Prmt7 Downregulation in Mouse Spermatogonia Functions through miR-877-3p/Col6a3
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatment
2.2. RNA Extraction and qRT-PCR
2.3. Protein Extraction and Western Blotting
2.4. miRNA Sequencing
2.5. miRNA Sequencing Data Analysis
2.6. Prediction and Functional Enrichment Analyses of miRNA Target Genes
2.7. Statistical Analysis
3. Results
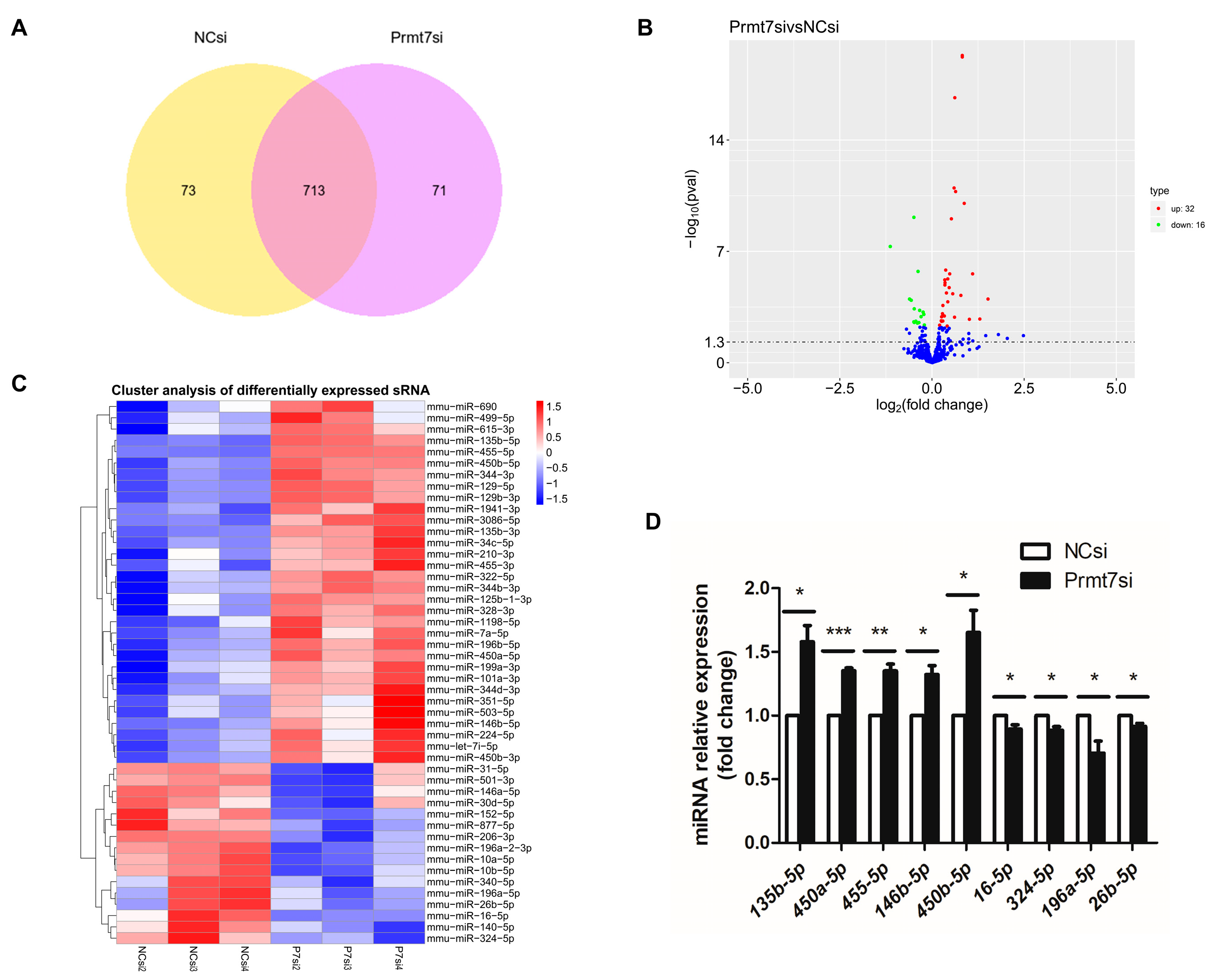
3.1. Validation of Prmt7 Knockdown in GC-1 Cells before miRNA Sequencing
3.2. Analysis of Differentially Expressed miRNAs upon Prmt7 Downregulation
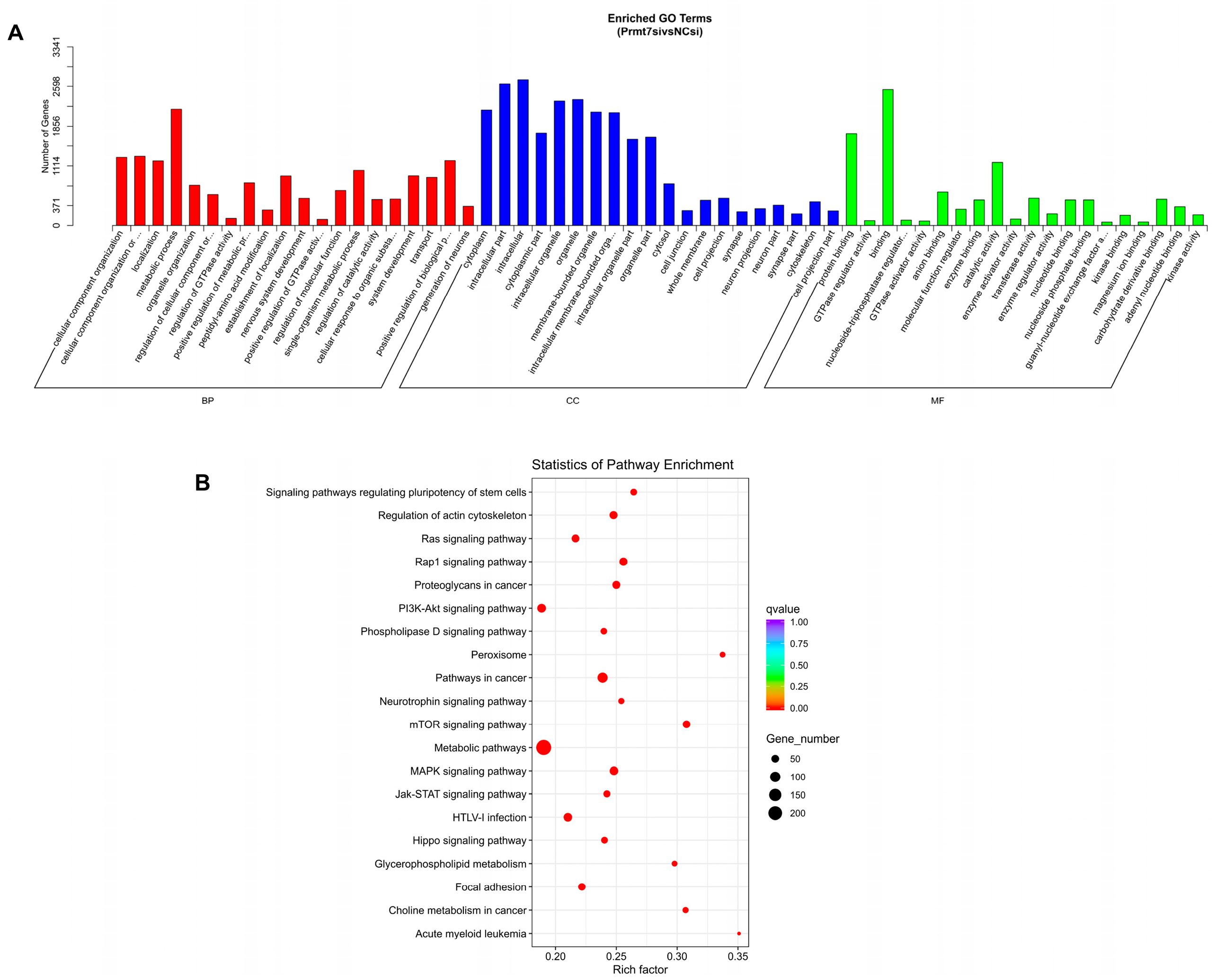
3.3. Functional Enrichment Analysis of Target Genes of DEmiRNAs
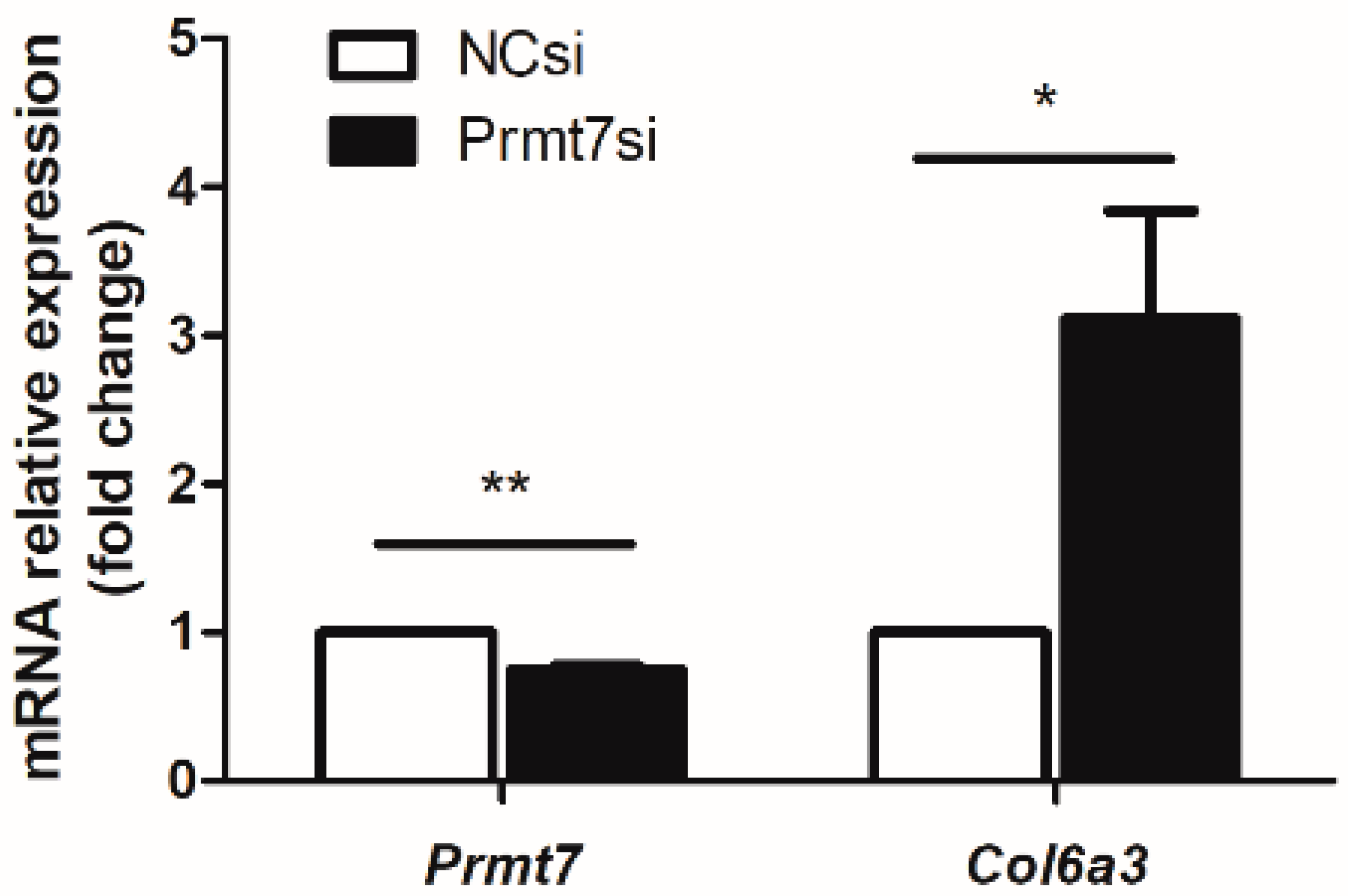
3.4. The Possible Molecular Mechanisms Underlying the GC-1 Proliferation Regulated by Prmt7/miRNA/Target Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Griswold, M.D. Spermatogenesis: The Commitment to Meiosis. Physiol. Rev. 2016, 96, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubota, H.; Brinster, R.L. Spermatogonial stem cells. Biol. Reprod. 2018, 99, 52–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, X.X.; Wang, J.; Wu, J. Extrinsic and intrinsic factors controlling spermatogonial stem cell self-renewal and differentiation. Asian J. Androl. 2015, 17, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Pfeiffer, M.J.; Drexler, H.C.; Fuellen, G.; Boiani, M. Proteomic Analysis of Mouse Oocytes Identifies PRMT7 as a Reprogramming Factor that Replaces SOX2 in the Induction of Pluripotent Stem Cells. J. Proteome Res. 2016, 15, 2407–2421. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Zhang, M.; Guo, J.; Liu, Z.; Zhou, R.; Guo, F.; Mu, Y. The Effects of Flavonoid Apigenin on Male Reproductive Health: Inhibition of Spermatogonial Proliferation through Downregulation of Prmt7/Akt3 Pathway. Int. J. Mol. Sci. 2021, 22, 12209. [Google Scholar] [CrossRef]
- Jelinic, P.; Stehle, J.C.; Shaw, P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006, 4, e355. [Google Scholar] [CrossRef]
