Evolution of Intrinsic Disorder in Protein Loops
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
4.1. The Evolutionary Centrality of Loop Disorder and Flexibility
4.2. The Unexpected Evolutionary Rise of Order in Loop Structure
4.3. Percolation of Evolutionary Constraints from Higher to Lower Levels of Organization
4.4. Evolutionary Percolation and Trade-Offs between Flexibility and Rigidity in Loop Behavior
4.5. Evolutionary Impact on Loop Structure and Geometry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uversky, V.N. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013, 22, 693–724. [Google Scholar] [CrossRef]
- Uversky, V.N. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta-Proteins Proteom. 2013, 1834, 932–951. [Google Scholar] [CrossRef]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Showing your ID: Intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005, 18, 343–384. [Google Scholar] [CrossRef]
- Liu, J.; Tan, H.; Rost, B. Loopy proteins appear conserved in evolution. J. Mol. Biol. 2002, 322, 53–64. [Google Scholar] [CrossRef]
- Daughdrill, G.W.; Narayanaswami, P.; Gilmore, S.H.; Belczyk, A.; Brown, C.J. Dynamic behavior of an intrinsically unstructured linker domain is conserved in the face of negligible amino acid sequence conservation. J. Mol. Evol. 2007, 65, 277–288. [Google Scholar] [CrossRef]
- Marsh, J.A.; Teichmann, S.A. Protein flexibility facilitates quaternary structure assembly and evolution. PLoS Biol. 2014, 12, e1001870. [Google Scholar] [CrossRef] [PubMed]
- Feller, S.M.; Lewitzky, M. What’s in a loop? Cell Commun. Signal 2012, 10, 31. [Google Scholar] [CrossRef]
