1. Introduction
Sleep is an indispensable physiological phenomenon for human beings, which acts as preventive medicine for physical and mental diseases and mood improvement [
1]. However, due to social competition, work pressure, and the accelerated aging of the population, sleep disorders have become health risks that cannot be ignored; these disorders are mainly manifested as insomnia, circadian rhythm disorders, and obstructive sleep apnea (OSA) syndrome [
2,
3]. The incidence and characteristics of various sleep disorders vary at different sleep stages. In order to make diagnoses, sleep specialists have introduced polysomnogram (PSG) [
4] to monitor and record data from the body. PSG is a biological signal obtained through various sensors on different parts of the body, including an electroencephalogram (EEG), electrooculogram (EOG), electromyogram (EMG), and electrocardiogram (ECG). EEG is a cost-effective and, typically, a non-invasive test for monitoring and recording electrical activity during sleep. Moreover, EMGs and EOGs have been used as two important switches for detecting rapid eye movement (REM) sleep [
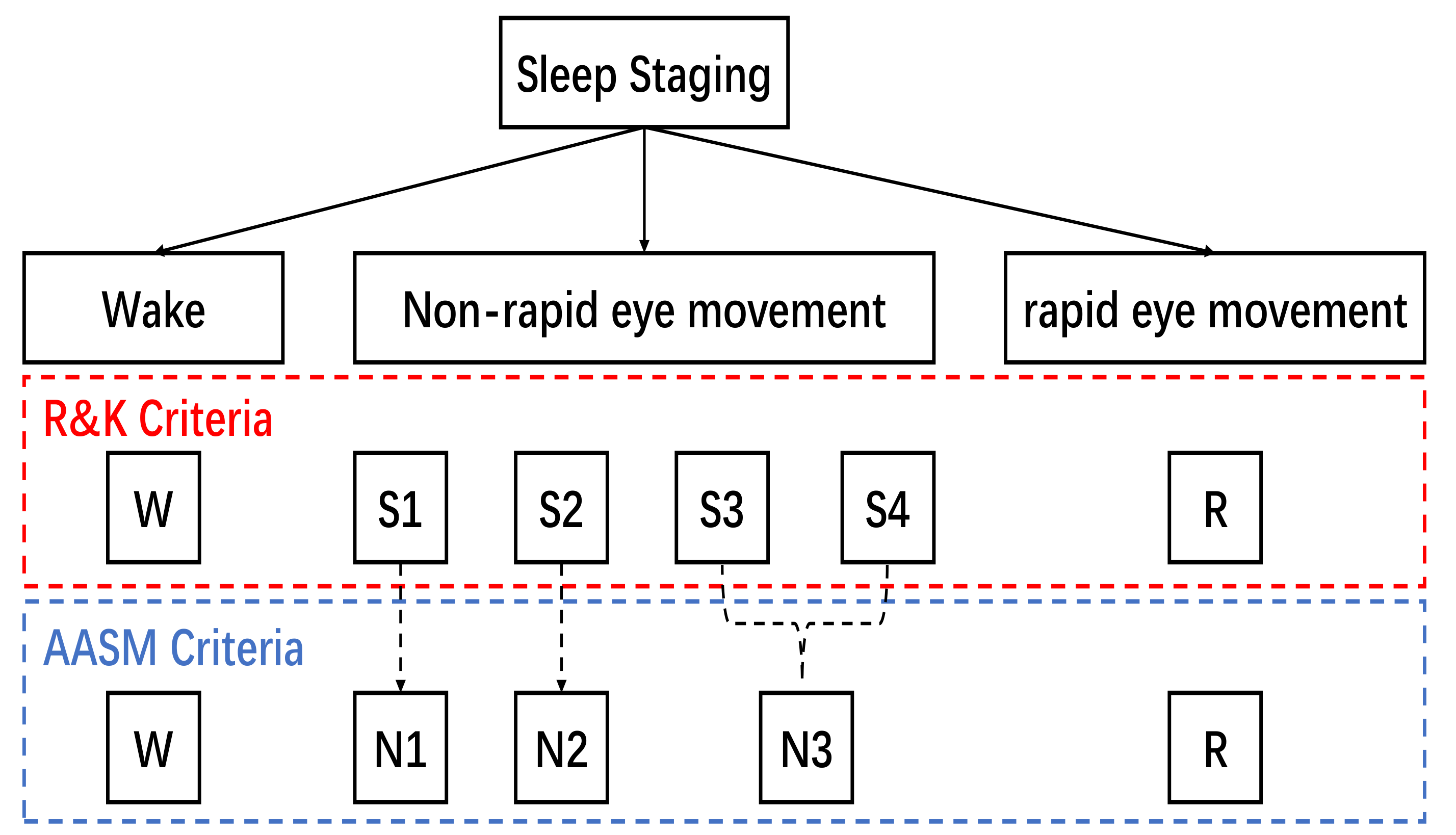
5]. Therefore, human experts need to combine other biological signals (such as EEG, EOG, and EMG) to achieve manual sleep stage classification. Rechtschaffen and Kales (R&K) [
6] delineated six sleep stages during sleep using early PSG. They categorize non-rapid eye movements (NREMs) into four sleep stages (
,
,
, and
). For standardization, the American Academy of Sleep Medicine (AASM) [
7] has defined the sleep staging criteria to achieve sleep scoring. According to the AASM manual, sleep experts use consecutive 30-s epochs of PSG data to classify five stages. These are wake, rapid eye movement (also referred to as stage
R), and three NREMs,
,
, and
. Based on the R&K criteria or the AASM criteria, sleep stages are shown in
Figure 1. Manual sleep stage classification is a laborious task [
8]. Therefore, automatic sleep stage classification with rapid and high accuracy based on EEG signals is of great research interest.
Looking back on the past decades, the various methods in the relevant studies on sleep stage classification have been proposed. According to studies [
1], sleep stage research has far-reaching implications for biomedical practice. In the early days, researchers used the hand-engineered feature-based methods to extract features in the time and frequency domains for sleep stage analysis. For example, Tsinalis et al. [
9] made the precision of sleep stage classification up to 78.9% via the extracted features in the time-frequency domains. Lee et al. [