- Chen, M.; Wang, Y.; Lin, L.; Dong, F.; Wu, H.; Bao, S.; Gao, F. PRMT7 is involved in regulation of germ cell proliferation during embryonic stage. BioChem. Biophys. Res. Commun. 2020, 533, 938–944. [Google Scholar] [CrossRef]
- Buhr, N.; Carapito, C.; Schaeffer, C.; Kieffer, E.; Van Dorsselaer, A.; Viville, S. Nuclear proteome analysis of undifferentiated mouse embryonic stem and germ cells. ElectrophoResis 2008, 29, 2381–2390. [Google Scholar] [CrossRef]
- Chen, T.Y.; Lee, S.H.; Dhar, S.S.; Lee, M.G. Protein arginine methyltransferase 7-mediated microRNA-221 repression maintains Oct4, Nanog, and Sox2 levels in mouse embryonic stem cells. J. Biol. Chem. 2018, 293, 3925–3936. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Li, X.; Guo, J.; Zhang, P.; Zeng, W. The roles of microRNAs in regulation of mammalian spermatogenesis. J. Anim. Sci. Biotechnol. 2017, 8, 35. [Google Scholar] [CrossRef]
- Bueno, M.J.; Malumbres, M. MicroRNAs and the cell cycle. Biochim. Biophys. Acta 2011, 1812, 592–601. [Google Scholar] [CrossRef] [Green Version]
- Lakshmipathy, U.; Love, B.; Goff, L.A.; Jörnsten, R.; Graichen, R.; Hart, R.P.; Chesnut, J.D. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem. Cells Dev. 2007, 16, 1003–1016. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Li, H.; Zeng, X.; Yang, P.; Liu, X.; Zhao, X.; Liang, S. Roles of microRNA on cancer cell metabolism. J. Transl. Med. 2012, 10, 228. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.W.; Mendell, J.T. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 2007, 96, R40–R44. [Google Scholar] [CrossRef]
- Liu, W.; Niu, Z.; Li, Q.; Pang, R.T.; Chiu, P.C.; Yeung, W.S. MicroRNA and Embryo Implantation. Am. J. Reprod. Immunol. 2016, 75, 263–271. [Google Scholar] [CrossRef]
- Kotaja, N. MicroRNAs and spermatogenesis. Fertil. Steril. 2014, 101, 1552–1562. [Google Scholar] [CrossRef]