- Espadaler, J.; Querol, E.; Aviles, F.X.; Oliva, B. Identification of function-associated loop motifs and application to protein function prediction. Bioinformatics 2006, 22, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Berezovsky, I.N.; Grosberg, A.Y.; Trifonov, E.N. Closed loops of nearly standard size: Common basic element of protein structure. FEBS Lett. 2000, 466, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Goncearenco, A.; Berezovsky, I.N. Prototypes of elementary functional loops unravel evolutionary connections between protein functions. Bioinformatics 2010, 26, i497–i503. [Google Scholar] [CrossRef]
- Goncearenco, A.; Berezovsky, I.N. Exploring the evolution of protein function in Archaea. BMC Evol. Biol. 2012, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.F.; Caetano-Anollés, K.; Caetano-Anollés, G. The early history and emergence of molecular functions and modular scale-free network behavior. Sci. Rep. 2016, 6, 25058. [Google Scholar] [CrossRef]
- Schad, E.; Tompa, P.; Hegyi, H. The relationship between proteome size, structural disorder and organism complexity. Genome Biol. 2011, 12, R120. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunker, A.K.; Uversky, V.N. Orderly order in protein intrinsic disorder distribution: Disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 2012, 30, 137–149. [Google Scholar] [CrossRef]
- Xue, B.; Williams, R.W.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N. Archaic chaos: Intrinsically disordered proteins in Archaea. BMC Syst. Biol. 2010, 4, S1. [Google Scholar] [CrossRef]
- Basile, W.; Salvatore, M.; Bassot, C.; Elofsson, A. Why do eukaryotic proteins contain more intrinsically disordered regions? PLoS Comput. Biol. 2019, 15, e1007186. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.W.; Romero, P.; Uversky, V.N.; Dunker, A.K. Conservation of intrinsic disorder in protein domains and families: II. functions of conserved disorder. J. Proteome Res. 2006, 5, 888–898. [Google Scholar] [CrossRef]
