Growth Traits and Sperm Proteomics Analyses of Myostatin Gene-Edited Chinese Yellow Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Construction of CRISPR/Cas9 Vectors
2.3. Transfection of Bovine Fetal Fibroblast Cells with CRISPR/Cas9 Cutting Vector and Screening of Positive Gene-Edited Cells
2.4. Production of MSTN Gene-Edited Bulls by SCNT
2.5. Growth Parameters Detection
2.6. Blood Biochemical Analysis of Cattle
2.7. Semen and Sperm Analysis
2.8. Sperm Protein Extraction and Digestion
2.9. LC-MS/MS Analysis
2.10. Bioinformatics Analysis
2.11. Statistical Analysis
3. Results
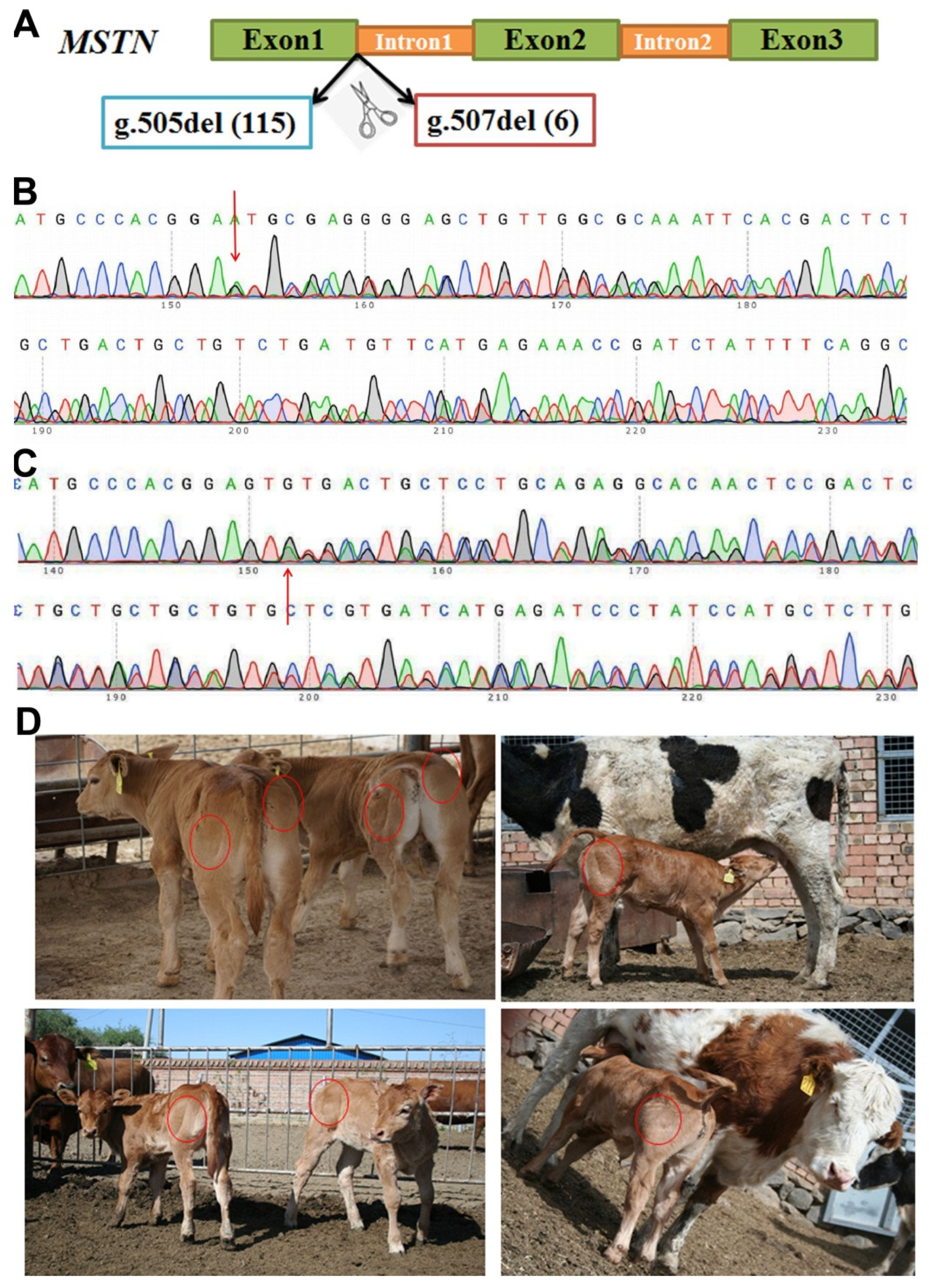
3.1. Generation and Identification of MSTN Gene-Edited Chinese Yellow Cattle
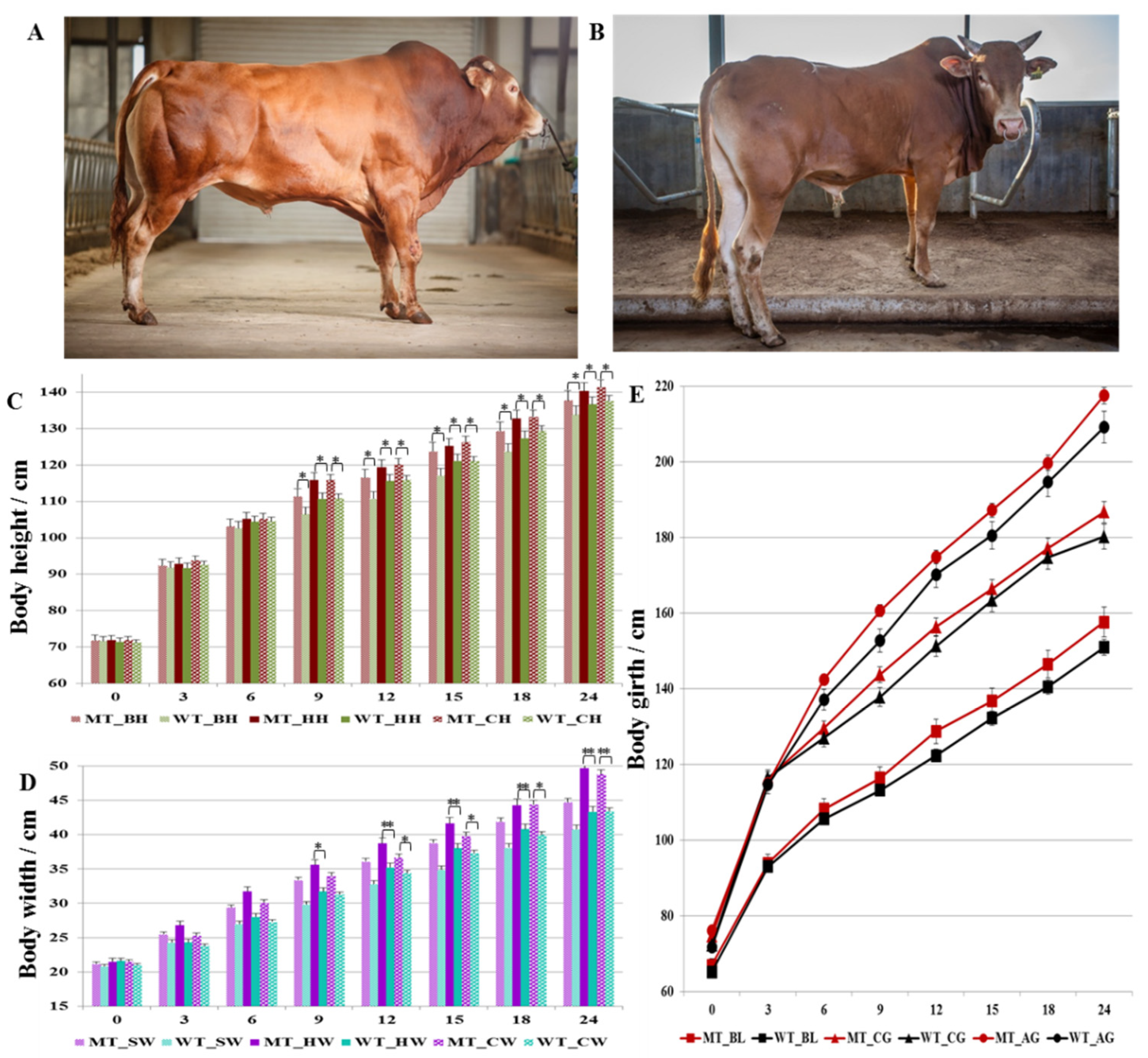
3.2. Analysis of Growth Traits of MSTN Gene-Edited Cattle
3.2.1. Body Weight of MSTN Gene-Edited Cattle
3.2.2. Body Size of MSTN Gene-Edited Cattle
3.3. The Physiological and Biochemical Indexes of MSTN Gene-Edited Cattle
3.4. Semen Characteristics of MSTN Gene Edited Cattle
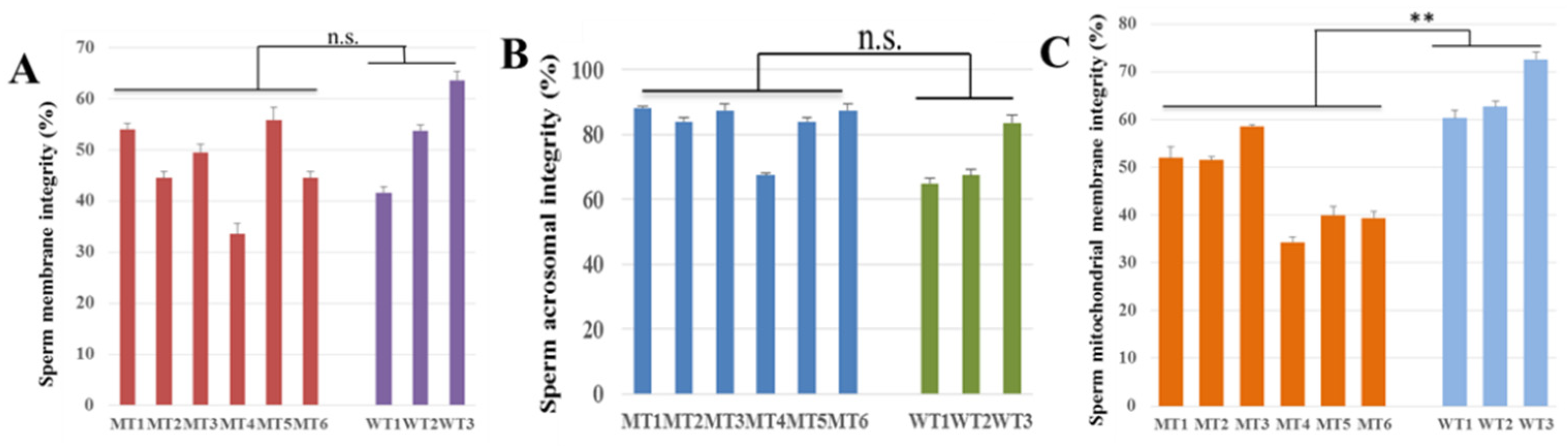
3.5. Characteristics of Cryopreserved Spermatozoa of MSTN Gene-Edited Cattle
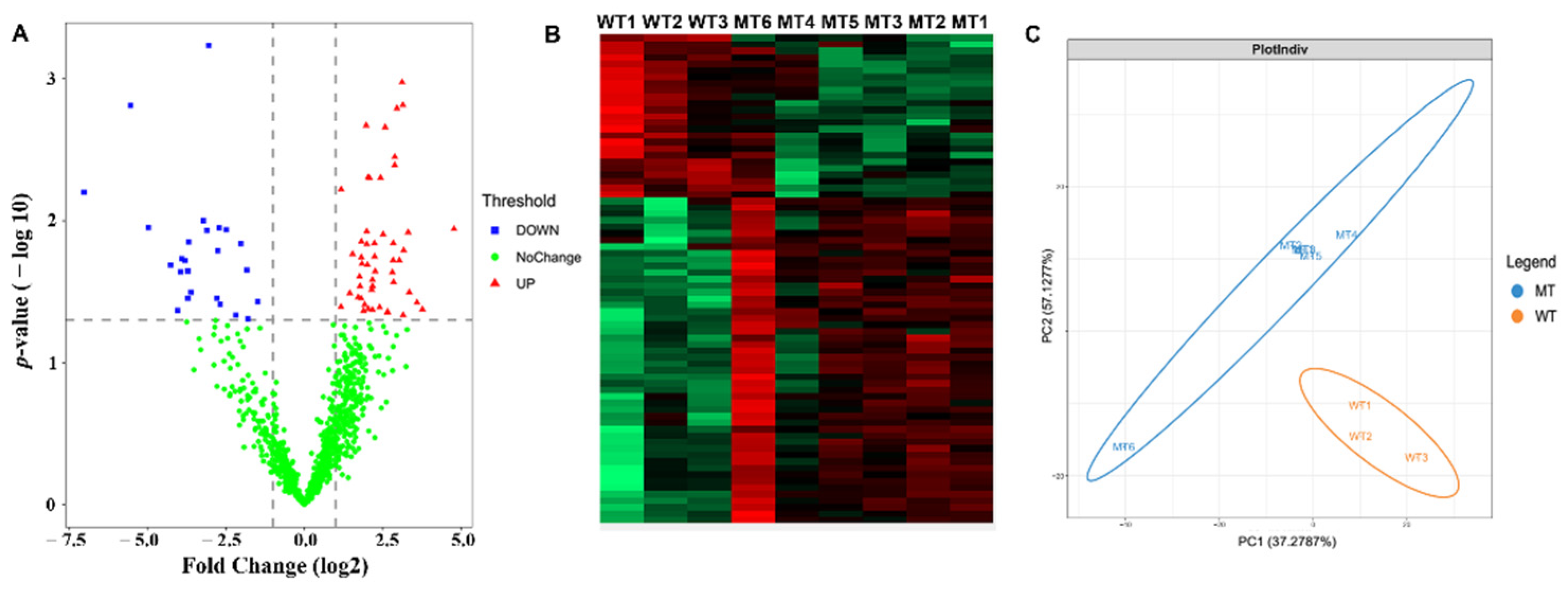
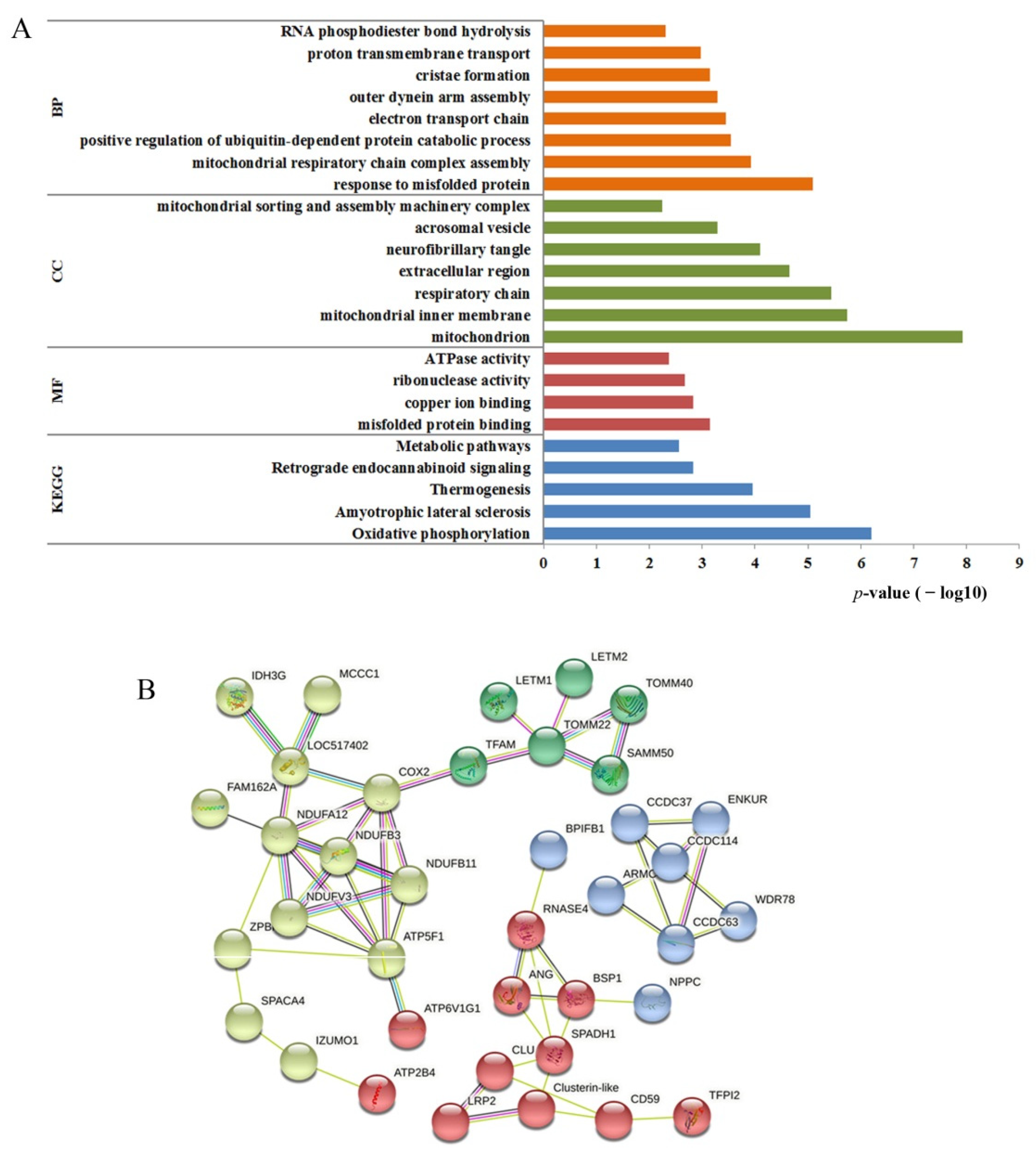
3.6. Proteomic Analysis of Cryopreserved Spermatozoa of MSTN Gene-Edited Cattle
3.7. The Fertilization Ability of Cryopreserved Semen of MSTN Gene-Edited Cattle
4. Discussion
4.1. Effects of MSTN Mutation on Bovine Growth Traits
4.2. Effects of MSTN Mutation on Physiological and Biochemical Indexes of Cattle
4.3. Effects of MSTN Mutation on Bovine Semen and Spermatozoa
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kambadur, R.; Sharma, M.; Smith, T.P.; Bass, J. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997, 7, 910–915. [Google Scholar] [CrossRef] [PubMed]
- Phocas, F. Genetic analysis of breeding traits in a Charolais cattle population segregating an inactive myostatin allele. J. Anim. Sci. 2009, 87, 1865–1871. [Google Scholar] [CrossRef]
- Esmailizadeh, A.; Bottema, C.; Sellick, G.; Verbyla, A.; Morris, C.; Cullen, N.; Pitchford, W. Effects of the myostatin F94L substitution on beef traits. J. Anim Sci. 2008, 86, 1038–1046. [Google Scholar] [CrossRef] [PubMed]
- Bouyer, C.; Forestier, L.; Renand, G.; Oulmouden, A. Deep intronic mutation and pseudo exon activation as a novel muscular hypertrophy modifier in cattle. PLoS ONE 2014, 9, e97399. [Google Scholar] [CrossRef] [PubMed]
- Dierks, C.; Eder, J.; Glatzer, S.; Lehner, S.; Distl, O. A novel myostatin mutation in double-muscled German Gelbvieh. Anim. Genet. 2015, 46, 91–92. [Google Scholar] [CrossRef] [PubMed]
- Arthur, P. Double muscling in cattle: A review. Aus. J. Agric. Res. 1995, 46, 1493–1515. [Google Scholar] [CrossRef]
- Casas, E.; Keele, J.W.; Shackelford, S.D.; Koohmaraie, M.; Sonstegard, T.S.; Smith, T.P.; Kappes, S.M.; Stone, R.T. Association of the muscle hypertrophy locus with carcass traits in beef cattle. J. Anim. Sci. 1998, 76, 468–473. [Google Scholar] [CrossRef][Green Version]
- Casas, E.; Keele, J.W.; Fahrenkrug, S.C.; Smith, T.P.; Cundiff, L.V.; Stone, R.T. Quantitative analysis of birth, weaning, and yearling weights and calving difficulty in Piedmontese crossbreds segregating an inactive myostatin allele. J. Anim. Sci. 1999, 77, 1686–1692. [Google Scholar] [CrossRef]
- Wheeler, T.; Cundiff, L.; Shackelford, S.; Koohmaraie, M. Characterization of biological types of cattle (Cycle V): Carcass traits and longissimus palatability. J. Anim. Sci. 2001, 79, 1209–1222. [Google Scholar] [CrossRef]