- Wang, L.; Xu, C. Role of microRNAs in mammalian spermatogenesis and testicular germ cell tumors. Reproduction 2015, 149, R127–R137. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.; Hao, L.; Hu, F.; Dong, Y.; Gou, L.; Zhang, W.; Wang, X.; Zhao, Y.; Jia, M.; Hu, S.; et al. MicroRNA profiles and potential regulatory pattern during the early stage of spermatogenesis in mice. Sci. China Life Sci. 2015, 58, 442–450. [Google Scholar] [CrossRef] [Green Version]
- Huszar, J.M.; Payne, C.J. MicroRNA 146 (Mir146) modulates spermatogonial differentiation by retinoic acid in mice. Biol. Reprod. 2013, 88, 15. [Google Scholar] [CrossRef]
- Curry, E.; Safranski, T.J.; Pratt, S.L. Differential expression of porcine sperm microRNAs and their association with sperm morphology and motility. Theriogenology 2011, 76, 1532–1539. [Google Scholar] [CrossRef]
- He, Z.; Jiang, J.; Kokkinaki, M.; Tang, L.; Zeng, W.; Gallicano, I.; Dobrinski, I.; Dym, M. MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem. Cells 2013, 31, 2205–2217. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Tang, D.; Gao, J.; Dou, X.; Cheng, P.; Peng, D.; Zhang, Y.; Mao, J.; Zhang, L.; Zhang, X. miR-34c disrupts spermatogonial stem cell homeostasis in cryptorchid testes by targeting Nanos2. Reprod. Biol. Endocrinol. 2018, 16, 97. [Google Scholar] [CrossRef]
- Niu, Z.; Goodyear, S.M.; Rao, S.; Wu, X.; Tobias, J.W.; Avarbock, M.R.; Brinster, R.L. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA 2011, 108, 12740–12745. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.E.; Racicot, K.E.; Kaucher, A.V.; Oatley, M.J.; Oatley, J.M. MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development 2013, 140, 280–290. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Chen, T.Y.; Dhar, S.S.; Gu, B.; Chen, K.; Kim, Y.Z.; Li, W.; Lee, M.G. A feedback loop comprising PRMT7 and miR-24-2 interplays with Oct4, Nanog, Klf4 and c-Myc to regulate stemness. Nucleic. Acids Res. 2016, 44, 10603–10618. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Nishiyama, T.; Shimizu, K.; Kadota, K. TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinform. 2013, 14, 219. [Google Scholar] [CrossRef] [Green Version]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Akawi, N.; McRae, J.; Ansari, M.; Balasubramanian, M.; Blyth, M.; Brady, A.F.; Clayton, S.; Cole, T.; Deshpande, C.; Fitzgerald, T.W.; et al. Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nat. Genet. 2015, 47, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, B.; Pan, N.; Zhang, H.; Xing, C.; Ji, W. miR-146b-5p inhibits tumorigenesis and metastasis of gallbladder cancer by targeting Toll-like receptor 4 via the nuclear factor-κB pathway. Oncol. Rep. 2021, 45, 15. [Google Scholar] [CrossRef]
- Peng, Y.; Fang, X.; Yao, H.; Zhang, Y.; Shi, J. MiR-146b-5p Regulates the Expression of Long Noncoding RNA MALAT1 and Its Effect on the Invasion and Proliferation of Papillary Thyroid Cancer. Cancer Biother. Radiopharm. 2021, 36, 433–440. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Dong, Y.; Fan, Y.; Li, Y.; Zhao, C.; Wang, C.; Liu, J.; Li, X.; Dong, M.; et al. MiR-146b-5p functions as a suppressor miRNA and prognosis predictor in non-small cell lung cancer. J. Cancer 2017, 8, 1704–1716. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Wu, G.; Yan, W.; Zhan, H.; Sun, P. miR-146b-5p regulates cell growth, invasion, and metabolism by targeting PDHB in colorectal cancer. Am. J. Cancer Res. 2017, 7, 1136–1150. [Google Scholar]
- Lin, F.; Wang, X.; Jie, Z.; Hong, X.; Li, X.; Wang, M.; Yu, Y. Inhibitory effects of miR-146b-5p on cell migration and invasion of pancreatic cancer by targeting MMP16. J. Huazhong Univ. Sci. Technol. Med. Sci. 2011, 31, 509. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, M.; Dai, M.; Chen, C.; Tang, Q.; Jing, W.; Wang, H.; Tian, W. miR-450a-5p within rat adipose tissue exosome-like vesicles promotes adipogenic differentiation by targeting WISP2. J. Cell Sci. 2017, 130, 1158–1168. [Google Scholar] [CrossRef] [Green Version]
- Cheng, D.; He, Z.; Zheng, L.; Xie, D.; Dong, S.; Zhang, P. PRMT7 contributes to the metastasis phenotype in human non-small-cell lung cancer cells possibly through the interaction with HSPA5 and EEF2. Onco. Targets 2018, 11, 4869–4876. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Wan, L.; Zou, H.; Pan, Z.; Zhou, W.; Lu, X. PRMT7 promotes the growth of renal cell carcinoma through modulating the β-catenin/C-MYC axis. Int. J. Biochem. Cell Biol. 2020, 120, 105686. [Google Scholar] [CrossRef]
