From Parasitized to Healthy-Looking Ants (Hymenoptera: Formicidae): Morphological Reconstruction Using Algorithmic Processing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
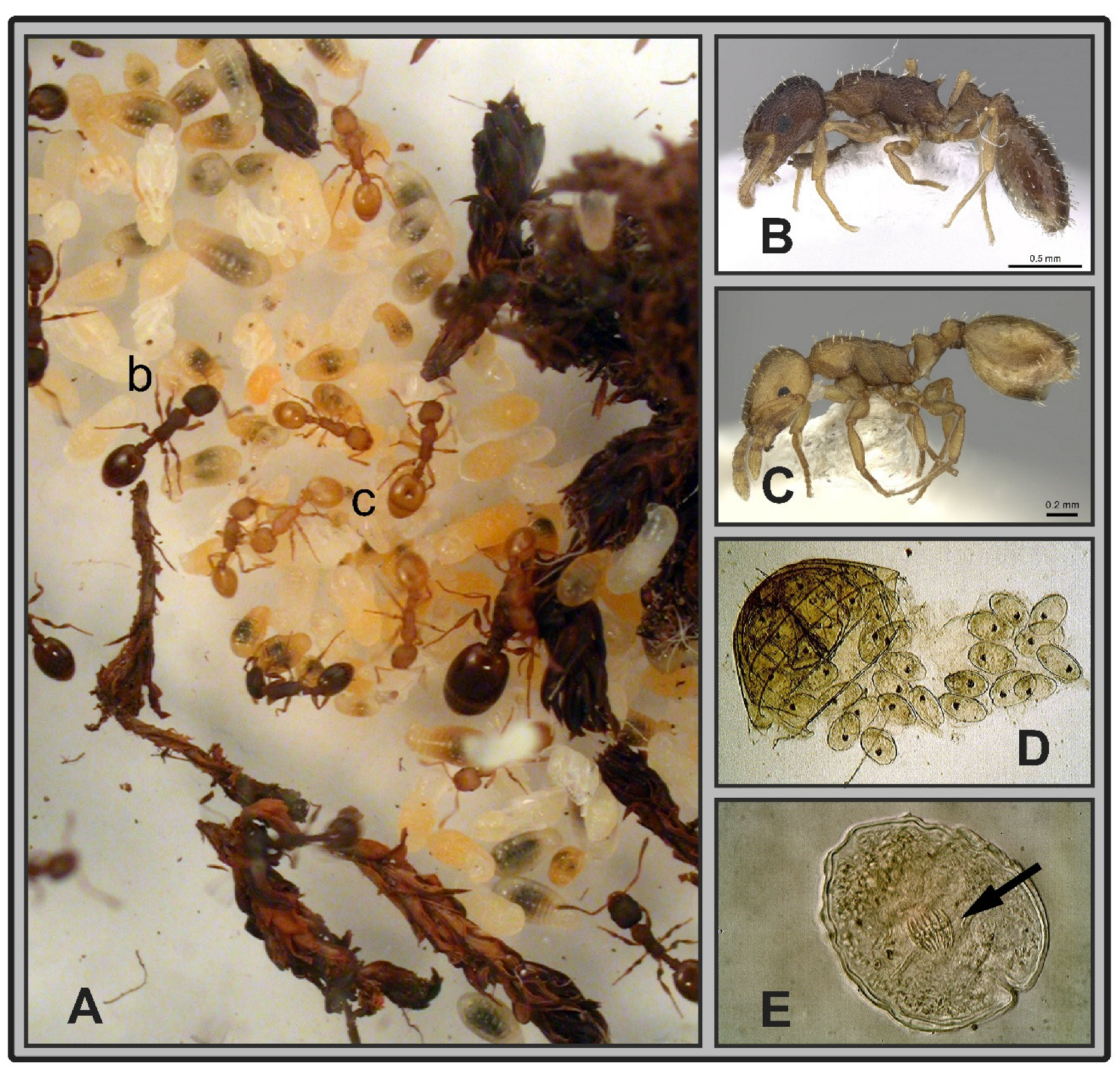
2.2. Identification of Infected Specimens
2.3. Morphometrics
3. Data Analyses
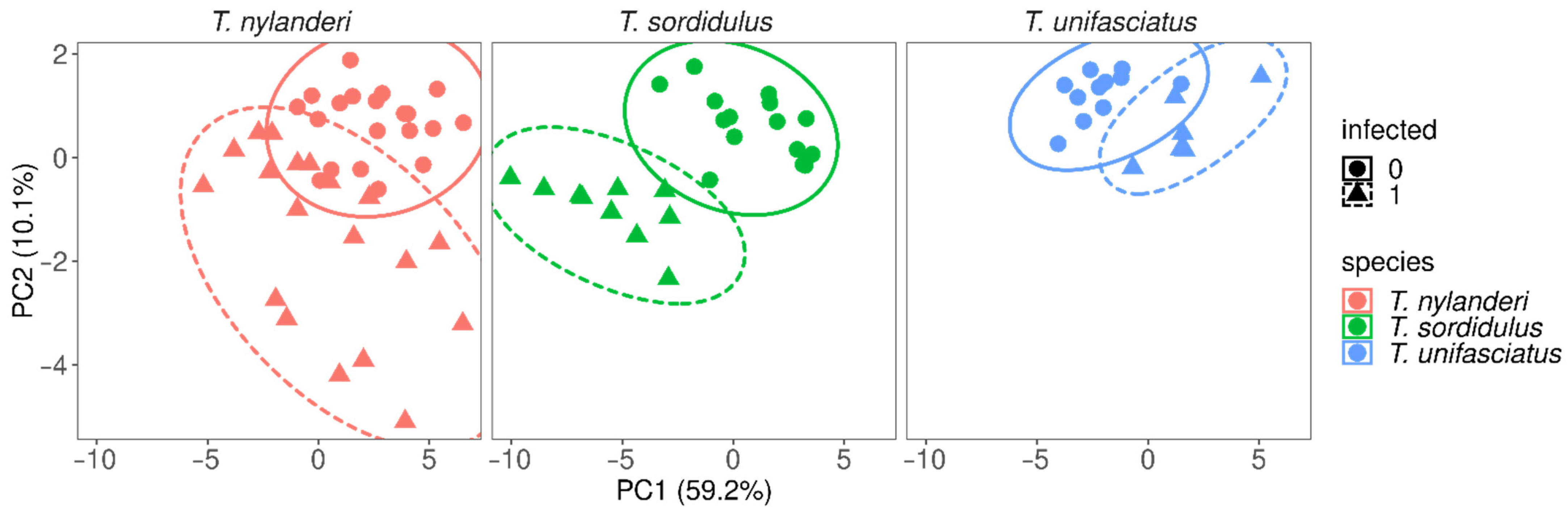
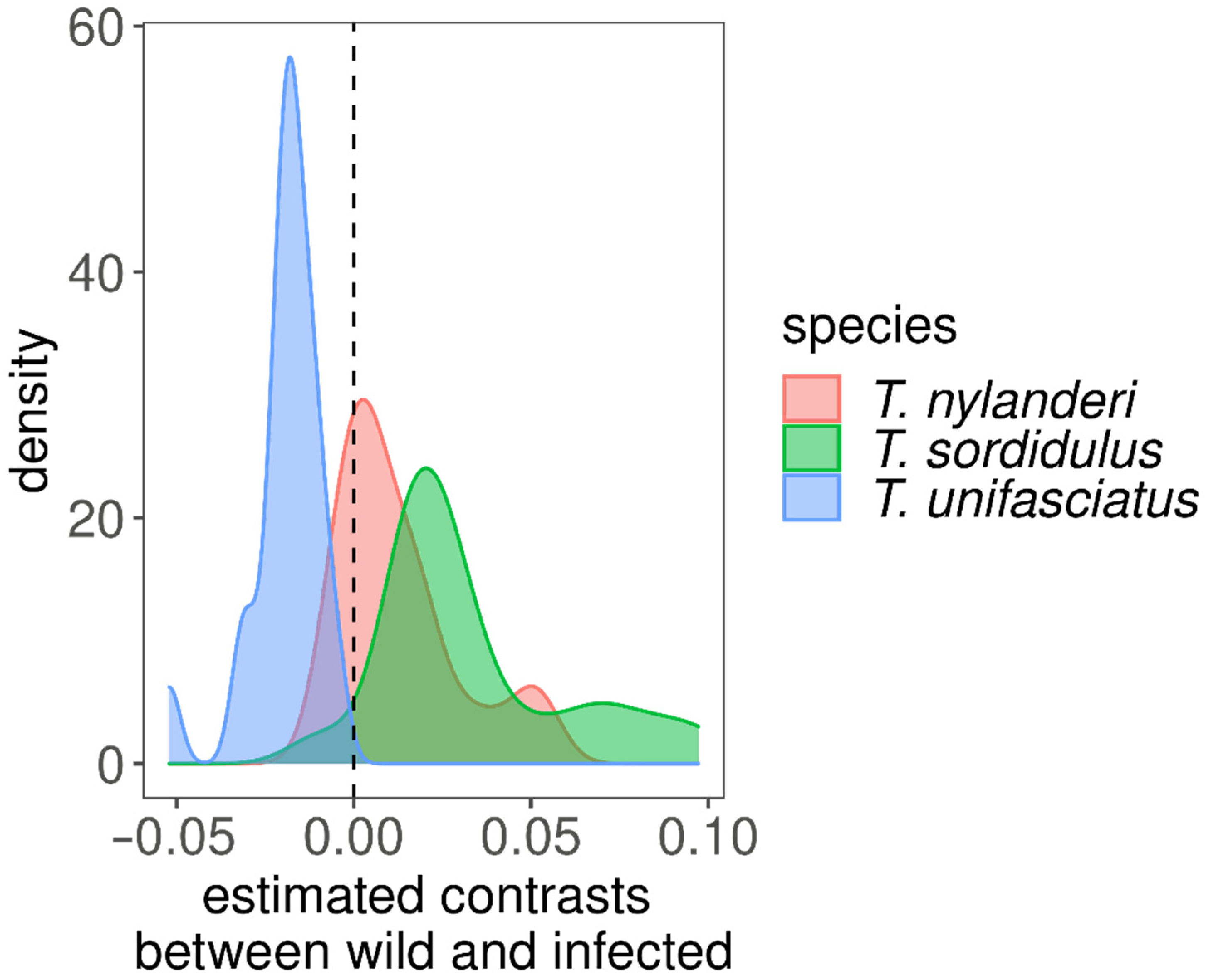
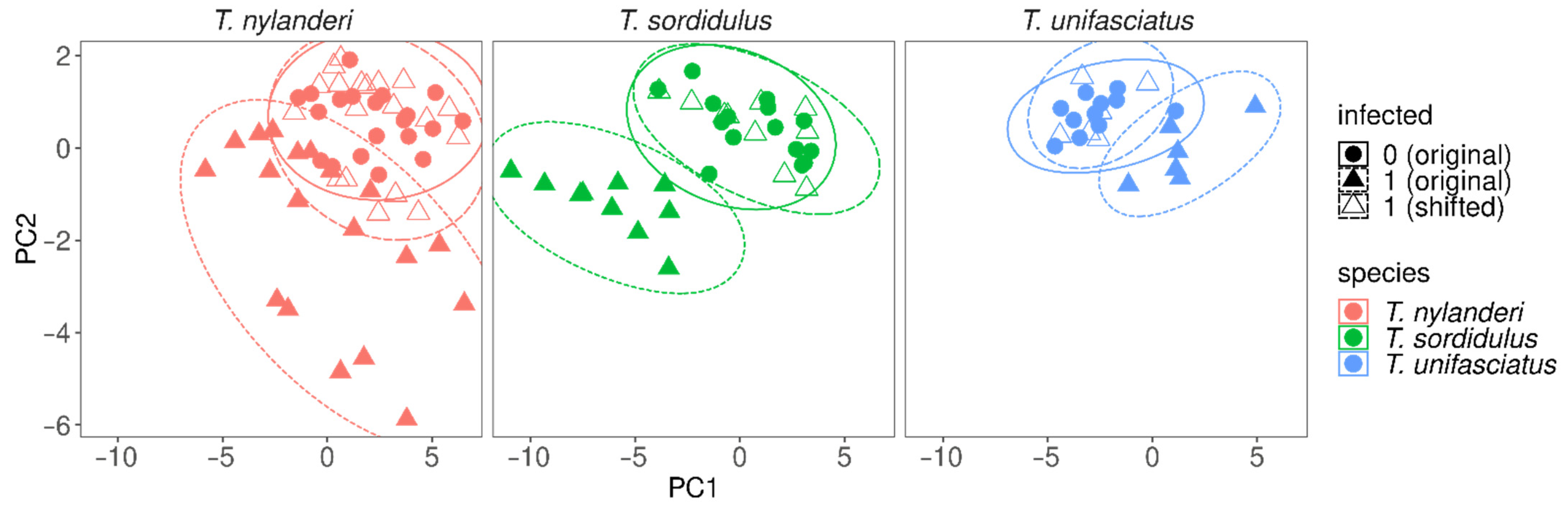
4. Results
5. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Lim, G.S.; Balke, M.; Meier, R. Determining species boundaries in a world FULL of rarity: Singletons, species delimitation methods. Syst. Biol. 2012, 61, 165–169. [Google Scholar] [CrossRef] [Green Version]
- Sogin, M.L.; Morrison, H.G.; Huber, J.A.; Welch, M.D.; Huse, S.M.; Neal, P.R.; Arrieta, J.M.; Herndl, G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 2006, 103, 12115–12120. [Google Scholar] [CrossRef] [Green Version]
- Thessen, A.E.; Patterson, D.J.; Murray, S.A. The taxonomic significance of species that have only been observed once: The genus Gymnodinium (Dinoflagellata) as an example. PLoS ONE 2012, 7, e44015. [Google Scholar] [CrossRef]
- Garcia, F.H.; Fischer, G.; Kück, P.; Thormann, B.; Peters, M.K. Tetramorium boehmei sp. n.–a new ant (Hymenoptera: Formicidae) species from the Kakamega Forest, Western Kenya. Bonn. Zool. Bull. 2010, 57, 359–366. [Google Scholar]
- Bharti, H.; Wachkoo, A.A. Cerapachys browni and Cerapachys costatus, two new rare ant species (Hymenoptera: Formicidae) from India. Biologia 2013, 68, 1189–1192. [Google Scholar] [CrossRef]
- Wong, M.K.; Yong, G.W. Notes on the habitat and biology of the rare ant genus Tyrannomyrmex (Fernández, 2003). Asian Myrmecol. 2017, 9, e009007. [Google Scholar]