- Short, R.; MacNeil, M.; Grosz, M.; Gerrard, D.; Grings, E. Pleiotropic effects in Hereford, Limousin, and Piedmontese F2 crossbred calves of genes controlling muscularity including the Piedmontese myostatin allele. J. Anim. Sci. 2002, 80, 1–11. [Google Scholar] [CrossRef]
- Wiener, P.; Smith, J.A.; Lewis, A.M.; Woolliams, J.A.; Williams, J.L. Muscle-related traits in cattle: The role of the myostatin gene in the South Devon breed. Genet. Sel. Evol. 2002, 34, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Casas, E.; Bennett, G.; Smith, T.; Cundiff, L. Association of myostatin on early calf mortality, growth, and carcass composition traits in crossbred cattle. J. Anim. Sci. 2004, 82, 2913–2918. [Google Scholar] [CrossRef] [PubMed]
- Casas, E.; Keele, J.W.; Shackelford, S.D.; Koohmaraie, M.; Stone, R.T. Identification of quantitative trait loci for growth and carcass composition in cattle. Anim. Genet. 2004, 35, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Casas, E.; Lunstra, D.D.; Stone, R.T. Quantitative trait loci for male reproductive traits in beef cattle. Anim. Genet. 2004, 35, 451–453. [Google Scholar] [CrossRef] [PubMed]
- Sellick, G.S.; Pitchford, W.; Morris, C.; Cullen, N.; Crawford, A.; Raadsma, H.; Bottema, C. Effect of myostatin F94L on carcass yield in cattle. Anim. Genet. 2007, 38, 440–446. [Google Scholar] [CrossRef]
- Allais, S.; Levéziel, H.; Hocquette, J.-F.; Rousset, S.; Denoyelle, C.; Journaux, L.; Renand, G. Fine mapping of quantitative trait loci underlying sensory meat quality traits in three French beef cattle breeds. J. Anim. Sci. 2014, 92, 4329–4341. [Google Scholar] [CrossRef]
- Li, G.P.; Bunch, T.D.; White, K.L.; Aston, K.I.; Meerdo, L.N.; Pate, B.J.; Sessions, B.R. Development, chromosomal composition, and cell allocation of bovine cloned blastocyst derived from chemically assisted enucleation and cultured in conditioned media. Mol. Reprod. Dev. 2004, 68, 189–197. [Google Scholar] [CrossRef]
- Li, G.P.; White, K.L.; Aston, K.I.; Bunch, T.D.; Hicks, B.; Liu, Y.; Sessions, B. Colcemid-treatment of heifer oocytes enhances nuclear transfer embryonic development, establishment of pregnancy and development to term. Mol. Reprod. Dev. 2009, 76, 620–628. [Google Scholar] [CrossRef]
- Wu, D.; Gu, M.; Wei, Z.; Bai, C.; Su, G.; Liu, X.; Zhao, Y.; Yang, L.; Li, G. Myostatin Knockout Regulates Bile Acid Metabolism by Promoting Bile Acid Synthesis in Cattle. Animals 2022, 12, 205. [Google Scholar] [CrossRef]
- Su, G.; Wu, S.; Wu, M.; Wang, L.; Yang, L.; Du, M.; Zhao, X.; Su, X.; Liu, X.; Bai, C.; et al. Melatonin improves the quality of frozen bull semen and influences gene expression related to embryo genome activation. Theriogenology 2021, 176, 54–62. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, X.; Li, X.; Liu, C.; Yang, M.; Han, B.; Hai, C.; Su, G.; Li, G.; Zhao, Y. Cysteine is highly enriched in the canonical N-linked glycosylation motif of bovine spermatozoa N-Glycoproteome. Theriogenology 2022, 184, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bibikova, M.; Golic, M.; Golic, K.G.; Carroll, D. Targeted-chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 2002, 161, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Huang, S.; Jiang, W.Z.; Wright, D.; Spalding, M.H.; Weeks, D.P.; Yang, B. TAL nucleases (TALNs): Hybrid proteins com- posed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011, 39, 359–372. [Google Scholar] [CrossRef]