- Debès, C.; Wang, M.; Caetano-Anollés, G.; Gräter, F. Evolutionary optimization of protein folding. PLoS Comput. Biol. 2013, 9, e1002861. [Google Scholar] [CrossRef]
- Aziz, M.F.; Mughal, F.; Caetano-Anollés, G. Tracing the birth of structural domains from loops during protein eolution. Sci. Rep. 2023, 13, 14688. [Google Scholar] [CrossRef]
- Bonet, J.; Planas-Iglesias, J.; Garcia-Garcia, J.; Marín-López, M.A.; Fernandez-Fuentes, N.; Oliva, B. ArchDB 2014: Structural classification of loops in proteins. Nucleic Acids Res. 2014, 42, D315–D319. [Google Scholar] [CrossRef] [PubMed]
- Conte, L.L.; Ailey, B.; Hubbard, T.J.; Brenner, S.E.; Murzin, A.G. SCOP: A structural classification of proteins database. Nucleic Acids Res. 2000, 28, 257–259. [Google Scholar] [CrossRef]
- Caetano-Anollés, G.; Nasir, A. Benefits of using molecular structure and abundance in phylogenomic analysis. Front. Genet. 2012, 3, 172. [Google Scholar] [CrossRef]
- Mughal, F.; Nasir, A.; Caetano-Anollés, G. The origin and evolution of viruses inferred from fold family structure. Arch. Virol. 2020, 165, 2177–2191. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Caetano-Anollés, G. The evolutionary history of protein fold families and proteomes confirms that the archaeal ancestor is more ancient than the ancestors of other superkingdoms. BMC Evol. Biol. 2012, 12, 13. [Google Scholar] [CrossRef]
- Kim, K.M.; Qin, T.; Jiang, Y.-Y.; Chen, L.L.; Xiong, M.; Caetano-Anollés, D.; Zhang, H.-Y.; Caetano-Anollés, G. Protein domain structure uncovers the origin of aerobic metabolism and the rise of planetary oxygen. Structure 2012, 20, 67–76. [Google Scholar] [CrossRef]
- Bao, Y.; Federhen, S.; Leipe, D.; Pham, V.; Resenchuk, S.; Rozanov, M.; Tatusov, R.; Tatusova, T. National center for biotechnology information viral genomes project. J. Virol. 2004, 78, 7291–7298. [Google Scholar] [CrossRef]