- Liu, Y.; Li, L.; Liu, X.; Wang, Y.; Liu, L.; Peng, L.; Zhang, Y. Arginine methylation of SHANK2 by PRMT7 promotes human breast cancer metastasis through activating endosomal FAK signalling. eLife 2020, 9, e57617. [Google Scholar] [CrossRef]
- Jeong, H.J.; Lee, H.J.; Vuong, T.A.; Choi, K.S.; Choi, D.; Koo, S.H.; Cho, S.C.; Cho, H.; Kang, J.S. Prmt7 Deficiency Causes Reduced Skeletal Muscle Oxidative Metabolism and Age-Related Obesity. Diabetes 2016, 65, 1868–1882. [Google Scholar] [CrossRef] [Green Version]
- Agolini, E.; Dentici, M.L.; Bellacchio, E.; Alesi, V.; Radio, F.C.; Torella, A.; Musacchia, F.; Tartaglia, M.; Dallapiccola, B.; Nigro, V.; et al. Expanding the clinical and molecular spectrum of PRMT7 mutations: 3 additional patients and review. Clin. Genet. 2018, 93, 675–681. [Google Scholar] [CrossRef] [Green Version]
- Leem, Y.E.; Bae, J.H.; Jeong, H.J.; Kang, J.S. PRMT7 deficiency enhances adipogenesis through modulation of C/EBP-β. BioChem. Biophys. Res. Commun. 2019, 517, 484–490. [Google Scholar] [CrossRef]
- Fiorica, P.N.; Wheeler, H.E. Transcriptome association studies of neuropsychiatric traits in African Americans implicate PRMT7 in schizophrenia. PeerJ 2019, 7, e7778. [Google Scholar] [CrossRef] [Green Version]
- Yao, R.; Jiang, H.; Ma, Y.; Wang, L.; Wang, L.; Du, J.; Hou, P.; Gao, Y.; Zhao, L.; Wang, G.; et al. PRMT7 induces epithelial-to-mesenchymal transition and promotes metastasis in breast cancer. Cancer Res. 2014, 74, 5656–5667. [Google Scholar] [CrossRef] [Green Version]
- Alzahrani, A.S. PI3K/Akt/mTOR inhibitors in cancer: At the bench and bedside. Semin. Cancer Biol. 2019, 59, 125–132. [Google Scholar] [CrossRef]
- Schaefer, T.; Steiner, R.; Lengerke, C. SOX2 and p53 Expression Control Converges in PI3K/AKT Signaling with Versatile Implications for Stemness and Cancer. Int. J. Mol. Sci. 2020, 21, 4902. [Google Scholar] [CrossRef]
