Profiling Levels of Serum microRNAs and Soluble ACE2 in COVID-19 Patients
Abstract
:1. Introduction
2. Subjects, Materials, and Methods
2.1. Subjects
2.2. Serum Collection and RNA Extraction
2.3. Human Serum miRNA Quantification
2.4. Quantification of sACE2 Using ELISA
2.5. Statistical Analysis
3. Results
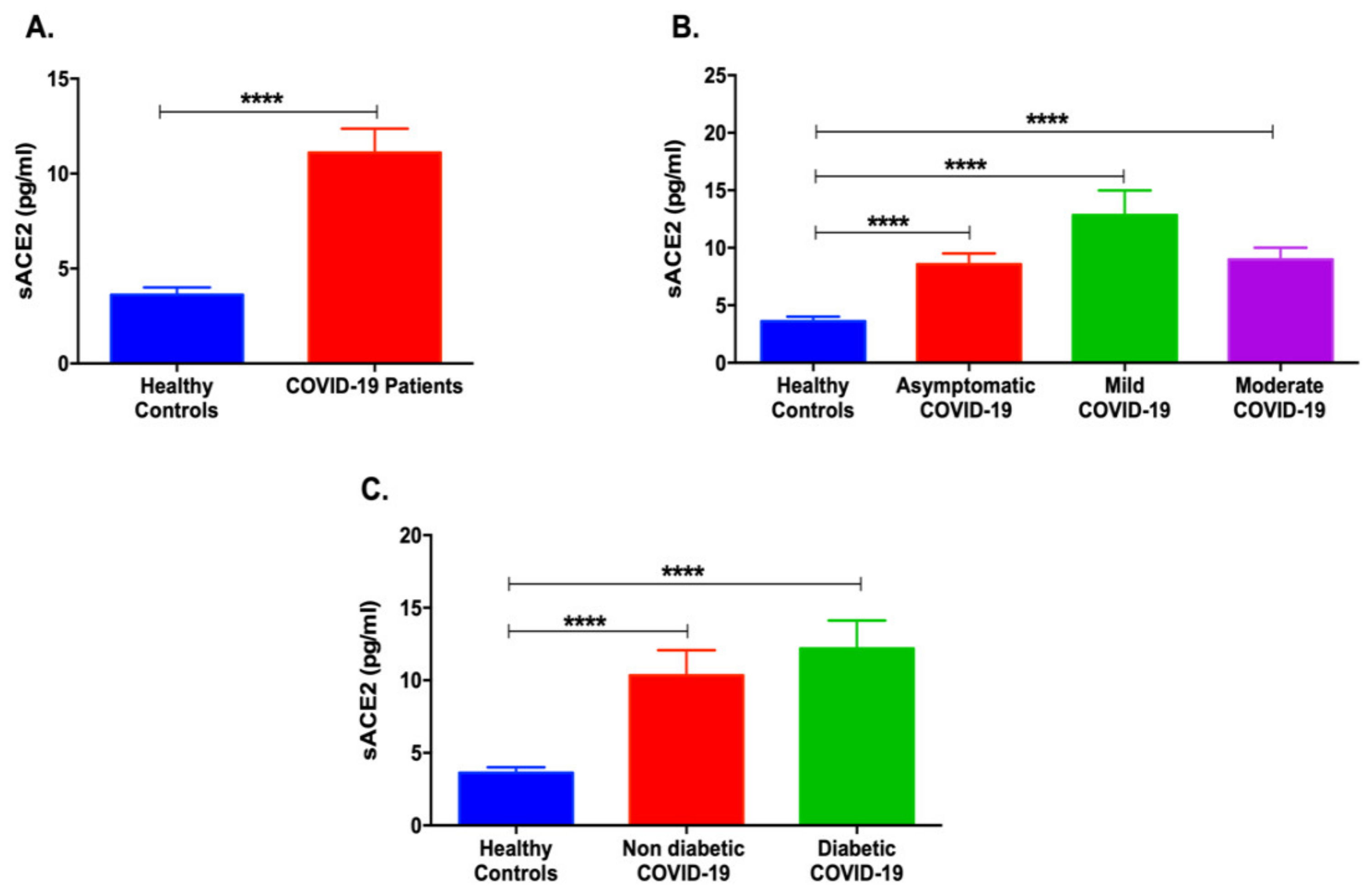
3.1. Serum sACE2 Levels Are Increased in COVID-19 Patients with Different Severities
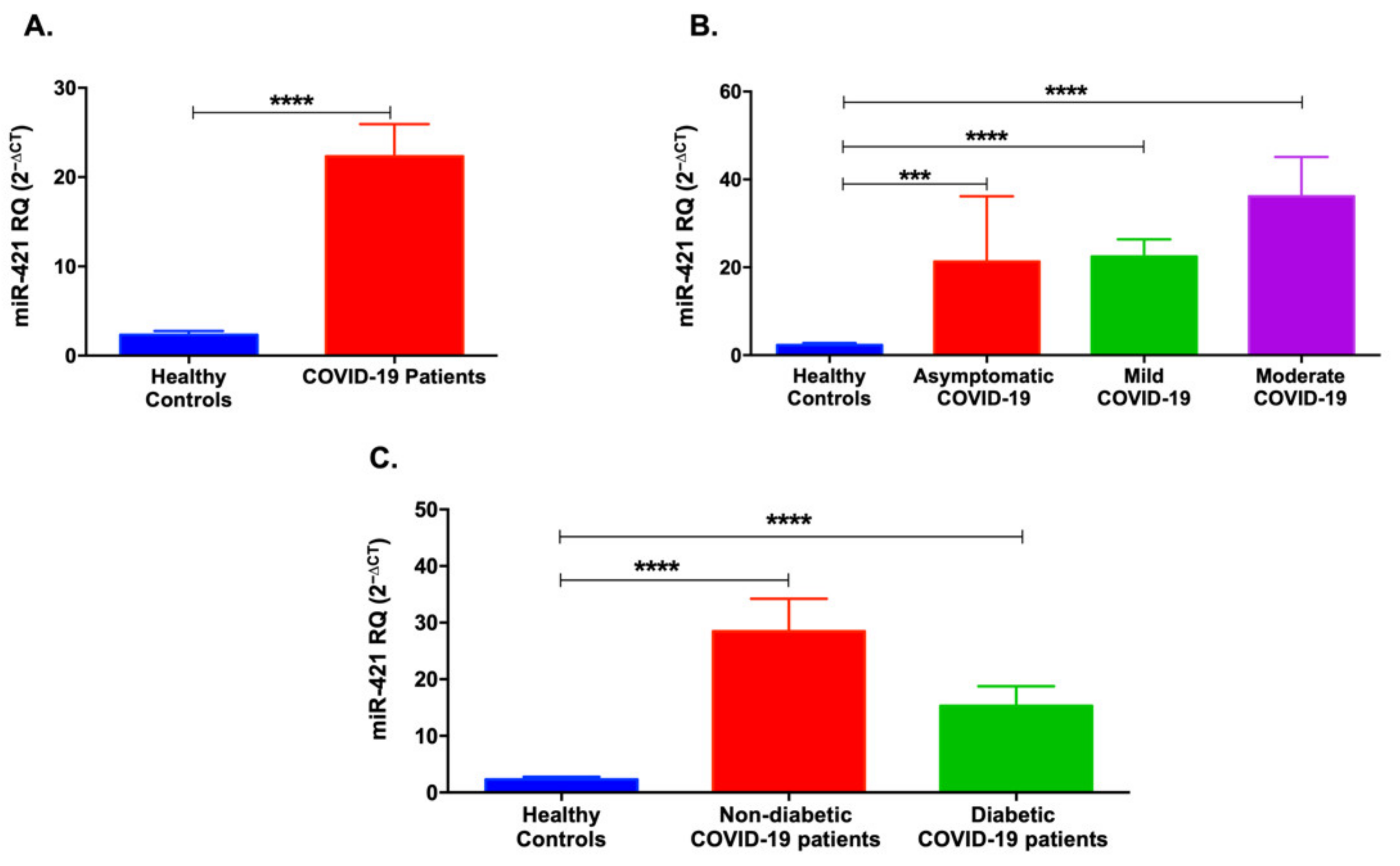
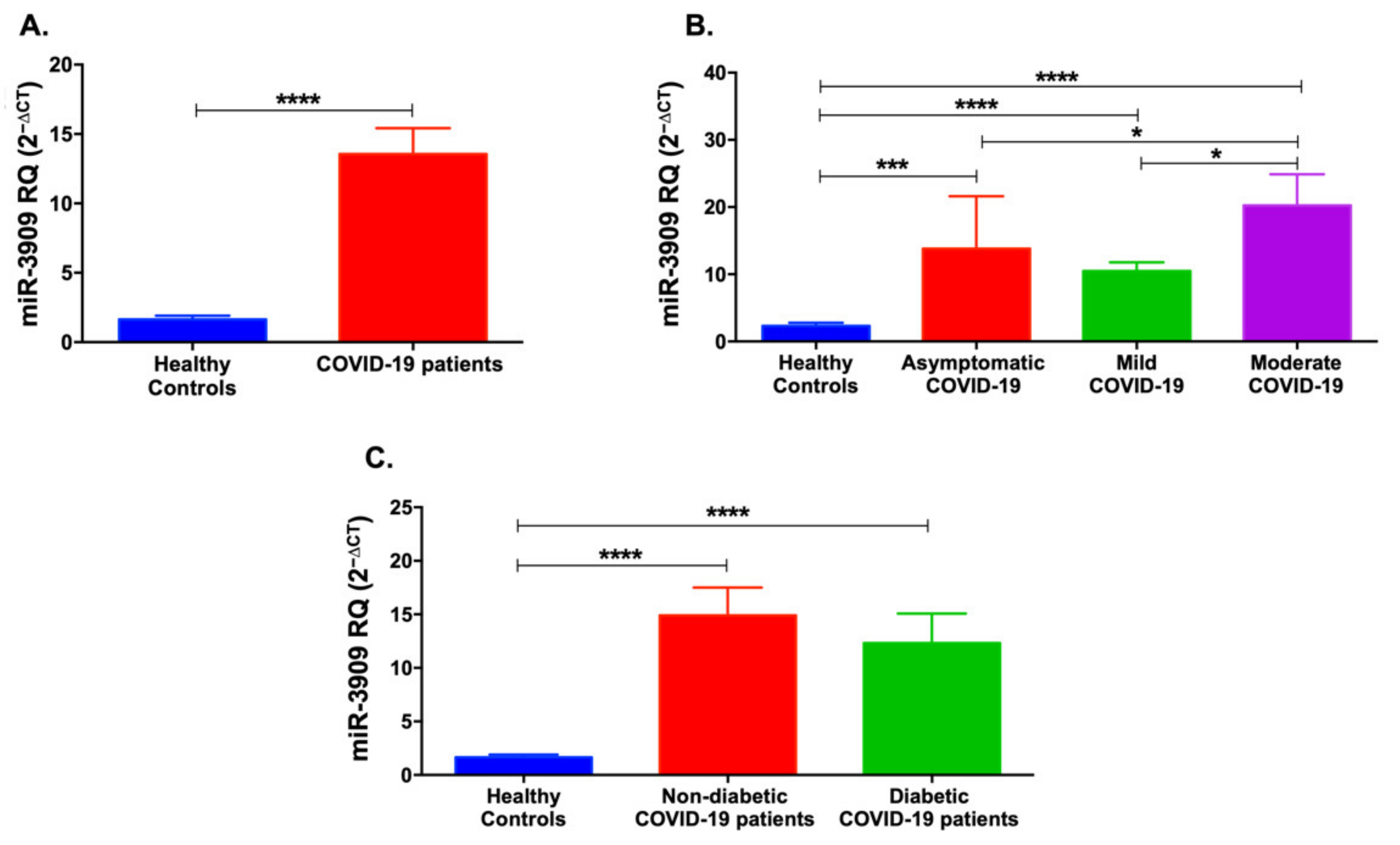
3.2. Serum Levels of miR-421 and miR-3909 Are Higher in COVID-19 Patients
3.3. Serum Levels of miR-212-5p and miR-4677-3p Are Higher in COVID-19 Patients and Are Affected by Disease Severity
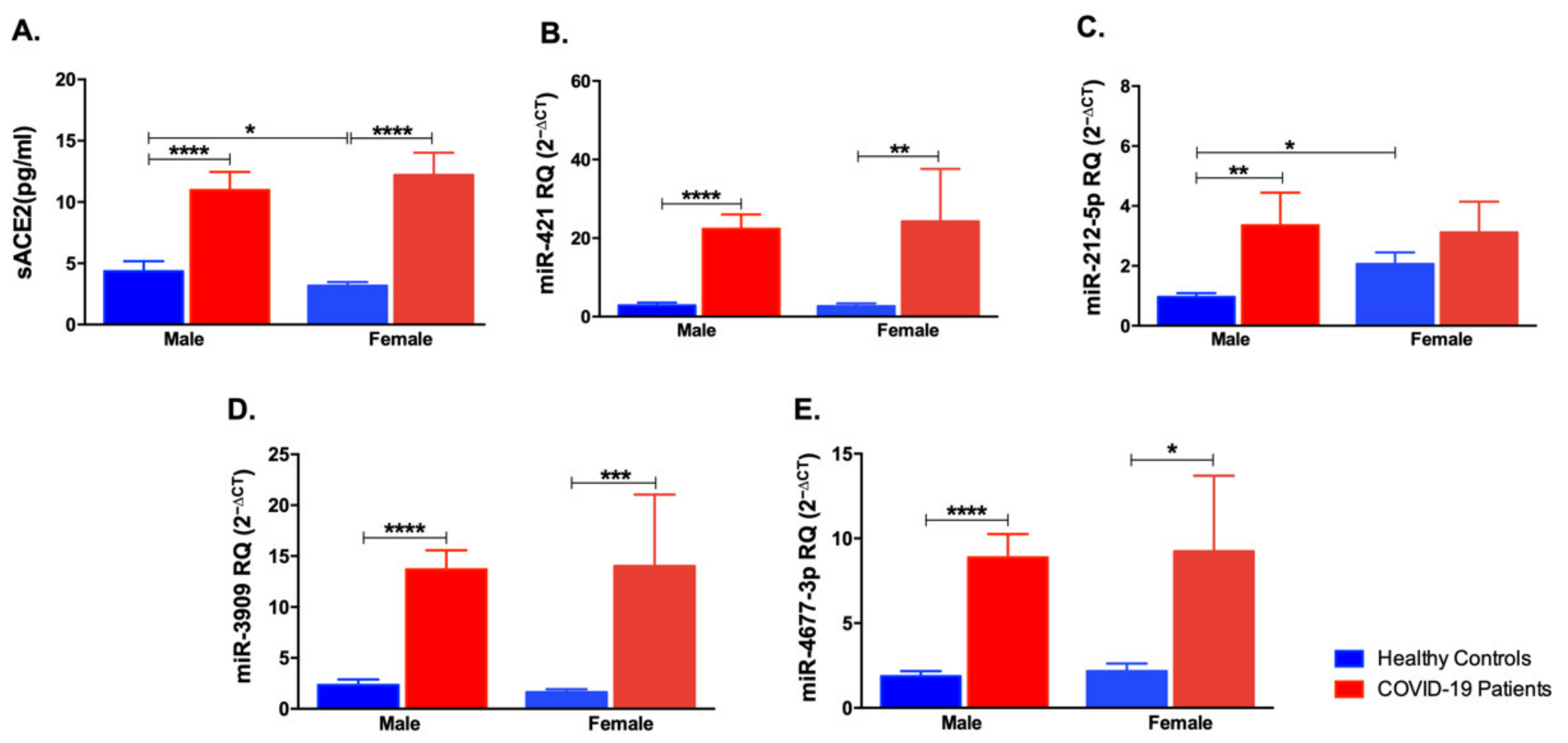
3.4. Impact of Gender Variation on the Expression of sACE2, miR-421, miR-212-5p, miR-3909, and miR-4677-3p
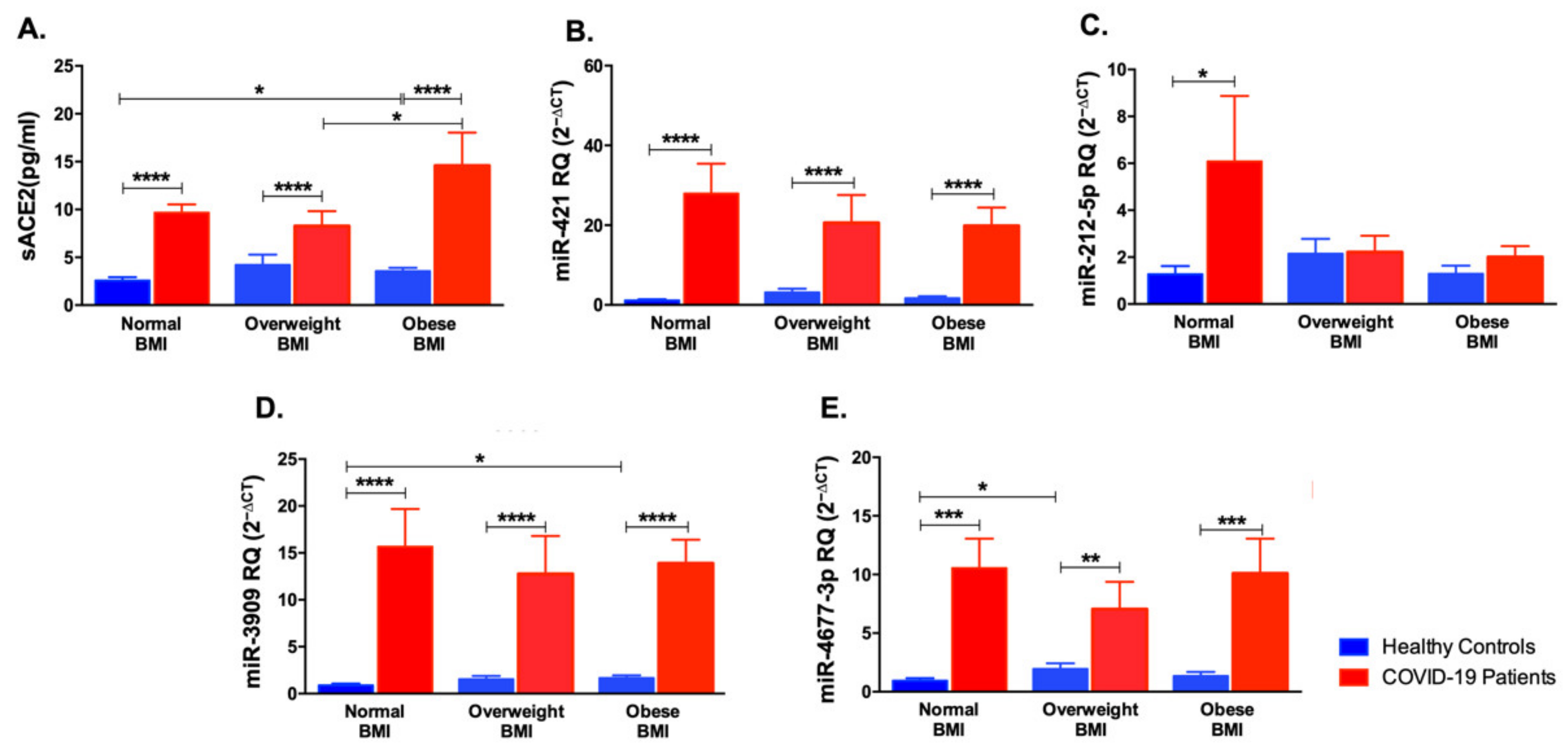
3.5. Impact of Body Mass Index Variation on the Expression of sACE2, miR-421, miR-212-5p, miR-3909, and miR-4677-3p
3.6. Correlation Analysis between sACE2, miRNAs and the Clinical and Laboratory Investigations of COVID-19 Patients
4. Discussion
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sardu, C.; Gargiulo, G.; Esposito, G.; Paolisso, G.; Marfella, R. Impact of diabetes mellitus on clinical outcomes in patients affected by Covid-19. Cardiovasc. Diabetol. 2020, 19, 76. [Google Scholar] [CrossRef] [PubMed]