- Dosztányi, Z. Prediction of protein disorder based on IUPred. Protein Sci. 2018, 27, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yafremava, L.S.; Caetano-Anollés, D.; Mittenthal, J.E.; Caetano-Anollés, G. Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world. Genome Res. 2007, 17, 1572–1585. [Google Scholar] [CrossRef]
- Nasir, A.; Caetano-Anollés, G. A phylogenomic data-driven exploration of viral origins and evolution. Sci. Adv. 2015, 1, e1500527. [Google Scholar] [CrossRef]
- Kim, K.M.; Nasir, A.; Hwang, K.; Caetano-Anollés, G. A tree of cellular life inferred from a genomic census of molecular functions. J. Mol. Evol. 2014, 79, 240–262. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G.; Caetano-Anollés, D. An evolutionarily structured universe of protein architecture. Genome Res. 2003, 13, 1563–1571. [Google Scholar] [CrossRef]
- Wang, M.; Caetano-Anollés, G. The evolutionary mechanics of domain organization in proteomes and the rise of modularity in the protein world. Structure 2009, 17, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jiang, Y.-Y.; Kim, K.M.; Qu, G.; Ji, H.-F.; Mittenthal, J.E.; Zhang, H.-Y.; Caetano-Anollés, G. A universal molecular clock of protein folds and its power in tracing the early history of aerobic metabolism and planet oxygenation. Mol. Biol. Evol. 2011, 28, 567–582. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Conover, W.J. Rank tests for one sample, two samples, and k samples without the assumption of a continuous distribution function. Ann. Stat. 1973, 1, 1105–1125. [Google Scholar] [CrossRef]
- Zeng, C.; Zhan, W.; Deng, L. SDADB: A functional annotation database of protein structural domains. Database 2018, 2018, bay064. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, E.; Saladino, G.; Lambrughi, M.; Lindorff-Larsen, K.; Gervasion, F.L.; Nussinoc, R. The role of protein loops and linkers in conformational dynamics and allostery. Chem. Rev. 2016, 116, 6391–6423. [Google Scholar] [CrossRef]