10] developed an automatic sleep staging system with a mean percentage agreement of 75.52% for diagnosing OSA, using single-channel frontal EEG to classify wake, light sleep, deep sleep, and REM sleep. In order to achieve sleep stage classification, some machine learning-based methods [
11,
12] have been introduced in sleep stage classifications, e.g., support vector machine (SVM) [
13] and random forest [
14]. However, these methods have some limitations, such as the need to observe each PSG epoch for extracting features with a prior knowledge. For the time being, more studies are focusing on deep learning-based methods. Owing to the availability of high-quality datasets of EEG signals, deep learning-based methods are widely used to extract features from EEG signals for sleep stage classification. In our opinion, the latest deep learning-based methods for sleep stage classification can be split into two categories: non-GCN-based methods and GCN-based methods.
- 1.
Non-GCN-based Methods
More studies are solving the task of sleep stage classification based on recurrent neural networks (RNNs) and convolutional neural networks (CNNs). RNNs are commonly used to model the temporal dynamics of EEG signals [
15]. In SeqSleepNet [
16], a hierarchical RNN is used to model sleep staging and achieve accuracy up to 87.1%. In RNN, there are two kinds of the most representative structures, long short-term memory (LSTM) [
17] and gated recurrent unit (GRU) [
18]. For example, IITnet [
19] is proposed to automatically score sleep stages via BiLSTM. However, the problem of gradient disappearance or explosion occurs during RNN training, which makes it difficult to train a deep RNN model. Compared to RNNs, CNNs have high performance in parallel computing. To extract local and global features, Tsinalis et al. [
20] proposed an automatic classification approach for sleep stage scoring based on single-channel EEG. Phan et al. [
21] used a simple yet efficient CNN to extract sleep features from EEG signals. In addition, SleepEEGNet [
22] employs deep CNNs as the backbone network for sleep stage classification, achieving an accuracy of 84.26 %. Chanbon et al. [
23] introduce an end-to-end deep learning approach for sleep stage classification using multivariate and multimodal EEG signals. Furthermore, there are some works that combine CNN with RNN to simultaneously extract spatial and temporal features for sleep stage classification, e.g., DeepSleepNet [
24] and TinySleepNet [
25]. However, EEGs are non-Euclidean data, which naturally results in CNNs and RNNs being limited in feature extractions. Furthermore, their development potential is further hindered by the enormous parameter overhead.
- 2.
GCN-based methods.