- Bailey, E.L. The Effects of Urbanization on Insect Morphology: A Meta-Analysis; B.I.S, University of Tennessee at Chattanooga: Chattanooga, TN, USA, 2021. [Google Scholar]
- Norton, T.A.; Mathieson, A.C.; Neushul, M. Morphology and Environment. In The Biology of Seaweeds; Lobban, C.S., Wynne, M.J., Eds.; University of California Press: Berkley, MI, USA; Los Angeles, CA, USA, 1981; pp. 421–451. [Google Scholar]
- Lucius, R.; Loos-Frank, B.; Lane, R.P.; Poulin, R.; Roberts, C.; Grencis, R.K. The Biology of Parasites; Wiley-VCG VErlag GmbH & Co. KGaA: Weinheim, Germany, 2017; pp. 1–451. [Google Scholar]
- Laciny, A. Among the shapeshifters: Parasite-induced morphologies in ants (Hymenoptera, Formicidae) and their relevance within the EcoEvoDevo framework. EvoDevo 2021, 12, 1–21. [Google Scholar] [CrossRef]
- Londe, S.; Monnin, T.; Cornette, R.; Debat, V.; Fisher, B.L.; Molet, M. Phenotypic plasticity and modularity allow for the production of novel mosaic phenotypes in ants. EvoDevo 2015, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Csősz, S.; Majoros, G. Ontogenetic origin of mermithogenic Myrmica phenotypes (Hymenoptera, Formicidae). Insect. Soc. 2009, 56, 70–76. [Google Scholar] [CrossRef]
- Trabalon, M.; Plateaux, L.; Péru, L.; Bagnères, A.G.; Hartmann, N. Modification of morphological characters and cuticular compounds in worker ants Leptothorax nylanderi induced by endoparasites Anomotaenia Brevis. J. Insect Physiol. 2000, 46, 169–178. [Google Scholar] [CrossRef]
- Wheeler, W.M. Mermis parasitism and intercastes among ants. J. Exp. Zool. 1928, 50, 165–237. [Google Scholar] [CrossRef] [Green Version]
- Menozzi, C. Res Mutinenses. Formicidae (Hymenoptera). Atti. Soc. Nat. Mat. Modena 1925, 3, 22–47. [Google Scholar]