- Eck, R.V.; Dayhoff, M.O. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science 1966, 152, 363–366. [Google Scholar] [CrossRef]
- Romero Romero, M.L.; Rabin, A.; Dan, S.; Tawfik, D.S. Functional Proteins from Short Peptides: Dayhoff’s Hypothesis Turns 50. Angew. Chem. Int. Ed. Engl. 2016, 55, 15966–15971. [Google Scholar] [CrossRef]
- Berezovsky, I.N. Towards descriptor of elementary functions for protein design. Curr. Opin. Struct. Biol. 2019, 58, 159–165. [Google Scholar] [CrossRef]
- Tee, W.-V.; Guarnera, E.; Berezovsky, I.N. Disorder driven allosteric control of protein activity. Curr. Res. Struct. Biol. 2020, 2, 191–203. [Google Scholar] [CrossRef]
- Tee, W.-V.; Tan, Z.W.; Guarnera, E.; Berezovsky, I.N. Conservation and diversity in allosteric fingerprints of proteins for evolutionary-inspired engineering and design. J. Mol. Biol. 2022, 434, 167577. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Fuentes, N.; Dybas, J.M.; Fiser, A. Structural characteristics of novel protein folds. PLoS Comput. Biol. 2010, 6, e1000750. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G.; Wang, M.; Caetano-Anollés, G.; Mittenthal, J.E. The origin, evolution and structure of the protein world. Biochem. J. 2009, 417, 621–637. [Google Scholar] [CrossRef]
- Goubau, D.; Deddouche, S.; Reis e Sousa, C. Cytosolic sensing of viruses. Immunity 2013, 38, 855–869. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Peisley, A.; Richards, C.; Yao, H.; Zeng, X.; Lin, C.; Chu, F.; Walz, T.; Hur, S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 2013, 152, 276–289. [Google Scholar] [CrossRef]
- Biesiadka, J.; Bujacz, G.; Sikorski, M.M.; Jaskolski, M. Crystal structures of two homologous pathogenesis-related proteins from yellow lupine. J. Mol. Biol. 2002, 319, 1223–1234. [Google Scholar] [CrossRef]