- Mauvais-Jarvis, F. Aging, Male Sex, Obesity, and Metabolic Inflammation Create the Perfect Storm for COVID-19. Diabetes 2020, 69, 1857–1863. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.J.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Wijnant, S.R.; Jacobs, M.; Van Eeckhoutte, H.P.; Lapauw, B.; Joos, G.F.; Bracke, K.R.; Brusselle, G.G. Expression of ACE2, the SARS-CoV-2 Receptor, in Lung Tissue of Patients With Type 2 Diabetes. Diabetes 2020, 69, 2691. [Google Scholar] [CrossRef]
- Taneera, J.; El-Huneidi, W.; Hamad, M.; Mohammed, A.K.; Elaraby, E.; Hachim, M.Y. Expression Profile of SARS-CoV-2 Host Receptors in Human Pancreatic Islets Revealed Upregulation of ACE2 in Diabetic Donors. Biology 2020, 9, 215. [Google Scholar] [CrossRef]
- Lambert, D.W.; Yarski, M.; Warner, F.J.; Thornhill, P.; Parkin, E.T.; Smith, A.I.; Hooper, N.M.; Turner, A.J. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). J. Biol. Chem. 2005, 280, 30113–30119. [Google Scholar] [CrossRef] [Green Version]
- Elemam, N.; Hasswan, H.; Aljaibeji, H.; Sulaiman, N. Circulating Soluble ACE2 and Upstream microRNA Expressions in Serum of Type 2 Diabetes Mellitus Patients. Int. J. Mol. Sci. 2021, 22, 5263. [Google Scholar] [CrossRef]
- Osman, I.O.; Melenotte, C.; Brouqui, P.; Million, M.; Lagier, J.-C.; Parola, P.; Stein, A.; La Scola, B.; Meddeb, L.; Mege, J.-L.; et al. Expression of ACE2, Soluble ACE2, Angiotensin I, Angiotensin II and Angiotensin-(1-7) Is Modulated in COVID-19 Patients. Front. Immunol. 2021, 12, 625732. [Google Scholar] [CrossRef]