- Wasmann, E. Nachträge zum sozialen Parasitismus und der Sklaverei bei Ameisen. Biol. Zbl. 1910, 30, 515–524. [Google Scholar]
- Nishiura, N.; Kaneko, K. Evolution of phenotypic fluctuation under host-parasite interactions. PLoS Comput. Biol. 2021, 17, e1008694. [Google Scholar] [CrossRef]
- Csősz, S. Nematode infection as significant source of unjustified taxonomic descriptions in ants (Hymenoptera: Formicidae). Myrmecol. News 2012, 17, 27–31. [Google Scholar]
- Poulin, R.; Thomas, F. Phenotypic Variability Induced by Parasites: Extent and Evolutionary Implications. Parasitol. Today 1999, 15, 28–32. [Google Scholar] [CrossRef]
- Beros, S.; Jongepier, E.; Hagemeier, F.; Foitzik, S. The parasite’s long arm: A tapeworm parasite induces behavioural changes in uninfected group members of its social host. P. Roy. Soc. B-Biol. Sci. 2015, 282, 20151473. [Google Scholar] [CrossRef] [Green Version]
- Feldmeyer, B.; Mazur, J.; Beros, S.; Lerp, H.; Binder, H.; Foitzik, S. Gene expression patterns underlying parasite-induced alterations in host behaviour and life history. Mol. Ecol. 2016, 25, 648–660. [Google Scholar] [CrossRef]
- Stoldt, M.; Klein, L.; Beros, S.; Butter, F.; Jongepier, E.; Feldmeyer, B.; Foitzik, S. Parasite presence induces gene expression changes in an ant host related to immunity and longevity. Genes 2021, 12, 95. [Google Scholar] [CrossRef]
- Heinze, J.; Rüppell, O.; Foitzik, S.; Buschinger, A. First records of Leptothorax rugatulus (Hymenoptera: Formicidae) with cysticercoids of tapeworms (Cestoda: Dilepididae) from the southwestern United States. Fla. Entomol. 1998, 81, 122–125. [Google Scholar] [CrossRef]