- Miller, J.C.; Tan, S.; Qiao, G.J.; Barlow, K.A.; Wang, J.B.; Xia, D.F.; Meng, X.D.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011, 29, 143–148. [Google Scholar] [CrossRef]
- Mahfouz, M.M.; Li, L.X.; Shamimuzzaman, M.; Wibowo, A.; Fang, X.Y.; Zhu, J.K. De novo-engineered transcription acti- vator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc. Natl. Acad. Sci. USA 2011, 108, 2623–2628. [Google Scholar] [CrossRef]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. Elife 2013, 2, e00471. [Google Scholar] [CrossRef]
- Cong, L.; Ann Ran, F.; Cox, D.; Lin, S.L.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.B.; Jiang, W.Y.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.H.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome en- gineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Genicot, B.; Mouligneau, F.; Lekeux, P. Effect of an oral sedative on the behavior and the zootechnologic performance of fattening bulls of the white-blue Belgian breed. Zent. Vet. A 1991, 38, 668–675. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lee, S.J. Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. USA 1997, 94, 12457–12461. [Google Scholar] [CrossRef] [PubMed]
- Coopman, F.; Krafft, A.; Dewulf, J.; Van Zeveren, A.; Gengler, N. Estimation of phenotypic and genetic parameters for weight gain and weight at fixed ages in the double-muscled Belgian Blue Beef breed using field records. J. Anim. Breed. Genet. 2007, 124, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Bai, C.; Wei, Z.; Su, G.; Wu, Y.; Han, L.; Yang, L.; Liu, X.; Zhao, Y.; Song, E.; et al. Study of Myostatin Gene-editing in Chinese Yellow Cattle. J. Inn. Mong. Univ. 2020, 51, 12–32. [Google Scholar]
- Cundiff, L.V.; Gregory, K.; Koch, R.M. Germplasm evaluation in beef cattle-cycle IV: Birth and weaning traits. J. Anim. Sci. 1998, 76, 2528–2535. [Google Scholar] [CrossRef]
- Xue, J.; Zhu, B.; Zhang, W.; Xia, J.; Gao, H.; Li, J.; Zhang, G. Selection of Objective Traits and Estimation of Economic Weights for Chinese Simmental Beef Cattle. Chin. Anim. Husb. Vet. Med. 2016, 43, 2120–2127. [Google Scholar]
- Zhou, X.; Gu, M.; Zhu, L.; Wu, D.; Yang, M.; Gao, Y.; Wang, X.; Bai, C.; Wei, Z.; Yang, L.; et al. Comparison of microbial community and metabolites in four stomach compartments of MSTN-gene-edited and non-edited cattle. Front. Microbiol. 2022, 13, 844962. [Google Scholar] [CrossRef]
- Gu, M.; Zhou, X.; Zhu, L.; Gao, Y.; Gao, L.; Bai, C.; Yang, L.; Li, G. Myostatin Mutation Promotes Glycolysis by Increasing Phosphorylation of Phosphofructokinase via Activation of PDE5A-cGMP-PKG in Cattle Heart. Front. Cell Dev. Biol. 2022, 9, 774185. [Google Scholar] [CrossRef]
- Gao, L.; Yang, M.; Wei, Z.; Gu, M.; Yang, L.; Bai, C.; Wu, Y.; Li, G. MSTN Mutant Promotes Myogenic Differentiation by Increasing Demethylase, T.ET1 Expression via the SMAD2/SMAD3 Pathway. Int. J. Biol. Sci. 2020, 16, 1324–1334. [Google Scholar] [CrossRef]
- Mirnamniha, M.; Faroughi, F.; Tahmasbpour, E.; Ebrahimi, P.; Beigi Harchegani, A. An overview on role of some trace elements in human reproductive health, sperm function and fertilization process. Rev. Environ. Health 2019, 34, 339–348. [Google Scholar] [CrossRef]