- Patel, S.K.; Juno, J.A.; Lee, W.S.; Wragg, K.M.; Hogarth, P.M.; Kent, S.J.; Burrell, L.M. Plasma ACE2 activity is persistently elevated following SARS-CoV-2 infection: Implications for COVID-19 pathogenesis and consequences. Eur. Respir. J. 2021, 57, 2003730. [Google Scholar] [CrossRef]
- Kragstrup, T.W.; Singh, H.S.; Grundberg, I.; Nielsen, A.L.-L.; Rivellese, F.; Mehta, A.; Goldberg, M.B.; Filbin, M.R.; Qvist, P.; Bibby, B.M. Plasma ACE2 predicts outcome of COVID-19 in hospitalized patients. PLoS ONE 2021, 16, e0252799. [Google Scholar] [CrossRef]
- Bhalla, V.; Blish, C.A.; South, A.M. A historical perspective on ACE2 in the COVID-19 era. J. Hum. Hypertens. 2021, 35, 935–939. [Google Scholar] [CrossRef]
- Han, Y.; Král, P. Computational Design of ACE2-Based Peptide Inhibitors of SARS-CoV-2. ACS Nano 2020, 14, 5143–5147. [Google Scholar] [CrossRef] [Green Version]
- Peter, E.K.; Schug, A. The inhibitory effect of a coronavirus spike protein fragment with ACE2. Biophys. J. 2021, 120, 1001–1010. [Google Scholar] [CrossRef]
- Li, C.H.; Chen, Y. Small and Long Non-Coding RNAs: Novel Targets in Perspective Cancer Therapy. Curr Genom. 2015, 16, 319–326. [Google Scholar] [CrossRef] [Green Version]
- Hum, C.; Loiselle, J.; Ahmed, N.; Shaw, T.A.; Toudic, C.; Pezacki, J.P. MicroRNA Mimics or Inhibitors as Antiviral Therapeutic Approaches Against COVID-19. Drugs 2021, 81, 517–531. [Google Scholar] [CrossRef]
- Sabbatinelli, J.; Giuliani, A.; Matacchione, G.; Latini, S.; Laprovitera, N.; Pomponio, G.; Ferrarini, A.; Baroni, S.S.; Pavani, M.; Moretti, M.; et al. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech. Ageing Dev. 2021, 193, 111413. [Google Scholar] [CrossRef]