- Csősz, S.; Heinze, J.; Mikó, I. Taxonomic synopsis of the Ponto-Mediterranean ants of Temnothorax nylanderi species-group. PLoS ONE 2015, 10, e0140000. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing, R version 4.0.2; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 22 June 2020).
- Lenth, R. Emmeans: Estimated Marginal Means, aka Least-Squares Means (Version 1.3.4). 2019. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 27 March 2022).
- Wäldchen, J.; Mäder, P. Machine learning for image based species identification. Methods Ecol. Evol. 2018, 9, 2216–2225. [Google Scholar] [CrossRef]
- Sosiak, C.E.; Barden, P. Multidimensional trait morphology predicts ecology across ant lineages. Funct. Ecol. 2021, 35, 139–152. [Google Scholar] [CrossRef]
- Jones, D.T. Setting the standards for machine learning in biology. Nat. Rev. Mol. Cell Biol. 2019, 20, 659–660. [Google Scholar] [CrossRef]
- Beros, S.; Enders, C.; Menzel, F.; Foitzik, S. Parasitism and queen presence interactively shape worker behaviour and fertility in an ant host. Anim. Behav. 2019, 148, 63–70. [Google Scholar] [CrossRef]
- Czaplinski, B.; Vaucher, C. Family Hymenolepididae Ariola, 1899. In Keys to the Cestode Parasites of Vertebrates; Khalil, L.F., Jones, A., Bray, R.A., Eds.; CAB International: Wallingford, UK, 1994; pp. 595–663. [Google Scholar]
- Kotelnikov, G.A. The intermediate hosts of Hymenolepididae in birds. Tr. Vsesoyuznogo Inst. Gel’mintologii 1968, 14, 196–205. [Google Scholar]
- Mayhew, R.L. Studies on the Avian Species of the Cestode Family Hymenolepididae; Johnson Reprint Corporation: Urbana, IL, USA, 1925; Volume 10, pp. 1–520. [Google Scholar]
- Buschinger, A. Ameisen des Tribus Leptothoracini (Hym. Formicidae) als Zwischenwirte von Cestoden. Zool. Anz. 1973, 191, 369–380. [Google Scholar]