- Van Der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Marsh, J.A.; Teichmann, S.A. Parallel dynamics and evolution: Protein conformational fluctuations and assembly reflect evolutionary changes in sequence and structure. BioEssays 2014, 36, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Kempf, J.G.; Jung, J.-J.; Ragain, C.; Sampson, N.S.; Loria, J.P. Dynamic requirements for a functional protein hinge. J. Mol. Biol. 2007, 368, 131–149. [Google Scholar] [CrossRef]
- Goh, G.K.M.; Dunker, A.K.; Uversky, V.N. Protein intrinsic disorder toolbox for comparative analysis of viral proteins. BMC Genomics. 2008, 9, S4. [Google Scholar] [CrossRef]
- Marín, M.; Uversky, V.N.; Ott, T. Intrinsic disorder in pathogen effectors: Protein flexibility as an evolutionary hallmark in a molecular arms race. Plant Cell 2013, 25, 3153–3157. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.; Takayama, S.; Campen, A.M.; Vise, P.; Marshall, T.W.; Oldfield, C.J.; Williams, C.J.; Dunker, A.K. Evolutionary rate heterogeneity in proteins with long disordered regions. J. Mol. Evol. 2002, 55, 104–110. [Google Scholar] [CrossRef]
- Mughal, F.; Caetano-Anollés, G. MANET 3.0: Hierarchy and modularity in evolving metabolic networks. PLoS ONE 2019, 14, e0224201. [Google Scholar] [CrossRef]
- Romero Romero, M.L.; Yang, F.; Lin, Y.-R.; Tawfik, D.S. Simple yet functional phosphate-loop proteins. Proc. Natl. Acad. Sci. USA 2018, 115, E11943–E11950. [Google Scholar] [CrossRef]
- Mészáros, B.; Erdos, G.; Dosztányi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef] [PubMed]
- Mohan, A.; Sullivan, W.J.; Radivojac, P.; Dunker, A.K.; Uversky, V.N. Intrinsic disorder in pathogenic and non-pathogenic microbes: Discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol. Biosyst. 2008, 4, 328–340. [Google Scholar] [CrossRef]
