | Colon | ND | - |
| sample9 | Colon | ND | - |
| sample10 | Colon | Detected | Concordant with clinical data |
| sample11 | Colon | Detected | Not concordant with clinical data |
| sample12 | Colon | ND | - |
| sample13 | Colon | Detected | Concordant with clinical data |
| sample15 | Pancreas | ND | - |
| sample16 | Pancreas | ND | - |
| sample17 | Pancreas | Detected | Concordant with clinical data |
| sample18 | Pancreas | ND | - |
| sample19 | Pancreas | ND | - |
| Follow-Up | PIK3CA (c.1624G>A) | TP53 (c.403T>C) | Date | Clinical |
|---|---|---|---|---|
| 1 | 1.7% | 0.6% | April 2019 | Surgery of primary tumor |
| 2 | 0.8% | 0.3% (ND) | May 2019 | Metastasis surgery |
| 3 | 0.9% | 0.7% | August 2020 | 2 metastasis surgeries |
| 4 | ND | ND | October 2020 | 2 months after surgery/therapy |
| 5 | 1.7% | 1.2% | January 2022 | Possible relapse but not identified in imaging (December 2022) Tumor identified via imaging (May 2022) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radefeldt, M.; Stellmacher-Kaiser, S.; Krake, S.; Kragl, B.; Lemke, S.; Beetz, C.; Bauer, P.; Junghanß, C.; Al-Ali, R. Basic ctDNA Panel Promises Affordable Clinical Validity in Colon Cancer Patients but Not in Pancreas Cancer Patients. Life 2023, 13, 2274. https://doi.org/10.3390/life13122274
Radefeldt M, Stellmacher-Kaiser S, Krake S, Kragl B, Lemke S, Beetz C, Bauer P, Junghanß C, Al-Ali R. Basic ctDNA Panel Promises Affordable Clinical Validity in Colon Cancer Patients but Not in Pancreas Cancer Patients. Life. 2023; 13(12):2274. https://doi.org/10.3390/life13122274
Chicago/Turabian StyleRadefeldt, Mandy, Silke Stellmacher-Kaiser, Susann Krake, Brigitte Kragl, Sabrina Lemke, Christian Beetz, Peter Bauer, Christian Junghanß, and Ruslan Al-Ali. 2023. "Basic ctDNA Panel Promises Affordable Clinical Validity in Colon Cancer Patients but Not in Pancreas Cancer Patients" Life 13, no. 12: 2274. https://doi.org/10.3390/life13122274
APA StyleRadefeldt, M., Stellmacher-Kaiser, S., Krake, S., Kragl, B., Lemke, S., Beetz, C., Bauer, P., Junghanß, C., & Al-Ali, R. (2023). Basic ctDNA Panel Promises Affordable Clinical Validity in Colon Cancer Patients but Not in Pancreas Cancer Patients. Life, 13(12), 2274. https://doi.org/10.3390/life13122274