- Hashemi, M.M.; Behnampour, N.; Nejabat, M.; Tabandeh, A.; Ghazi-Moghaddam, B.; Joshaghani, H.R. Impact of Seminal Plasma Trace Elements on Human Sperm Motility Parameters. Rom. J. Intern. Med. 2018, 56, 15–20. [Google Scholar] [CrossRef]
- Hamad, A.R.; Al-Daghistani, H.A.; Shquirat, W.D.; Abdel-Dayem, M.; Al-Swaif, M. Sodium, Potassium, Calcium and Copper Levels in Seminal Plasma are Associated with Sperm Quality in Fertile and Infertile Men. Biochem. Pharmacol. 2014, 3, 1–7. [Google Scholar] [CrossRef]
- Górski, R.; Kotwicka, M.; Skibińska, I.; Jendraszak, M.; Wosiński, S. Effect of low-frequency electric field screening on motility of human sperm. Ann. Agric. Environ. Med. 2020, 27, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Boe-Hansen, G.B.; Fortes, M.R.S.; Satake, N. Morphological defects, sperm DNA integrity, and protamination of bovine spermatozoa. Andrology 2018, 6, 627–633. [Google Scholar] [CrossRef]
- McFarlane, C.; Plummer, E.; Thomas, M.; Hennebry, A.; Ashby, M.; Ling, N.; Smith, H.; Sharma, M.; Kambadur, R. Myostatin induces cachexia by activating the ubiquitin proteolytic system through an NF-kappaB-independent, FoxO1-dependent mechanism. J. Cell Physiol. 2006, 209, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Mosher, D.S.; Quignon, P.; Bustamante, C.D.; Sutter, N.B.; Mellersh, C.S.; Parker, H.G.; Ostrander, E.A. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 2007, 3, e79. [Google Scholar] [CrossRef]
- Sheng, H.; Guo, Y.; Zhang, L.; Zhang, J.; Miao, M.; Tan, H.; Hu, D.; Li, X.; Ding, X.; Li, G.; et al. Proteomic Studies on the Mechanism of Myostatin Regulating Cattle Skeletal Muscle Development. Front. Genet. 2021, 12, 752129. [Google Scholar] [CrossRef]
- Xin, X.B.; Yang, S.P.; Li, X.; Liu, X.F.; Zhang, L.L.; Ding, X.B.; Zhang, S.; Li, G.P.; Guo, H. Proteomics insights into the effects of MSTN on muscle glucose and lipid metabolism in genetically edited cattle. Gen. Comp. Endocrinol. 2020, 291, 113237. [Google Scholar] [CrossRef]
- Yang, S.; Li, X.; Liu, X.; Ding, X.; Xin, X.; Jin, C.; Zhang, S.; Li, G.; Guo, H. Parallel comparative proteomics and phosphoproteomics reveal that cattle myostatin regulates phosphorylation of key enzymes in glycogen metabolism and glycolysis pathway. Oncotarget 2018, 9, 11352–11370. [Google Scholar] [CrossRef]
- Lu, S.; Gu, Y.; Wu, Y.; Yang, S.; Li, C.; Meng, L.; Yuan, W.; Jiang, T.; Zhang, X.; Li, Y.; et al. Bi-allelic variants in human WDR63 cause male infertility via abnormal inner dynein arms assembly. Cell Discov. 2021, 7, 110. [Google Scholar] [CrossRef]
- Gao, Y.; Xu, C.; Tan, Q.; Shen, Q.; Wu, H.; Lv, M.; Li, K.; Tang, D.; Song, B.; Xu, Y.; et al. Case Report: Novel Biallelic Mutations in ARMC4 Cause Primary Ciliary Dyskinesia and Male Infertility in a Chinese Family. Front. Genet. 2021, 12, 715339. [Google Scholar] [CrossRef]
- Priyanka, P.P.; Yenugu, S. Coiled-Coil Domain-Containing (CCDC) Proteins: Functional Roles in General and Male Reproductive Physiology. Reprod. Sci. 2021, 28, 2725–2734. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Yadav, S.; Mehta, P.; Gupta, G.; Rajender, S. Histone Methylation Regulates Gene Expression in the Round Spermatids to Set the RNA Payloads of Sperm. Reprod. Sci. 2022, 29, 857–882. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Zhang, L.; Li, Y.; Wei, H.; Wu, S.; Liu, T.; Liu, L.; Xing, Q.; Wang, S.; Bao, Z. Expression of the Testis-Specific Serine/Threonine Kinases Suggests Their Role in Spermiogenesis of Bay Scallop Argopecten irradians. Front. Physiol. 2021, 12, 657559. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Chen, X.; Zhu, X.; He, B.; Lu, C.; Liu, Y.; Xu, X.; Wu, X. ATF6 deficiency damages the development of spermatogenesis in male Atf6 knockout mice. Andrologia 2022, 54, e14350. [Google Scholar] [CrossRef]