- Abdullah-Al-Kamran Khan, M.; Sany, M.R.U.; Islam, M.S.; Islam, A.B.M.M.K. Epigenetic Regulator miRNA Pattern Differences Among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 World-Wide Isolates Delineated the Mystery Behind the Epic Pathogenicity and Distinct Clinical Characteristics of Pandemic COVID-19. Front. Genet. 2020, 11, 765. [Google Scholar] [CrossRef]
- Fani, M.; Zandi, M.; Ebrahimi, S.; Soltani, S.; Abbasi, S. The role of miRNAs in COVID-19 disease. Future Virol. 2021, 16, 301–306. [Google Scholar] [CrossRef]
- Hamoudi, R.; Sharif-Askari, N.S.; Sharif-Askari, F.S.; Abusnana, S.; Aljaibeji, H.; Taneera, J.; Sulaiman, N. Prediabetes and diabetes prevalence and risk factors comparison between ethnic groups in the United Arab Emirates. Sci. Rep. 2019, 9, 17437. [Google Scholar] [CrossRef] [PubMed]
- Al Hosani, F.; Aden, B.; Al Memari, S.; Al Mazrouei, S.; Ajab, S.; Abid, M.; Alsuwaidi, A.R.; Grivna, M.; Paulo, M.S.; Sheek-Hussein, M. Epidemiology of asymptomatic and symptomatic Coronavirus Disease 2019 confirmed cases in the Emirate of Abu Dhabi, United Arab Emirates: Observational study. Medicine 2021, 100, e25219. [Google Scholar] [CrossRef] [PubMed]
- Nagy, B., Jr.; Fejes, Z.; Szentkereszty, Z.; Sütő, R.; Várkonyi, I.; Ajzner, É.; Kappelmayer, J.; Papp, Z.; Tóth, A.; Fagyas, M. A dramatic rise in serum ACE2 activity in a critically ill COVID-19 patient. Int. J. Infect. Dis. 2021, 103, 412–414. [Google Scholar] [CrossRef]