- Gabrion, C.; Plateaux, L.; Quentin, C. Anomotaenia brevis (Clerc, 1902) Führmann, 1908, Cestode Cyclophyllide parasite de Leptothorax nylanderi (Förster), Hyménoptère Formicidé. Ann. Parasit. Hum. Comp. 1976, 51, 407–420. [Google Scholar] [CrossRef]
- Plateaux, L. Sur les modifications produites chez une fourmi par la présence d’un parasite cestode. Ann. Sci. Nat. Zool. 1972, 14, 203–220. [Google Scholar]
- Shingleton, A.W.; Das, J.; Vinicius, L.; Stern, D.L. The temporal requirements for insulin signaling during development in Drosophila. PLoS Biol. 2005, 3, e289. [Google Scholar] [CrossRef]
- Shingleton, A.W.; Frankino, W.A.; Flatt, T.; Nijhout, H.F.; Emlen, D.J. Size and shape: The developmental regulation of static allometry in insects. Bioessays 2007, 29, 536–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tocco, C.; Roggero, A.; Rolando, A.; Palestrini, C. Interspecific shape divergence in Aphodiini dung beetles: The case of Amidorus obscurus and A. immaturus (Coleoptera: Scarabaeoidea). Org. Divers. Evol. 2011, 11, 263–273. [Google Scholar] [CrossRef]
- Rowland, J.M.; Emlen, D.J. Two thresholds, three male forms result in facultative male trimorphism in beetles. Science 2009, 323, 773–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Component | Df | Sum Sq | Mean Sq | F Value | Pr (>F) |
|---|---|---|---|---|---|
| trait | 20 | 48.798 | 2.440 | 8921.731 | 0.000 |
| as. factor (infected) | 1 | 0.056 | 0.056 | 205.174 | 0.000 |
| species | 2 | 0.076 | 0.038 | 139.383 | 0.000 |
| trait: as. factor (infected) | 20 | 0.086 | 0.004 | 15.711 | 0.000 |
| trait: species | 40 | 0.117 | 0.003 | 10.707 | 0.000 |
| as. factor (infected): species | 2 | 0.131 | 0.066 | 239.836 | 0.000 |
| trait: as. factor (infected): species | 40 | 0.059 | 0.001 | 5.394 | 0.000 |
| Residuals | 1575 | 0.431 | 0.000 | NA | NA |