The graph convolutional network (GCN) [
26] is an advanced neural network structure for processing graph structured data. Since EEG channels are structured data with temporal relations, each channel can be considered as a node in the graph. For this reason, GCN-based methods have been proven to be more powerful in processing EEGs. Joint analysis of EEG and eye-tracking recordings is raised by Zhang et al. [
27], whose strategy is to introduce GCN to fuse features. However, EEG channel signals include the temporal dynamic information of brain activity and the functional dependence between brain regions. To remedy the deficiency of the traditional spatiotemporal prediction model, the spatiotemporal graph convolutional network (ST-GCN) [
28] is proposed to model spatiotemporal relations and to learn the dynamic EEG for the task of sleep staging. For example, the GraphSleepNet [
29] is proposed to utilize brain spatial features and transition information among sleep stages to achieve more specific performance. However, the dependence on non-adjacent electrodes placed in different brain regions is often overlooked. Since then, Jia et al. [
30] propose an multi-view spatial-temporal graph convolutional network (MSTGCN) to extract the most relevant spatial and temporal information with superior performance. They introduce spatiotemporal attention to extract temporal and spatial information, respectively. However, this method makes it ineffective to capture the spatiotemporal dependencies on separated attention.
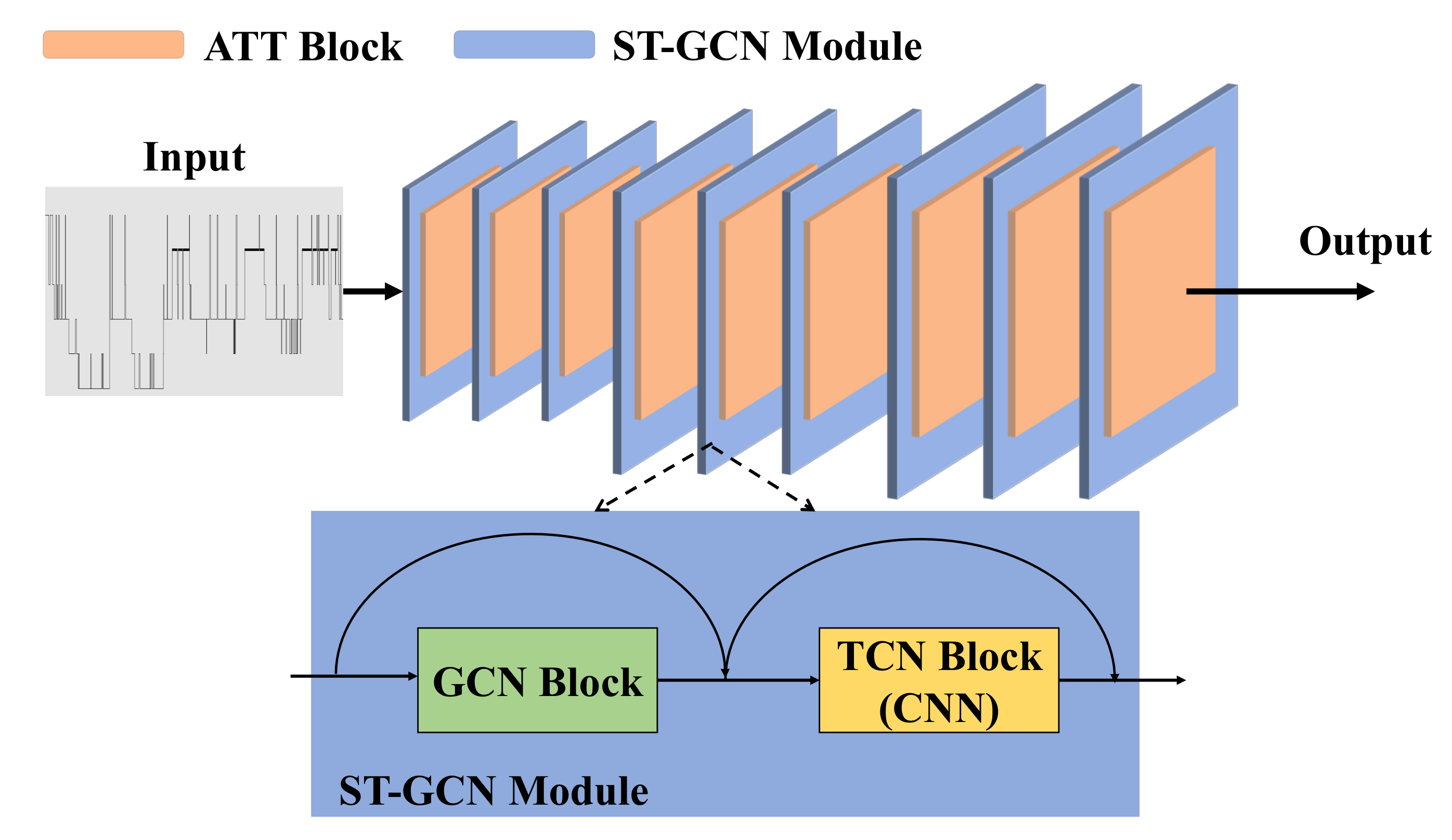
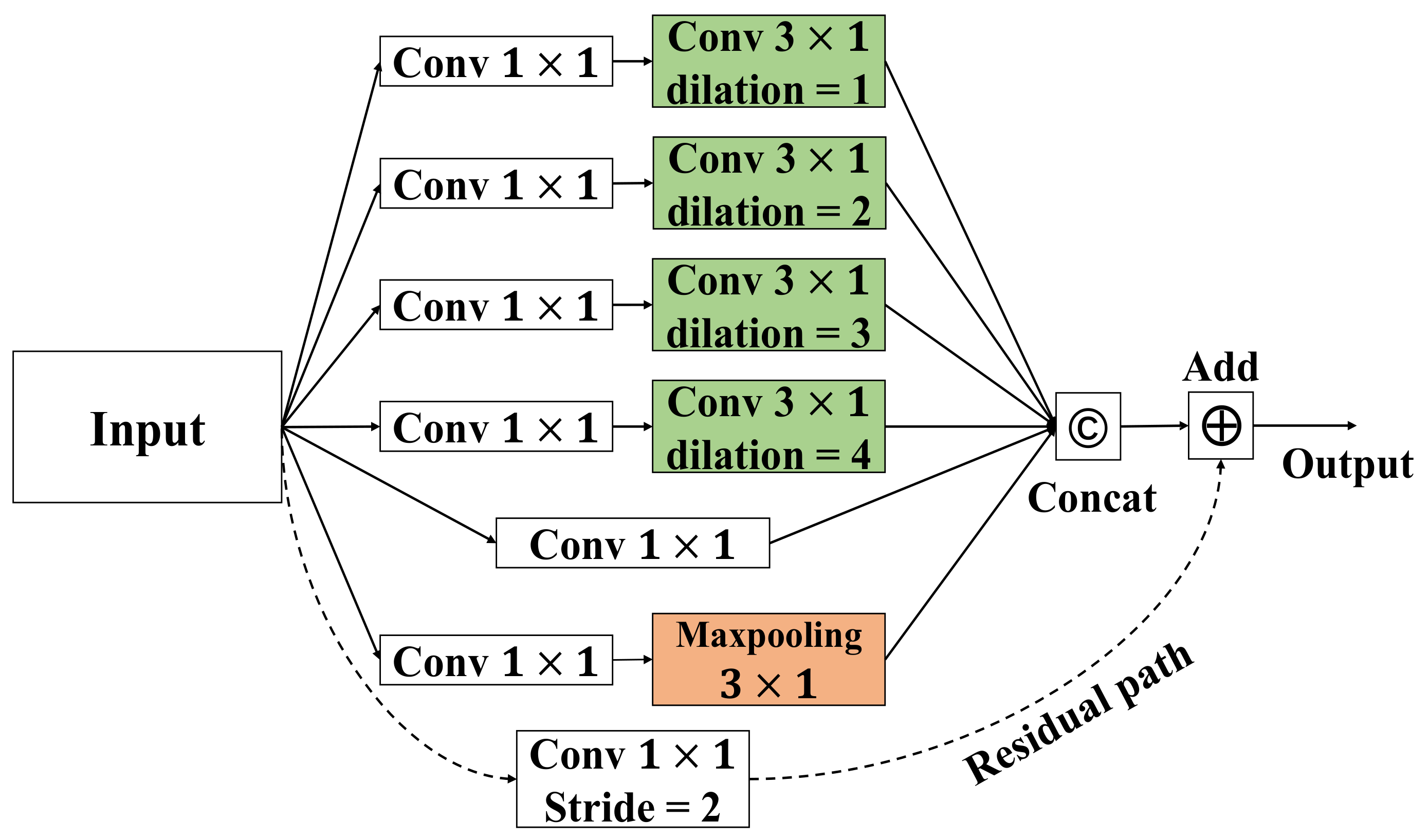
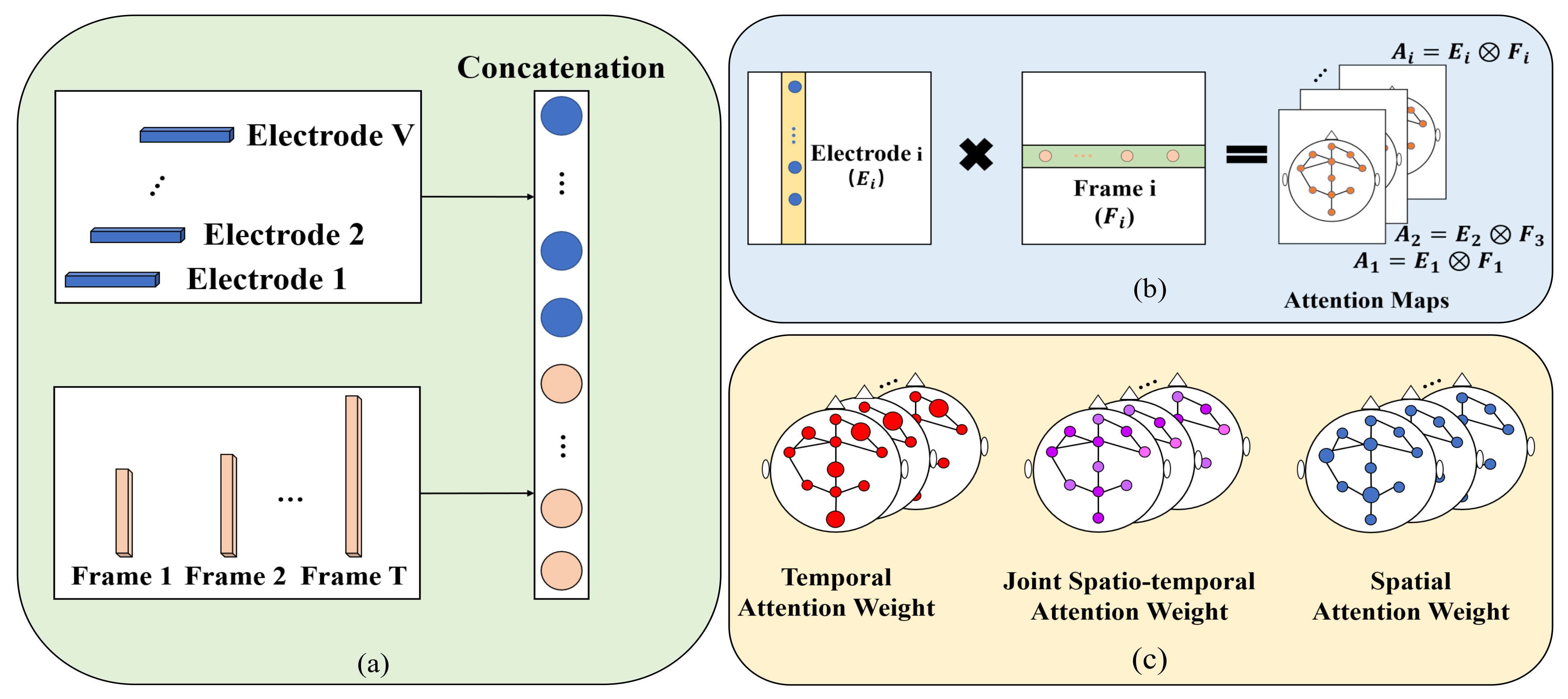
After summarizing the previous works, there are three shortcomings that need to be solved: (1) topological connections of electrodes in context are not well captured; (2) these methods force GCNs to aggregate features in different channels with the same topology, which limits the upper bound of model performance; and (3) attention weights are not sufficient to summarize long-range spatiotemporal characteristics. In order to address the aforementioned challenges, we propose a combination of dynamic and static ST-GCN with inter-temporal attention blocks for automatic sleep stage classification.
Overall, the main contributions of our proposed approach can be summarized as follows:
In previous work, sleep stage classification is achieved by complex modeling. In contrast, our proposed method is to leverage spatial graph convolutions along with interleaving temporal convolutions to achieve spatiotemporal modeling, which can be simpler yet efficient.