- Linsky, T.W.; Vergara, R.; Codina, N.; Nelson, J.W.; Walker, M.J.; Su, W.; Barnes, C.O.; Hsiang, T.Y.; Esser-Nobis, K.; Yu, K.; et al. De novo design of potent and resilient hACE2 decoys to neutralize SARS-CoV-2. Science 2020, 370, 1208–1214. [Google Scholar] [CrossRef] [PubMed]
- Zoufaly, A.; Poglitsch, M.; Aberle, J.H.; Hoepler, W.; Seitz, T.; Traugott, M.; Grieb, A.; Pawelka, E.; Laferl, H.; Wenisch, C.; et al. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir. Med. 2020, 8, 1154–1158. [Google Scholar] [CrossRef]
- Swärd, P.; Edsfeldt, A.; Reepalu, A.; Jehpsson, L.; Rosengren, B.E.; Karlsson, M.K. Age and sex differences in soluble ACE2 may give insights for COVID-19. Crit. Care 2020, 24, 221. [Google Scholar] [CrossRef]
- Kornilov, S.A.; Lucas, I.; Jade, K.; Dai, C.; Lovejoy, J.C.; Magis, A. Plasma levels of soluble ACE2are associated with sex, Metabolic Syndrome, and its biomarkers in a large cohort, pointing to a possible mechanism for increased severity in COVID-19. Crit. Care 2020, 24, 452. [Google Scholar] [CrossRef]
- Emilsson, V.; Gudmundsson, E.F.; Aspelund, T.; Jonsson, B.G.; Gudjonsson, A.; Launer, L.J.; Lamb, J.R.; Gudmundsdottir, V.; Jennings, L.L.; Gudnason, V. Serum levels of ACE2 are higher in patients with obesity and diabetes. Obes. Sci. Pract. 2020, 7, 239–243. [Google Scholar] [CrossRef]