- Schaefer, C.; Schlessinger, A.; Rost, B. Protein secondary structure appears to be robust under in silico evolution while protein disorder appears not to be. Bioinformatics 2010, 26, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Yeh, S.-W.; Liu, J.-W.; Yu, S.-H.; Shih, C.-H.; Hwang, J.-K.; Echave, J. Site-specific structural constraints on protein sequence evolutionary divergence: Local packing density versus solvent exposure. Mol. Biol. Evol. 2014, 31, 135–139. [Google Scholar] [CrossRef]
- Caetano-Anollés, G.; Aziz, M.F.; Mughal, F.; Caetano-Anollés, D. Tracing protein and proteome history with chronologies and networks: Folding recapitulates evolution. Expert Rev. Proteom. 2021, 18, 863–880. [Google Scholar] [CrossRef]
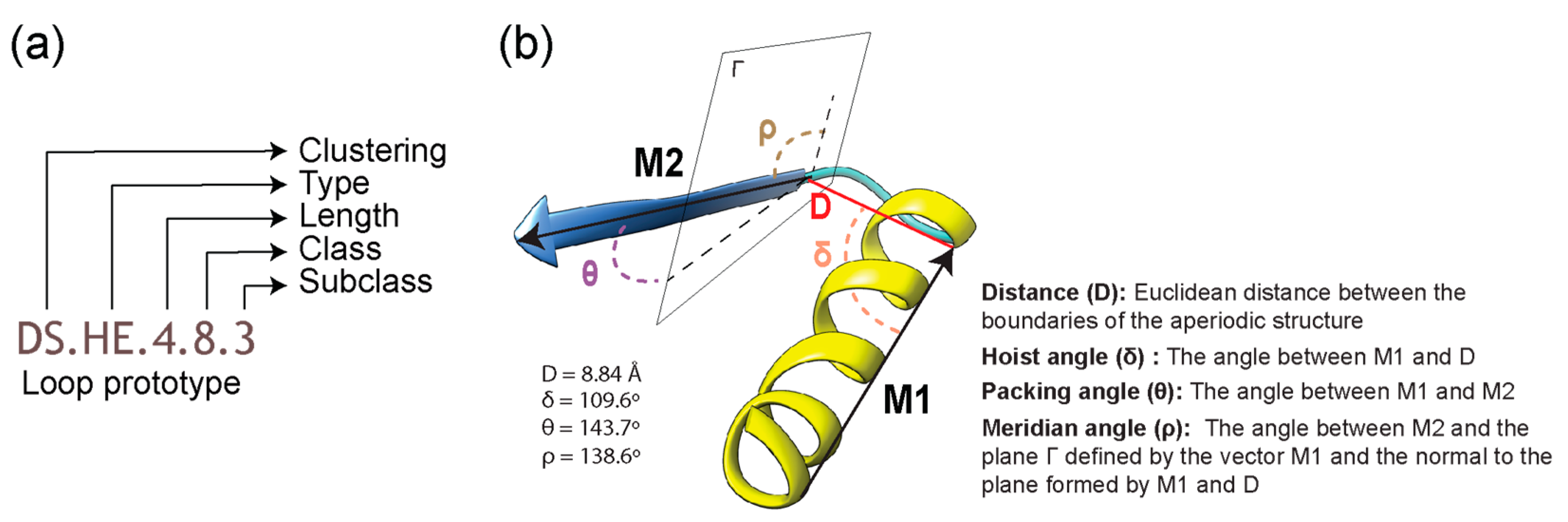
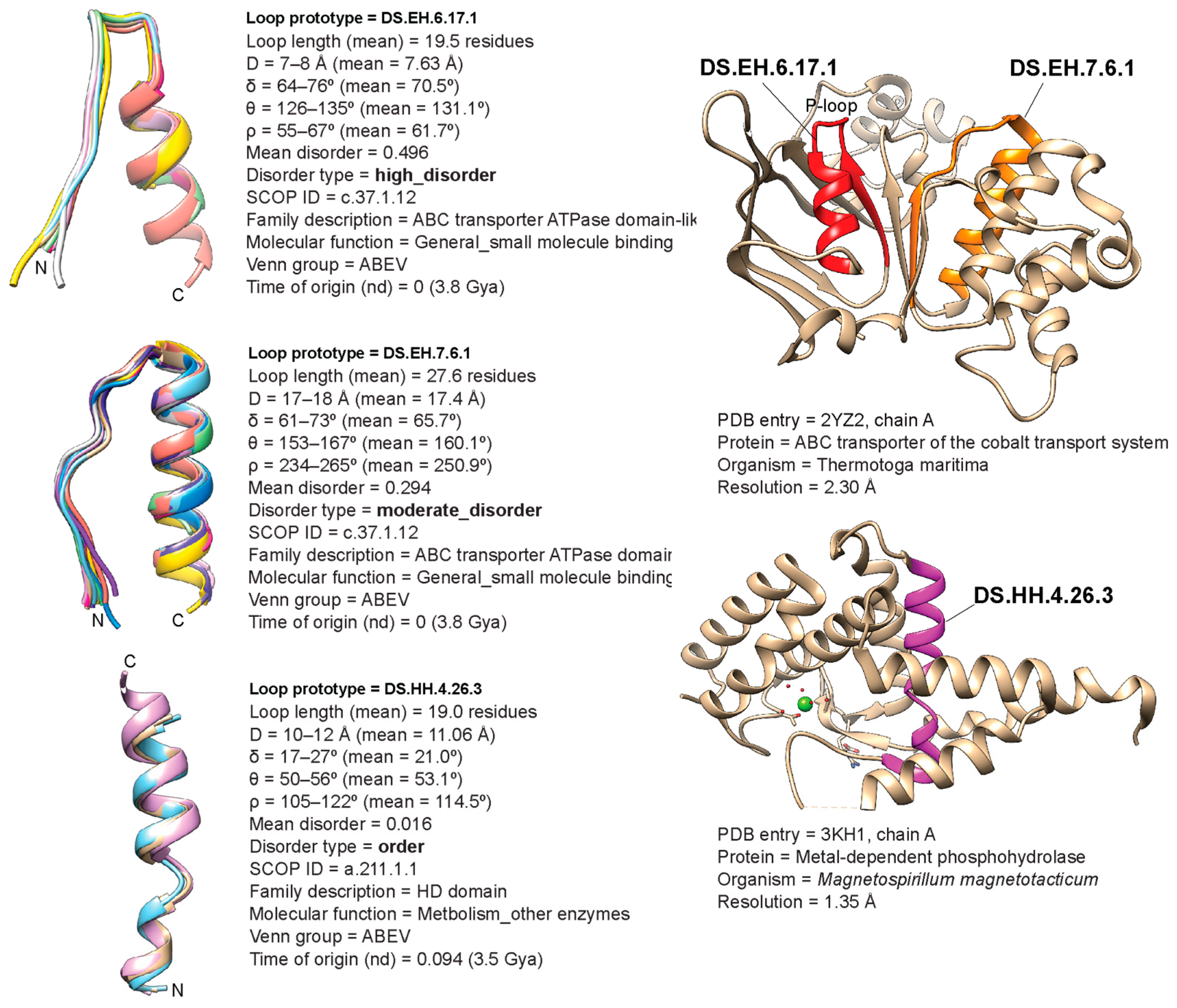
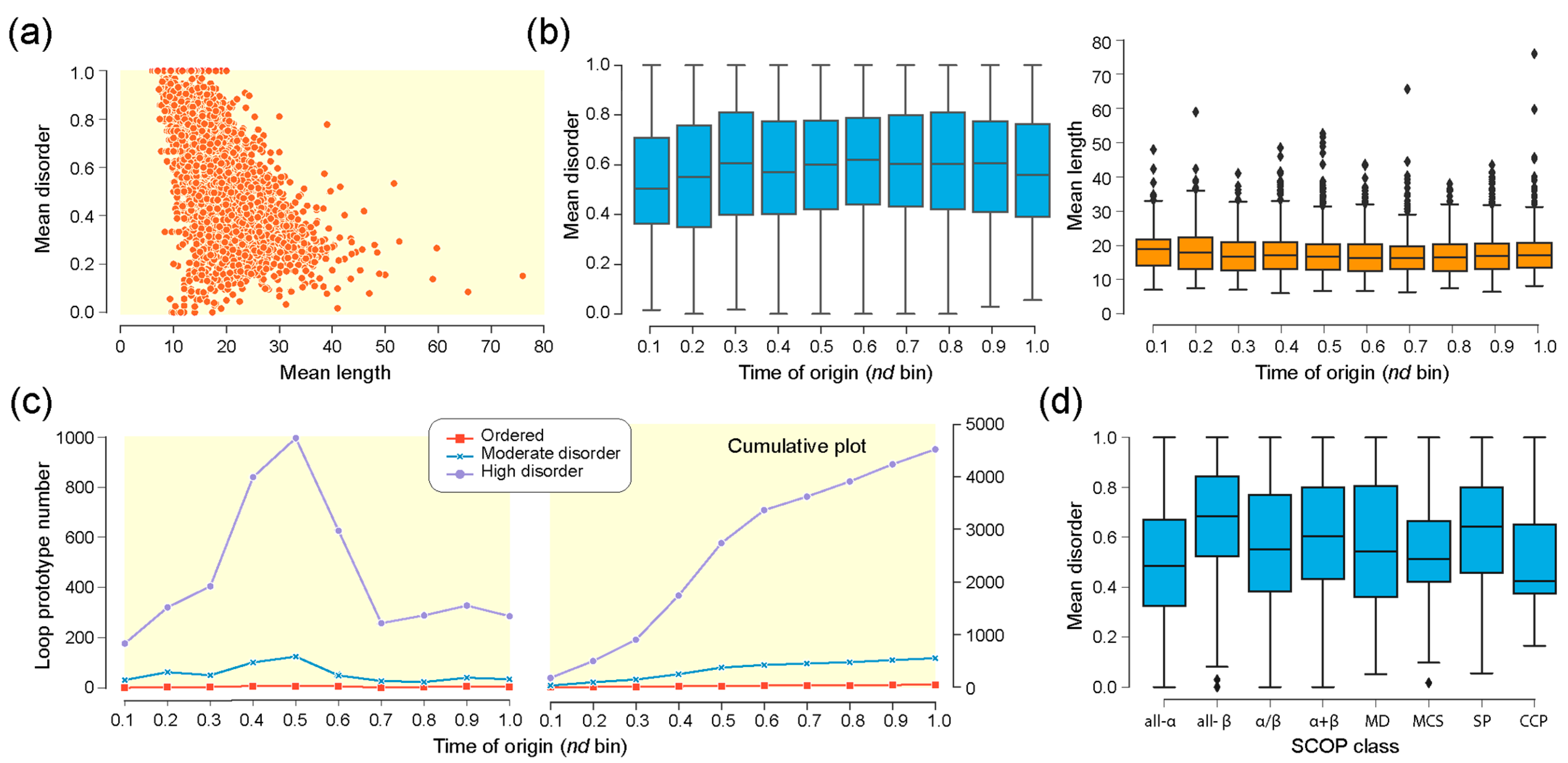
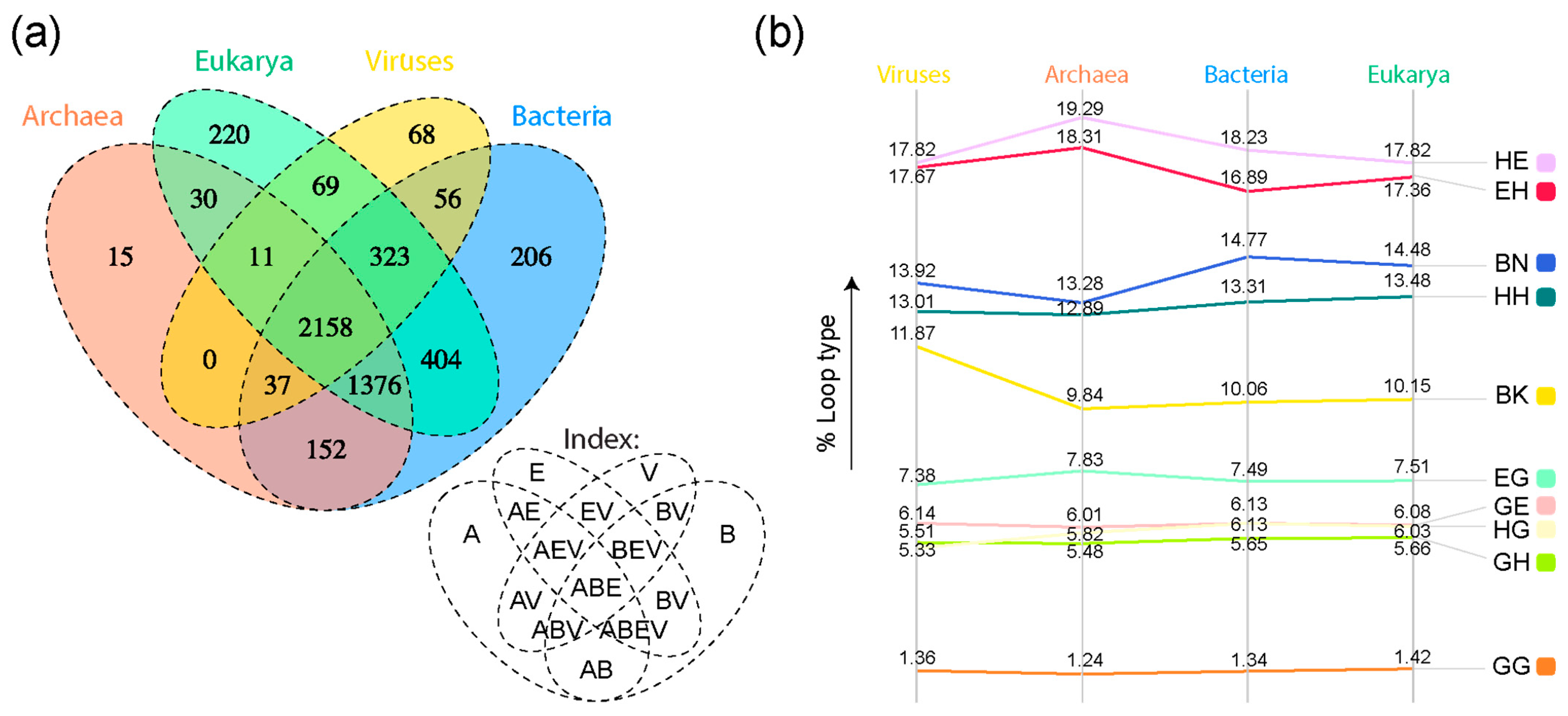
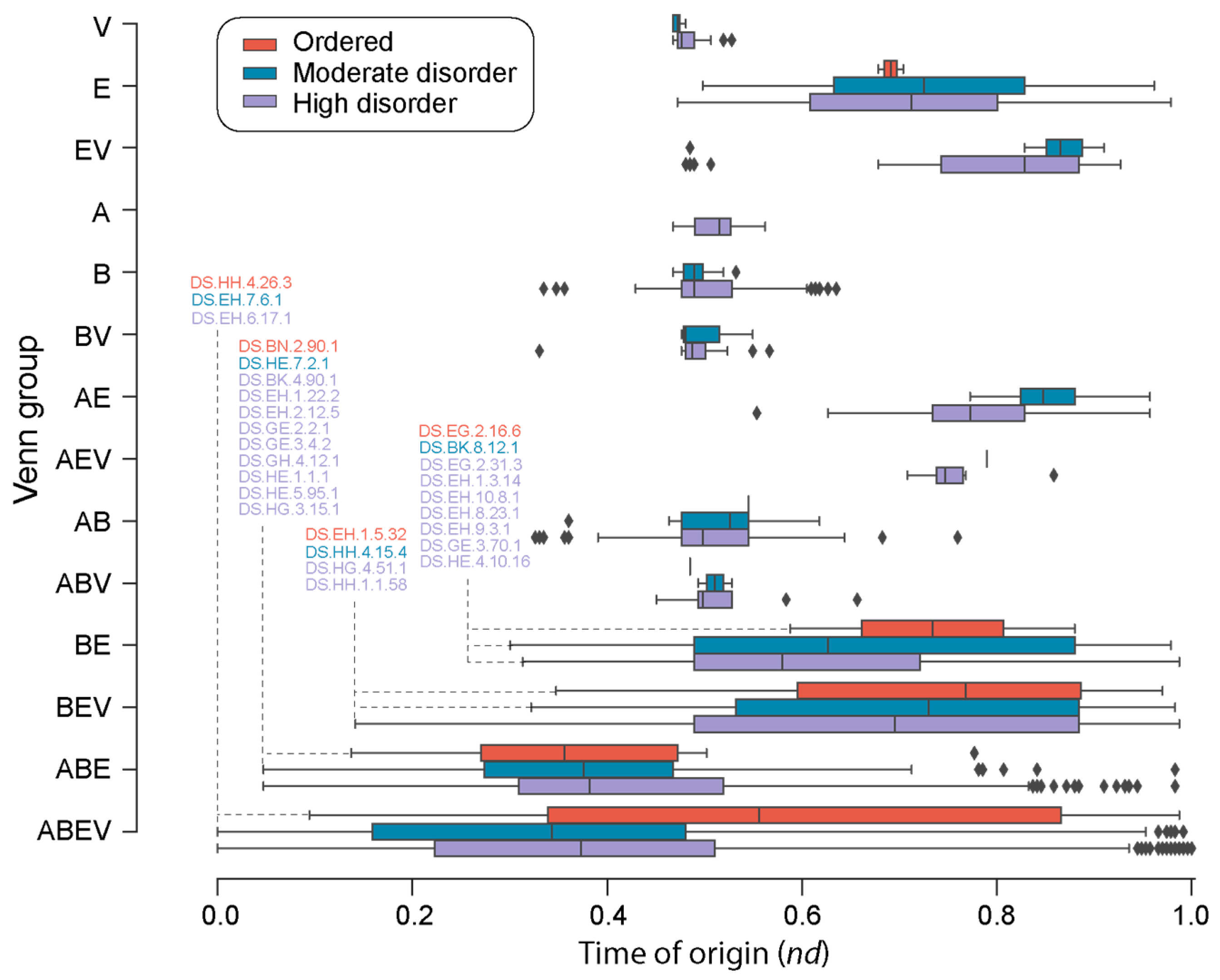

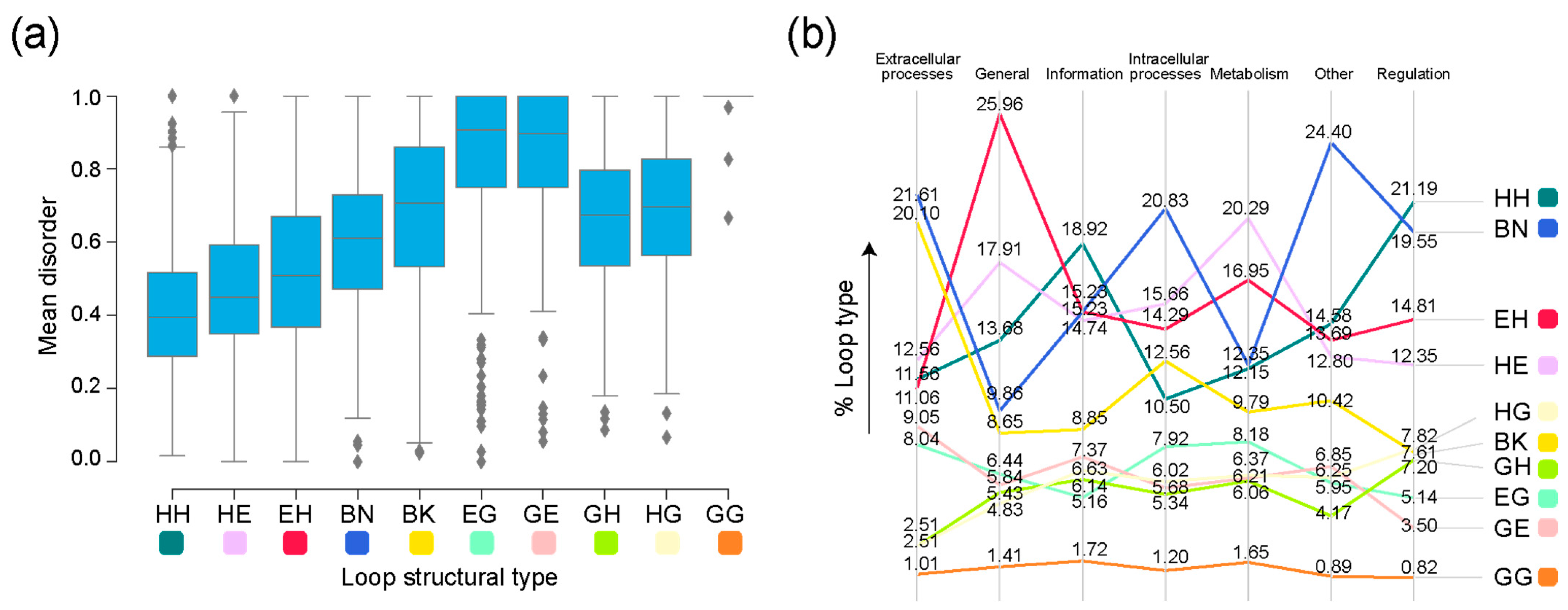
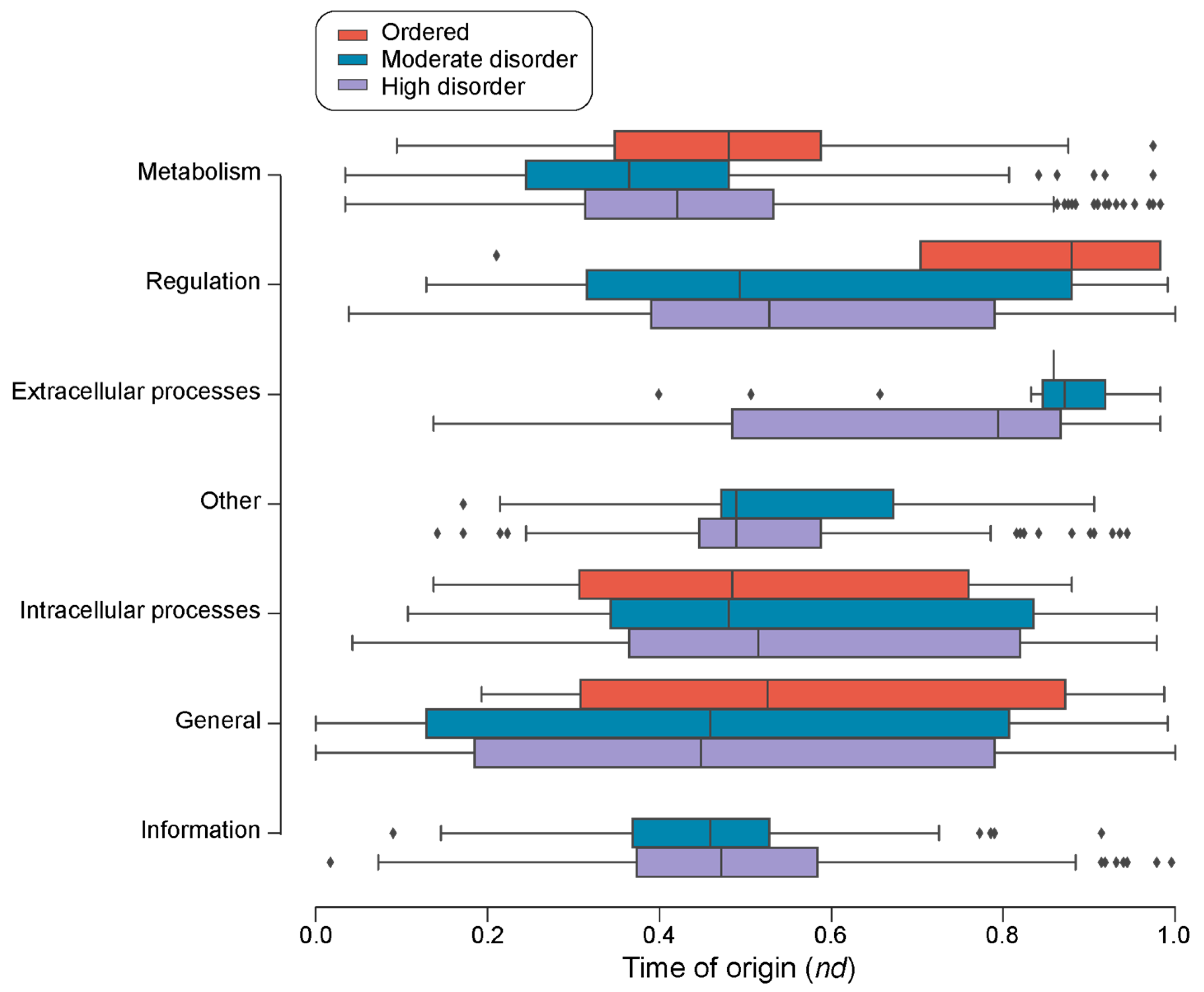

| Type | Bracing Secondary Structure |
|---|---|
| HH | alpha-alpha |
| HE | alpha-beta |
| EH | beta-alpha |
| BN | beta-beta hairpin |
| BK | beta-beta link |
| EG | beta-helix 310 |
| GE | helix 310-beta |
| GH | helix 310-helix |
| HG | helix-helix 310 |
| GG | helix 310-helix 310 |
| SCOP Class | All-α | All-β | α + β | α/β | CCP | MCS | MD | SP |
|---|---|---|---|---|---|---|---|---|
| all-α | −1 | 3.45 × 10−44 | 1.76 × 10−20 | 4.62 × 10−10 | 1 | 1 | 0.04501 | 4.66 × 10−5 |
| all-β | 3.45 × 10−44 | −1 | 1.21 × 10−8 | 4.36 × 10−24 | 0.31539 | 0.00021 | 0.00023 | 1 |
| α + β | 1.76 × 10−20 | 1.21 × 10−8 | −1 | 0.00038 | 1 | 0.32722 | 1 | 1 |
| α/β | 4.62 × 10−10 | 4.36 × 10−24 | 0.00038 | −1 | 1 | 1 | 1 | 0.48768 |
| CCP | 1 | 0.31539 | 1 | 1 | −1 | 1 | 1 | 1 |
| MCS | 1 | 0.00021 | 0.32722 | 1 | 1 | −1 | 1 | 0.35386 |