- Li, Y.F.; He, W.; Jha, K.N.; Klotz, K.; Kim, Y.H.; Mandal, A.; Pulido, S.; Digilio, L.; Flickinger, C.J.; Herr, J.C. FSCB, a novel protein kinase A-phosphorylated calcium-binding protein, is a CABYR-binding partner involved in late steps of fibrous sheath biogenesis. J. Biol. Chem. 2007, 282, 34104–34119. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, M.; Yu, R.; Liu, B.; Tian, Z.; Liu, S. FSCB phosphorylation regulates mouse spermatozoa capacitation through suppressing SUMOylation of ROPN1/ROPN1L. Am. J. Transl. Res. 2016, 8, 2776–2782. [Google Scholar]
- Fiedler, S.E.; Dudiki, T.; Vijayaraghavan, S.; Carr, D.W. Loss of R2D2 proteins ROPN1 and ROPN1L causes defects in murine sperm motility, phosphorylation, and fibrous sheath integrity. Biol. Reprod. 2013, 88, 41. [Google Scholar] [CrossRef]
- Sutton, K.A.; Jungnickel, M.K.; Wang, Y.; Cullen, K.; Lambert, S.; Florman, H.M. Enkurin is a novel calmodulin and TRPC channel binding protein in sperm. Dev. Biol. 2004, 274, 426–435. [Google Scholar] [CrossRef]
- Nsota Mbango, J.F.; Coutton, C.; Arnoult, C.; Ray, P.F.; Touré, A. Genetic causes of male infertility: Snapshot on morphological abnormalities of the sperm flagellum. Basic Clin. Androl. 2019, 29, 2. [Google Scholar] [CrossRef]
| Items | Value (% of Dry Matter) | |
|---|---|---|
| Special Feed for Calves | Special Feed for Bulls | |
| Crude protein | >18.0 | >16.0 |
| Crude ash | <14.0 | <12.0 |
| Crude fiber | <6.0 | <9.0 |
| Calcium | 0.6–1.2 | 0.5–1.8 |
| Phosphorus | >0.4 | >0.4 |
| NaCl | 0.5–1.0 | 0.8–1.5 |
| Lysine | >0.7 | >0.6 |
| Months of Age | MT (kg) | WT (kg) | Difference Value (kg) | Increase Rate (%) |
|---|---|---|---|---|
| 0 | 33.20 ± 1.23 | 30.67 ± 2.85 | 2.53 | 8.25 |
| 3 * | 111.40 ± 6.45 | 99.05 ± 7.46 | 12.35 | 12.47 |
| 6 * | 184.71 ± 12.16 | 167.20 ± 16.32 | 17.51 | 10.47 |
| 9 * | 252.82 ± 15.05 | 213.37 ± 20.05 | 39.45 | 18.49 |
| 12 * | 335.67 ± 16.27 | 280.13 ± 19.75 | 55.54 | 19.83 |
| 15 * | 412.76 ± 19.56 | 328.56 ± 24.05 | 84.20 | 25.63 |
| 18 * | 482.51 ± 18.94 | 402.25 ± 24.58 | 80.26 | 19.95 |
| 24 * | 593.59 ± 21.64 | 485.73 ± 28.17 | 107.86 | 22.21 |
| Index | MT | WT | ||||||
|---|---|---|---|---|---|---|---|---|
| 9 | 12 | 15 | 18 | 9 | 12 | 15 | 18 | |
| LYM% | 6.01 ± 1.38 | 6.81 ± 2.76 | 5.53 ± 1.60 | 5.12 ± 0.13 | 6.09 ± 3.43 | 6.87 ± 2.34 | 5.47 ± 2.19 | 5.53 ± 0.60 |
| LYM | 0.38 ± 0.14 | 0.43 ± 0.43 | 0.34 ± 0.13 | 0.38 ± 0.09 | 0.82 ± 0.25 | 0.46 ± 0.61 | 0.41 ± 0.22 | 0.37 ± 0.32 |
| NEUT | 1.09 ± 0.47 | 0.98 ± 0.43 | 0.69 ± 0.12 | 0.56 ± 0.13 | 1.10 ± 1.35 | 0.90 ± 0.81 | 0.66 ± 0.44 | 0.67 ± 0.61 |
| RBC | 9.10 ± 1.10 | 9.18 ± 0.080 | 9.01 ± 0.33 | 8.88 ± 0.34 | 9.20 ± 0.19 | 9.01 ± 0.21 | 8.87 ± 1.23 | 87.77 ± 0.32 |
| MCV | 3504 ± 254.21 | 3511 ± 345.30 | 3489 ± 309.30 | 3501 ± 280.34 | 3434 ± 234.01 | 3419 ± 276.07 | 3402 ± 151.67 | 3487 ± 154.32 |
| PLT | 286.31 ± 63.76 | 266.51 ± 72.52 | 254.12 ± 82.03 | 248.81 ± 81.09 | 278.39 ± 167.12 | 268.43 ± 30.01 | 243.42 ± 65.45 | 271.13 ± 51.22 |
| PCT | 0.23 ± 0.16 | 0.22 ± 0.13 | 0.22 ± 0.09 | 0.19 ± 0.14 | 0.22 ± 0.78 | 0.22 ± 0.56 | 0.21 ± 0.11 | 0.21 ± 0.12 |