- Farr, R.J.; Rootes, C.L.; Rowntree, L.C.; Nguyen, T.H.; Hensen, L.; Kedzierski, L.; Cheng, A.C.; Kedzierska, K.; Au, G.G.; Marsh, G.A.; et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009759. [Google Scholar] [CrossRef]
- Paul, S.; Vázquez, L.A.; Reyes-Pérez, P.R.; Estrada-Meza, C.; Alburquerque, R.A.; Pathak, S.; Banerjee, A.; Bandyopadhyay, A.; Chakraborty, S.; Srivastava, A. The role of microRNAs in solving COVID-19 puzzle from infection to therapeutics: A mini-review. Virus Res. 2022, 308, 198631. [Google Scholar] [CrossRef]
- Braga, A.A.; Bortolin, R.H.; E Graciano-Saldarriaga, M.; Hirata, T.D.; Cerda, A.; de Freitas, R.C.; Lin-Wang, H.T.; Borges, J.B.; França, J.I.; Masi, L.N.; et al. High serum miR-421 is associated with metabolic dysregulation and inflammation in patients with metabolic syndrome. Epigenomics 2021, 13, 423–436. [Google Scholar] [CrossRef]
- Abdolahi, S.; Hosseini, M.; Rezaei, R.; Mohebbi, S.R.; Rostami-Nejad, M.; Mojarad, E.N.; Mirjalali, H.; Yadegar, A.; Aghdaei, H.A.; Zali, M.R.; et al. Evaluation of miR-200c-3p and miR-421-5p levels during immune responses in the admitted and recovered COVID-19 subjects. Infect. Genet. Evol. 2022, 98, 105207. [Google Scholar] [CrossRef]
- Jiang, Z.; Ma, J.; Wang, Q.; Wu, F.; Ping, J.; Ming, L. Combination of Circulating miRNA-320a/b and D-Dimer Improves Diagnostic Accuracy in Deep Vein Thrombosis Patients. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 2031–2037. [Google Scholar] [CrossRef]