The inter-temporal attention blocks are introduced to achieve an automatic sleep stage classification, which can withdraw the most informative information across space and time, further proving that capture spatiotemporal relation plays an important role in sleep stage classification.
The proposed model significantly outperforms state-of-the-art methods on the sleep-EDF and the subgroup III of the ISRUC-SLEEP dataset. Our proposed method achieves better performance with 91.0% and 87.4% accuracy, both outperforming the state-of-the-art methods (86.4% and 82.1%).
The rest of this paper is organized as follows: In
Section 2, we present a series of preparatory works for our study. In
Section 3, we briefly describe the proposed network framework, including the dynamic and static ST-GCN and the inter-temporal attention block. The dataset used, the experiments, and the experimental results are presented in
Section 4. Moreover, finally, we conclude this work and provide an outlook on future work in
Section 5.
2. Preliminaries
A sleep stage network is described as an undirected graph
, where
is the collection of
N nodes representing electrodes in the brain, and the edge set
E represents the connection between nodes captured by an adjacency matrix
.
A is a matrix composed of 0 and 1, where 1 represents that the corresponding electrodes are connected, and 0 otherwise. Graph
G is made up of a 30-s EEG signal sequence
. The sleep feature matrix is the input of
G. We define the raw signal sequence as
, where
m denotes the number of samples,
Q means the number of electrodes, and
T is the time series length of each sample
. Inspired by Hyvräinen’s work [
31], we can extract the features of differential entropy (DE) on different frequency bands and define them on each sample feature matrix. Therefore, we can obtain a feature matrix at each sample
i, denoting the
features of the nodes
N.
Therein, denotes the features of electrode node n at sample i.
The objective of our study is to establish a mapping relationship between sleep signals and sleep stages using a spatiotemporal neural graph network. The issue of sleep stage is described as follows:
The given Equation (
2) can identify the current sleep stage
S. Therein,
denotes the temporal context of
,
S denotes the sleep stage class label defined by
,
indicates the length of sleep stage networks,
d denotes the temporal context coefficient, and
k is the number of intercepted time segments in a continuous EEG signal.
5. Discussion
Sleep disorders are highly prevalent in the world. Especially in the United States, nearly 25% of adults suffer from sleep disorders [
57]. Sleep disorders not only affect the quality of life, but also lead to health problems, such as heart disease and stroke. For people with sleep disorders to obtain adequate sleep, they may require the help of an appropriate method for sleep stage classification. In this work, we use a combination of dynamic and static ST-GCN with inter-temporal attention blocks to automatically classify sleep stages. We first consider that the distribution of brain electrodes is characteristic of non-Euclidean data. After the addition of ATT blocks, the sleep stage classification network achieves better performance. This confirms that spatial and temporal correlations play an important role in the sleep stage classification. The obtained results suggest that our method is promising in detecting new abnormalities in sleep and continuously improving our understanding of sleep mechanisms.
The NREM stages are divided into three sleep stages (
,
, and
) and are associated with the depth of sleep. Research shows that the stage
may affect the ability to learn new information and memory retention [
58]. In simple terms,
is the deepest sleep stage, which has the strongest repair function. Tafaro et al. [
59] report a positive relationship between sleep quality and survival in centenarians. From our experiment, the proposed method shows excellent performance in classifying the stage
compared with stages
and
. Therefore, accurate detection of the stage
provides an aid to long-term care, health and welfare services for the elderly. One study [
60] shows that patients with
in REM sleep had a significantly more collapsed airway and better ventilatory control stability compared with NREM sleep. Moreover, as it is suggested that the increased proportion of
stage may reveal a lower severity of OSA [
61], our method can be used as an ancillary treatment.
There are some challenges in more generic terms. First, since the stage is a transition period between wakefulness and sleep, it is difficult to detect this stage correctly. The system should be improved for the diagnosis of sleep fragmentation, such as obstructive sleep apnea. Second, the dataset is not perfect due to human errors. As far as we know, sleep scoring is defined by sleep experts. It is inevitable that similar sleep stages may be incorrectly marked. Therefore, the question for many sleep stage classification networks is how to use high-quality sleep stage datasets for the training process. In the future, we will develop a sleep stage system that provides more human-like performance to overcome the above challenges.