| MPV | 6.76 ± 0.34 | 6.77 ± 0.70 | 6.78 ± 0.08 | 6.76 ± 0.50 | 7.01 ± 1.09 | 7.02 ± 0.04 | 7.02 ± 0.32 | 7.00 ± 0.21 |
| BA | 0.11 ± 0.03 | 0.10 ± 0.05 | 0.10 ± 0.05 | 0.11 ± 0.03 | 0.11 ± 0.07 | 0.10 ± 0.07 | 0.10 ± 0.02 | 0.10 ± 0.03 |
| BA% | 1.02 ± 0.17 | 1.06 ± 0.09 | 0.99 ± 0.54 | 1.05 ± 0.35 | 1.07 ± 0.32 | 0.99 ± 0.87 | 0.98 ± 0.90 | 1.06 ± 0.11 |
| AL% | 0.88 ± 0.12 | 0.87 ± 0.49 | 0.79 ± 0.80 | 0.78 ± 0.10 | 1.04 ± 0.50 | 1.06 ± 0.21 | 0.88 ± 0.66 | 0.67 ± 0.16 |
| AL | 0.07 ± 0.04 | 0.07 ± 0.01 | 0.06 ± 0.04 | 0.06 ± 0.02 | 0.08 ± 0.04 | 0.08 ± 0.02 | 0.07 ± 0.08 | 0.07 ± 0.05 |
| HGB | 118.60 ± 10.43 | 116.44 ± 13.13 | 118.9 ± 10.33 | 116.4 ± 9.92 | 117.60 ± 9.76 | 117.43 ± 12.41 | 116.78 ± 8.48 | 116.35 ± 2.11 |
| Index | MT | WT | ||||||
|---|---|---|---|---|---|---|---|---|
| 9 | 12 | 15 | 18 | 9 | 12 | 15 | 18 | |
| GLU * | 3.87 ± 0.77 | 3.82 ± 1.04 | 3.84 ± 1.02 | 3.81 ± 0.70 | 4.47 ± 0.24 | 4.41 ± 0.33 | 4.39 ± 0.67 | 4.68 ± 0.27 |
| ASP | 58.64 ± 3.23 | 57.77 ± 12.54 | 60.65 ± 12.70 | 60.71 ± 8.24 | 57.83 ± 7.56 | 58.03 ± 7.21 | 59.06 ± 5.53 | 58.75 ± 4.59 |
| ALT | 24.86 ± 7.72 | 24.93 ± 3.43 | 25.01 ± 5.43 | 25.42 ± 5.18 | 25.12 ± 6.62 | 25.13 ± 3.44 | 24.31 ± 5.27 | 25.05 ± 4.01 |
| TP | 66.68 ± 1.66 | 65.79 ± 7.34 | 65.34 ± 7.41 | 66.07 ± 5.78 | 65.09 ± 8.34 | 65.77 ± 6.43 | 65.33 ± 5.34 | 66.08 ± 3.78 |
| ALB | 36.38 ± 2.35 | 35.85 ± 7.31 | 36.01 ± 7.43 | 36.07 ± 5.21 | 36 ± 3.07 | 36.17 ± 3.12 | 35.90 ± 7.84 | 36.45 ± 1.11 |
| CK | 193 ± 67.71 | 189 ± 54.32 | 192.70 ± 56.45 | 190.11 ± 52.43 | 189.87 ± 59.03 | 192.34 ± 61.01 | 187.47 ± 28.42 | 194.31 ± 16.41 |
| HDL * | 3.03 ± 0.76 | 3.01 ± 0.87 | 2.97 ± 0.69 | 3.08 ± 0.36 | 2.11 ± 0.43 | 2.24 ± 0.87 | 2.26 ± 0.52 | 2.09 ± 0.89 |
| LDL * | 1.55 ± 0.64 | 1.58 ± 0.43 | 1.54 ± 0.21 | 1.66 ± 0.31 | 0.79 ± 0.13 | 0.85 ± 0.56 | 0.85 ± 0.29 | 0.99 ± 0.65 |
| PAMY ** | 26.61 ± 5.12 | 26.44 ± 5.12 | 34.95 ± 6.67 | 35.19 ± 6.31 | 25.91 ± 7.12 | 23.21 ± 3.54 | 21.06 ± 7.45 | 22.27 ± 7.56 |
| CREA | 138.21 ± 40.01 | 139.21 ± 18.29 | 139.43 ± 25.33 | 138.92 ± 17.21 | 138.12 ± 23.30 | 137.54 ± 32.61 | 135.38 ± 17.54 | 138.22 ± 7.01 |
| LDH | 1108.34 ± 125.43 | 1108.76 ± 165.22 | 1110.32 ± 134.42 | 1103.87 ± 166.11 | 1110.16 ± 232.02 | 1113 ± 137.32 | 1094.54 ± 165.21 | 1096.24 ± 55.13 |
| AB | 22.17 ± 2.54 | 22.32 ± 4.77 | 22.88 ± 3.76 | 23.90 ± 2.10 | 21.82 ± 4.12 | 22.01 ± 3.05 | 21.25 ± 3.31 | 22.81 ± 1.89 |
| CHE | 143.57 ± 22.34 | 144.09 ± 24.32 | 143.21 ± 43.55 | 145.87 ± 22.42 | 144.15 ± 28.77 | 143.54 ± 18.04 | 144.33 ± 18.06 | 144.70 ± 35.23 |
| TC | 3.02 ± 0.78 | 3.03 ± 1.01 | 3.11 ± 1.04 | 3.07 ± 0.76 | 3.01 ± 0.12 | 3.01 ± 0.54 | 3.03 ± 0.87 | 3.06 ± 0.15 |
| LA * | 4.38 ± 2.66 | 3.97 ± 3.34 | 3.87 ± 2.79 | 3.96 ± 1.41 | 2.47 ± 0.70 | 2.44 ± 0.78 | 2.41 ± 0.43 | 2.35 ± 0.81 |
| TG | 0.31 ± 0.21 | 0.29 ± 0.12 | 0.30 ± 0.09 | 0.31 ± 0.21 | 0.31 ± 0.02 | 0.29 ± 0.09 | 0.29 ± 0.02 | 0.30 ± 0.11 |
| BUN ** | 2.53 ± 3.21 | 2.55 ± 1.87 | 2.65 ± 1.34 | 2.61 ± 0.36 | 1.30 ± 0.60 | 1.32 ± 0.43 | 1.28 ± 0.49 | 1.31 ± 0.54 |
| LIP ** | 15.72 ± 3.16 | 15.81 ± 5.06 | 15.59 ± 6.10 | 16.49 ± 9.10 | 11.54 ± 5.32 | 12.10 ± 4.14 | 12.06 ± 5.02 | 11.95 ± 6.21 |
| Bulls | Semen Volume (ml) | Sperm Density (106/mL) | Fresh Sperm Motility (%) | Frozen Sperm Motility (%) |
|---|---|---|---|---|
| MT1 | 5.6 ± 2.6 | 1538 ± 504 | 78.9 ± 9.7 | 49.1 ± 13.7 |
| MT2 | 5.1 ± 1.9 | 1344 ± 616 | 75.1 ± 14.2 | 37.6 ± 15.9 |
| MT3 | 4.1 ± 1.8 | 1258 ± 346 | 84.0 ± 4.8 | 62.1 ± 8.8 |