- Gutiérrez-Venegas, G.; González-Rosas, Z. Apigenin reduce lipoteichoic acid-induced inflammatory response in rat cardiomyoblast cells. Arch. Pharm. Res. 2017, 40, 240–249. [Google Scholar] [CrossRef]
- Duan, Y.; Liu, G.; Sun, Y.; Wu, J.; Xiong, Z.; Jin, T.; Chen, M. COL6A3 polymorphisms were associated with lung cancer risk in a Chinese population. Respir. Res. 2019, 20, 143. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Li, J.; Luo, H.; Song, W.; Yang, J. Differential Expression of COL1A1, COL1A2, COL6A3, and SULF1 as Prognostic Biomarkers in Gastric Cancer. Int. J. Gen. Med. 2021, 14, 5835–5843. [Google Scholar] [CrossRef]
- Das Bhowmik, A.; Karthik, T.; Uppin, M.; Sundaram, C.; Dalal, A. Targeted next generation sequencing reveals novel splice site mutations in COL6A3 gene in a patient with congenital muscular dystrophy. Neurol. India 2018, 66, 1812–1814. [Google Scholar]
- Pan, T.C.; Zhang, R.Z.; Markova, D.; Arita, M.; Zhang, Y.; Bogdanovich, S.; Khurana, T.S.; Bönnemann, C.G.; Birk, D.E.; Chu, M.L. COL6A3 protein deficiency in mice leads to muscle and tendon defects similar to human collagen VI congenital muscular dystrophy. J. Biol. Chem. 2013, 288, 14320–14331. [Google Scholar] [CrossRef] [Green Version]
- Dankel, S.N.; Svärd, J.; Matthä, S.; Claussnitzer, M.; Klöting, N.; Glunk, V.; Fandalyuk, Z.; Grytten, E.; Solsvik, M.H.; Nielsen, H.J.; et al. COL6A3 expression in adipocytes associates with insulin resistance and depends on PPARγ and adipocyte size. Obesity 2014, 22, 1807–1813. [Google Scholar] [CrossRef]
- McCulloch, L.J.; Rawling, T.J.; Sjöholm, K.; Franck, N.; Dankel, S.N.; Price, E.J.; Knight, B.; Liversedge, N.H.; Mellgren, G.; Nystrom, F.; et al. COL6A3 is regulated by leptin in human adipose tissue and reduced in obesity. Endocrinology 2015, 156, 134–146. [Google Scholar] [CrossRef] [Green Version]

| miRNA | Primer Sequences | |
|---|---|---|
| Prmt7 | Forward primer | GGAGATTGCCAGGTCATCCT |
| Reverse primer | GAAGTCAGCCCCTGCAGTAA | |