| kth Model | Accuracy | Precision | Sensitivity | Specificity | MCC |
|---|---|---|---|---|---|
| 1 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 2 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 3 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 4 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 5 | 0.98 | 0.95 | 1.00 | 0.98 | 0.97 |
| 6 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 7 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 8 | 0.98 | 0.95 | 1.00 | 0.98 | 0.97 |
| 9 | 0.97 | 0.95 | 0.95 | 0.98 | 0.93 |
| 10 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| averaged | 0.99 | 0.99 | 1.00 | 0.99 | 0.99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Csősz, S.; Báthori, F.; Molet, M.; Majoros, G.; Rádai, Z. From Parasitized to Healthy-Looking Ants (Hymenoptera: Formicidae): Morphological Reconstruction Using Algorithmic Processing. Life 2022, 12, 625. https://doi.org/10.3390/life12050625
Csősz S, Báthori F, Molet M, Majoros G, Rádai Z. From Parasitized to Healthy-Looking Ants (Hymenoptera: Formicidae): Morphological Reconstruction Using Algorithmic Processing. Life. 2022; 12(5):625. https://doi.org/10.3390/life12050625
Chicago/Turabian StyleCsősz, Sándor, Ferenc Báthori, Mathieu Molet, Gábor Majoros, and Zoltán Rádai. 2022. "From Parasitized to Healthy-Looking Ants (Hymenoptera: Formicidae): Morphological Reconstruction Using Algorithmic Processing" Life 12, no. 5: 625. https://doi.org/10.3390/life12050625
APA StyleCsősz, S., Báthori, F., Molet, M., Majoros, G., & Rádai, Z. (2022). From Parasitized to Healthy-Looking Ants (Hymenoptera: Formicidae): Morphological Reconstruction Using Algorithmic Processing. Life, 12(5), 625. https://doi.org/10.3390/life12050625