| MT4 | 3.9 ± 0.4 | 1024 ± 247 | 63.3 ± 8.4 | 18.8 ± 11.6 |
| MT5 | 5.5 ± 2.6 | 1242 ± 564 | 72.9 ± 10.8 | 42.3 ± 10.0 |
| MT6 | 4.8 ± 1.7 | 1109 ± 462 | 69.4 ± 9.1 | 34.9 ± 10.8 |
| Average of MT | 4.8 ± 0.7 | 1252 ± 180 | 73.9 ± 7.2 | 40.8 ± 14.5 |
| WT1 | 5.7 ± 1.1 | 1477 ± 612 | 74.6 ± 9.0 | 45.7 ± 9.2 |
| WT2 | 5.1 ± 1.2 | 1462 ± 632 | 78.3 ± 7.9 | 56.2 ± 5.8 |
| WT3 | 5.9 ± 1.1 | 2118 ± 695 | 81.9 ± 6.2 | 65.6 ± 9.8 |
| Average of WT | 5.6 ± 0.3 | 1686 ± 306 | 78.3 ± 3.0 | 55.8 ± 8.1 |
| p-value | 0.09 | 0.17 | 0.27 | 0.12 |
| Elements | MT1 | MT2 | MT3 | MT4 | MT5 | MT6 | WT1 | WT2 | WT3 |
|---|---|---|---|---|---|---|---|---|---|
| Na (mg/L) | 372.9 | 308.2 | 169.3 | 276.4 | 298.7 | 346.2 | 281.0 | 407.3 | 446.9 |
| K * (mg/L) | 202.7 | 65.8 | 395.9 | 246.8 | 223.0 | 89.9 | 105.4 | 75.3 | 79.3 |
| Ca (mg/L) | 381.1 | 249.2 | 212.2 | 164.1 | 235.0 | 224.2 | 259.2 | 328.6 | 229.8 |
| Mg (mg/L) | 49.0 | 45.4 | 46.8 | 26.6 | 39.2 | 43.2 | 26.4 | 77.4 | 46.0 |
| Fe (mg/L) | 6.8 | 4.3 | 3.0 | 2.4 | 3.3 | 3.1 | 3.9 | 7.2 | 3.9 |
| Zn (mg/L) | 6.3 | 4.4 | 3.4 | 2.1 | 3.1 | 2.6 | 2.2 | 8.6 | 4.3 |
| Se (μg/L) | 460.5 | 450.6 | 418.6 | 249.1 | 363.9 | 502.4 | ND | 985.2 | 151.9 |
| Cr (μg/L) | 433.8 | 386.5 | 271.4 | 172.9 | 274.9 | 306.1 | 85.6 | 420.5 | 174.7 |
| Ti (μg/L) | 170.1 | 180.6 | 114.1 | 68.3 | 79.2 | 83.1 | 136.8 | 159.3 | 121.1 |
| V ** (μg/L) | 135.3 | 121.4 | 88.1 | 59.4 | 88.8 | 98.6 | 442.6 | 515.0 | 385.2 |
| Al (μg/L) | 125.8 | 146.9 | 51.3 | ND | ND | ND | 1.5 | 59.3 | 17.2 |
| Hg ** (μg/L) | 112.2 | 88.2 | 80.9 | 82.6 | 82.3 | 81.4 | 0.7 | 1.7 | 0.9 |
| Sr ** (μg/L) | 99.1 | 86.8 | 88.0 | 114.1 | 102.3 | 78.2 | 33.0 | 38.0 | 25.4 |
| Ba (μg/L) | 52.3 | 73.8 | 32.1 | 90.9 | 40.7 | 44.5 | 105.5 | 241.0 | 101.6 |
| Cu * (μg/L) | 48.5 | 102.1 | 54.1 | 27.2 | 40.1 | 68.3 | 360.2 | 562.9 | 397.7 |
| Mn * (μg/L) | 22.3 | 19.7 | 16.2 | 99.1 | 8.6 | 7.0 | 70.2 | 172.1 | 115.5 |
| Ni (μg/L) | 37.1 | 26.0 | 18.9 | 15.0 | 20.5 | 19.8 | 28.1 | 14.7 | 9.6 |
| Pb (μg/L) | 11.3 | 13.7 | 9.5 | 9.9 | 10.8 | 9.4 | 1.3 | 33.1 | 7.4 |
| Sn * (μg/L) | 9.9 | 10.0 | 7.3 | 7.8 | 7.8 | 7.3 | 36.0 | 93.6 | 76.1 |
| Sb * (μg/L) | 8.1 | 8.0 | 4.9 | 0.1 | ND | ND | 80.8 | 175.2 | 84.7 |
| Co ** (μg/L) | 2.3 | 0.7 | 0.1 | 0.2 | ND | 0.2 | 9.6 | 15.0 | 12.6 |
| Indexes | MT1 | MT2 | MT3 | MT4 | MT5 | MT6 | WT1 | WT2 | WT3 |
|---|---|---|---|---|---|---|---|---|---|
| VCL (mm/s) | 77.6 | 61.7 | 79.9 | 63.8 | 73.0 | 51.4 | 66.2 | 52.7 | 64.0 |
| VSL (mm/s) | 37.2 | 24.7 | 29.6 | 27.4 | 27.7 | 20.8 | 35.1 | 25.2 | 26.0 |
| VAP (mm/s) | 48.0 | 35.0 | 47.0 | 38.2 | 39.9 | 29.7 | 44.5 | 32.3 | 36.9 |
| LIN (%) | 43.5 | 33.6 | 35.2 | 38.3 | 33.9 | 33.8 | 44.0 | 38.2 | 33.4 |
| STR (%) | 67.8 | 57.2 | 58.2 | 61.7 | 58.6 | 56.6 | 65.2 | 61.5 | 56.9 |
| WOB * (%) | 59.7 | 57.0 | 57.9 | 57.8 | 63.5 | 64.5 | 57.9 | 55.2 | 54.3 |
| ALH (um) | 3.2 | 2.9 | 3.4 | 2.8 | 3.3 | 2.4 | 2.6 | 2.3 | 2.9 |
| BCF * (Hz) | 8.4 | 6.8 | 7.7 | 6.6 | 6.3 | 6.3 | 6.4 | 6.3 | 5.8 |
| Accession. | Description | p-Value | FC | Regulated |
|---|---|---|---|---|
| Q58D96 | Isocitrate dehydrogenase [NAD] subunit, mitochondrial GN = IDH3G | 0.041 | 5.28 | UP |
| Q58DG1 | UPF0160 protein MYG1, mitochondrial GN = MYG1 | 0.005 | 4.12 | UP |
| P13619 | ATP synthase F(0) complex subunit B1, mitochondrial GN = ATP5F1 | 0.032 | 0.08 | DOWN |
| F1N3R4 | Mitochondrial proton/calcium exchanger protein GN = LETM1 | 0.012 | 5.72 | UP |