- Kriszta, G.; Kriszta, Z.; Váncsa, S.; Hegyi, P.J.; Frim, L.; Erőss, B.; Pethő, G.; Pintér, E. Effects of Angiotensin-Converting Enzyme Inhibitors and Angiotensin Receptor Blockers on Angiotensin-Converting Enzyme 2 Levels: A Comprehensive Analysis Based on Animal Studies. Front. Pharmacol. 2021, 12, 619524. [Google Scholar] [CrossRef]


| COVID-19 Patients (n = 59) | |
|---|---|
| Disease Severity | 34 Mild 17 Moderate 8 Asymptomatic |
| Gender (M/F) | |
| Male | 9/59 |
| Female | 50/59 |
| Age (Years) | 46.64 ± 14.20 |
| BMI (kg/m2) | 26.78 ± 4.865 |
| Comorbidities | |
| Diabetes | 27/59 |
| Hypertension | 19/59 |
| Dyslipidemia | 3/59 |
| Cardiovascular Diseases | 6/59 |
| Symptoms | |
| Fever | 37/59 |
| Headache | 3/56 |
| Flu | 1/59 |
| Dyspnea | 15/59 |
| Myalgia | 3/56 |
| Diarrhea | 5/59 |
| Cough | 28/59 |
| Vomiting | 6/59 |
| Nausea | 6/59 |
| Loss Of Smell | 0/59 |
| Loss Of Taste | 0/59 |
| Body ache | 3/59 |
| Sore Throat | 2/59 |
| Blood Tests | |
| ANC (×103 cells/(L) | 5.992 (3.64) |
| ALC (×103 cells/(L) | 1.596 (0.820) |
| ANC/ALC ratio | 5.698 (9.157) |
| Hb(g/dL) | 12.9 (2.65) |
| WBC (×103 cells/L) | 8.52 (4.13) |
| Liver Tests | |
| ALT (U/L) | 63.25 (108.7) |
| AST (U/L) | 46.522 (67.68) |
| Albumin (g/dL) | 4.37 (6.12) |
| Bilirubin (mg/dL) | 0.982 (1.655) |
| GGT (U/L) | 68.58 (54.47) |
| Inflammatory Markers | |
| CRP (mg/L) | 65.3 (76.0) |
| D-Dimer (µg/mL) | 1.35 (1.959) |
| Ferritin (ng/mL) | 832.8 (948.4) |
| LDH (U/L) | 312.3 (253.87) |
| Renal Function Tests | |
| Creatinine (mg/dL) | 0.924 (0.427) |
| BUN (mg/dL) | 27.53 (20.74) |
| Lipid Profile | |
| Triglycerides (mg/dL) | 109.75 (51.03) |