| MD | 0.04501 | 0.00023 | 1 | 1 | 1 | 1 | −1 | 1 |
| SP | 4.66 × 10−5 | 1 | 1 | 0.48768 | 1 | 0.35386 | 1 | −1 |
| GO ID | GO Description |
|---|---|
| GO:0000082 | G1/S transition of mitotic cell cycle |
| GO:0005975 | carbohydrate metabolic process |
| GO:0006099 | tricarboxylic acid cycle |
| GO:0006260 | DNA replication |
| GO:0006351 | transcription, DNA-templated |
| GO:0006355 | regulation of transcription, DNA-templated |
| GO:0006508 | proteolysis |
| GO:0006511 | ubiquitin-dependent protein catabolic process |
| GO:0006631 | fatty acid metabolic process |
| GO:0006633 | fatty acid biosynthetic process |
| GO:0006955 | immune response |
| GO:0007165 | signal transduction |
| GO:0009058 | biosynthetic process |
| GO:0009186 | deoxyribonucleoside diphosphate metabolic process |
| GO:0009405 | pathogenesis |
| GO:0015696 | ammonium transport |
| GO:0015976 | carbon utilization |
| GO:0019646 | aerobic electron transport chain |
| GO:0030245 | cellulose catabolic process |
| GO:0031388 | organic acid phosphorylation |
| GO:0043401 | steroid hormone mediated signaling pathway |
| GO:0055114 | oxidation–reduction process |
| GO:0072488 | ammonium transmembrane transport |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mughal, F.; Caetano-Anollés, G. Evolution of Intrinsic Disorder in Protein Loops. Life 2023, 13, 2055. https://doi.org/10.3390/life13102055
Mughal F, Caetano-Anollés G. Evolution of Intrinsic Disorder in Protein Loops. Life. 2023; 13(10):2055. https://doi.org/10.3390/life13102055
Chicago/Turabian StyleMughal, Fizza, and Gustavo Caetano-Anollés. 2023. "Evolution of Intrinsic Disorder in Protein Loops" Life 13, no. 10: 2055. https://doi.org/10.3390/life13102055
APA StyleMughal, F., & Caetano-Anollés, G. (2023). Evolution of Intrinsic Disorder in Protein Loops. Life, 13(10), 2055. https://doi.org/10.3390/life13102055