| E1BA21 | Transcription factor A, mitochondrial GN = TFAM | 0.042 | 13.64 | UP |
| E2GEZ1 | Translocase of outer mitochondrial membrane 22 GN = Tom22 | 0.017 | 2.91 | UP |
| E2GEZ2 | Translocase of outer mitochondrial membrane 40A GN = Tom40A | 0.023 | 4.78 | UP |
| Q58D49 | Cob(I)yrinic acid a,c-diamide adenosyltransferase, mitochondrial GN = MMAB | 0.042 | 4.12 | UP |
| Q8HXG5 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mitochondrial GN = NDUFB11 | 0.019 | 8.17 | UP |
| P25712 | NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial GN = NDUFV3 | 0.012 | 9.93 | UP |
| Q02365 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 GN=NDUFB3 | 0.032 | 2.74 | UP |
| E1B9Z2 | Leucine zipper and EF-hand containing transmembrane protein 2 GN = LETM2 | 0.014 | 7.14 | UP |
| O97725 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 GN = NDUFA12 | 0.023 | 7.00 | UP |
| D3K0R6 | Plasma membrane calcium-transporting ATPase 4 GN = ATP2B4 | 0.029 | 3.42 | UP |
| Q2HJ55 | Sorting and assembly machinery component 50 homolog GN = SAMM50 | 0.038 | 12.07 | UP |
| P68530 | Cytochrome c oxidase subunit 2 GN = MT-CO2 | 0.016 | 9.01 | UP |
| A5PKG4 | Cysteine desulfurase, mitochondrial GN = NFS1 | 0.015 | 3.98 | UP |
| F1MEM9 | Tetratricopeptide repeat domain 19 GN = TTC19 | 0.044 | 6.31 | UP |
| Q2NKR7 | Protein FAM162A GN = FAM162A | 0.001 | 8.70 | UP |
| E1BGC1 | Methylcrotonoyl-CoA carboxylase 1 GN = MCCC1 | 0.019 | 7.12 | UP |
| Q3ZBU7 | Tubulin beta-4A chain GN = TUBB4A | 0.044 | 6.27 | UP |
| E1B8W3 | Outer dynein arm docking complex subunit 2 GN = ARMC4 | 0.043 | 3.71 | UP |
| Q2T9N0 | Fibrous sheath CABYR-binding protein GN = FSCB | 0.020 | 4.04 | UP |
| Q2T9W3 | Coiled-coil domain-containing protein 63 GN = CCDC63 | 0.032 | 10.17 | UP |
| F1N2N9 | Coiled-coil domain-containing 114 GN = CCDC114 | 0.025 | 3.38 | UP |
| E1B836 | Enkurin, TRPC channel interacting protein GN = ENKUR | 0.035 | 3.28 | UP |
| E1B9S6 | Dynein axonemal intermediate chain 4 GN = WDR78 | 0.040 | 2.24 | UP |
| F6RLA2 | Testis specific serine kinase 4 GN = TSSK4 | 0.002 | 7.74 | UP |
| F6QYE2 | Cilia and flagella associated protein 100 GN = CFAP100 | 0.004 | 7.37 | UP |
| P10152 | Angiogenin-1 GN = ANG1 | 0.006 | 0.01 | DOWN |
| C6KH61 | Potassium calcium-activated channel subfamily U member 1 GN = KCNU1 | 0.005 | 4.23 | UP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Yang, L.; Su, G.; Wei, Z.; Liu, X.; Song, L.; Hai, C.; Wu, D.; Hao, Z.; Wu, Y.; et al. Growth Traits and Sperm Proteomics Analyses of Myostatin Gene-Edited Chinese Yellow Cattle. Life 2022, 12, 627. https://doi.org/10.3390/life12050627
Zhao Y, Yang L, Su G, Wei Z, Liu X, Song L, Hai C, Wu D, Hao Z, Wu Y, et al. Growth Traits and Sperm Proteomics Analyses of Myostatin Gene-Edited Chinese Yellow Cattle. Life. 2022; 12(5):627. https://doi.org/10.3390/life12050627
Chicago/Turabian StyleZhao, Yuefang, Lei Yang, Guanghua Su, Zhuying Wei, Xuefei Liu, Lishuang Song, Chao Hai, Di Wu, Zhenting Hao, Yunxi Wu, and et al. 2022. "Growth Traits and Sperm Proteomics Analyses of Myostatin Gene-Edited Chinese Yellow Cattle" Life 12, no. 5: 627. https://doi.org/10.3390/life12050627
APA StyleZhao, Y., Yang, L., Su, G., Wei, Z., Liu, X., Song, L., Hai, C., Wu, D., Hao, Z., Wu, Y., Zhang, L., Bai, C., & Li, G. (2022). Growth Traits and Sperm Proteomics Analyses of Myostatin Gene-Edited Chinese Yellow Cattle. Life, 12(5), 627. https://doi.org/10.3390/life12050627