| Col6a3 | Forward primer | GAGATGGGGTTGGCAGTGAA |
| Reverse primer | GCATTGATCAACGGAGTCGC | |
| β-actin | Forward primer | TGTGCTGTCCCTGTATGCCTCT |
| Reverse primer | TAGATGGGCACAGTGTGGGTGA | |
| mmu-miR-135b-5p | Forward primer | CGCGTATGGCTTTTCATTCCTATGTGA |
| mmu-miR-450a-5p | Forward primer | CGCGCTTTTGCGATGTGTTCCTAATAT |
| mmu-miR-455-5p | Forward primer | CGCTATGTGCCTTTGGACTACATCG |
| mmu-miR-146b-5p | Forward primer | CCGCTGAGAACTGAATTCCATAGGCT |
| mmu-miR-450b-5p | Forward primer | GCGCGTTTTGCAGTATGTTCCTGAATA |
| mmu-miR-16-5p | Forward primer | CCGTAGCAGCACGTAAATATTGGCG |
| mmu-miR-324-5p | Forward primer | TATATACGCATCCCCTAGGGCATTGG |
| mmu-miR-196a-5p | Forward primer | CGCGTAGGTAGTTTCATGTTGTTGGG |
| mmu-miR-26b-5p | Forward primer | GCGCGTTCAAGTAATTCAGGATAGGT |
| Intersection Gene | Log2 FC | miRNA | Log2 FC |
|---|---|---|---|
| Fut7 | −1.62 | mmu-miR-135b-3p | 1.29 |
| Atp2c2 | 1.25 | mmu-miR-140-5p | −0.22 |
| Gm14406 | 1.38 | mmu-miR-30d-5p | −0.40 |
| Cracdl | 1.03 | mmu-miR-31-5p | −0.57 |
| Hmcn2 | 1.12 | mmu-miR-324-5p | −0.50 |
| Lefty2 | 1.69 | mmu-miR-501-3p | −0.49 |
| Col6a3 | 1.08 | mmu-miR-877-5p | −0.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, H.; Zhang, M.; Guo, J.; Liu, Z.; Guo, F.; Wang, B.; Mu, Y. Prmt7 Downregulation in Mouse Spermatogonia Functions through miR-877-3p/Col6a3. Life 2022, 12, 1194. https://doi.org/10.3390/life12081194
Gao H, Zhang M, Guo J, Liu Z, Guo F, Wang B, Mu Y. Prmt7 Downregulation in Mouse Spermatogonia Functions through miR-877-3p/Col6a3. Life. 2022; 12(8):1194. https://doi.org/10.3390/life12081194
Chicago/Turabian StyleGao, Hongmei, Mingrui Zhang, Jiankang Guo, Zhiguo Liu, Fei Guo, Bingyuan Wang, and Yulian Mu. 2022. "Prmt7 Downregulation in Mouse Spermatogonia Functions through miR-877-3p/Col6a3" Life 12, no. 8: 1194. https://doi.org/10.3390/life12081194
APA StyleGao, H., Zhang, M., Guo, J., Liu, Z., Guo, F., Wang, B., & Mu, Y. (2022). Prmt7 Downregulation in Mouse Spermatogonia Functions through miR-877-3p/Col6a3. Life, 12(8), 1194. https://doi.org/10.3390/life12081194