| Coagulation Tests | |
| PT (seconds) | 14.535 (1.577) |
| aPTT (seconds) | 39.26 (6.65) |
| Platelets (×103 cells/µL) | 252.82 (122.98) |
| r; p Values | miR-421 | miR-212-5p | miR-3909 | miR-4677-3p |
|---|---|---|---|---|
| miR-421 | 0.484; 1.49 × 10−7 | 0.828; 3.73 × 10−31 | 0.824; 7.51 × 10−29 | |
| miR-212-5p | 0.484; 1.49 × 10−7 | 0.391; 3.43 × 10−5 | 0.460; 1.00 × 10−6 | |
| miR-3909 | 0.828; 3.73 × 10−31 | 0.391; 3.43 × 10−5 | 0.708; 2.58 × 10−18 | |
| miR-4677-3p | 0.824; 7.51 × 10−29 | 0.460; 1.00 × 10−6 | 0.708; 2.58 × 10−18 |
| miR-421 | miR-3909 | miR-4677-3p | sACE2 | |
|---|---|---|---|---|
| sACE2 | r = 0.581, p = 4.1x 10−12 | r = 0.670, p = 6.9 × 10−17 | r = 0.448, p = 7.1 × 10−7 | - |
| Age | r = 0.371, p = 7 × 10−5 | r = 0.450, p = 8.6 × 10−7 | r = 0.340, p = 0.00046 | r = 0.480, p = 1.19 × 10−7 |
| BMI | r = −0.587, p = 1.1 × 10−10 | r = −0.586, p = 1.2x 10−10 | r = -0.459, p = 2.89 × 10−6 | r = −0.599, p = 3.54 × 10−11 |
| COVID-19 Patients | |
| miR-212-5p and D-Dimer | r = −0.467, p = 0.0008 |
| sACE2 and aPTT | r = 0.350, p = 0.0146 |
| sACE2 and Platelets | r = 0.298, p = 0.033 |
| Non-diabetic COVID-19 Patients | |
| sACE2 and Platelets | r = 0.50, p = 0.01 |
| Diabetic COVID-19 Patients | |
| miR-421 and ANC | r = 0.452, p = 0.02 |
| miR-421 and ANC/ALC ratio | r = 0.471, p = 0.015 |
| miR-212-5p and D-Dimer | r = −0.617, p = 0.0013 |
| miR-3909 and ANC | r = 0.573, p = 0.00219 |
| miR-3909 and WBC | r = 0.441, p = 0.024 |
| miR-3909 and ANC/ALC ratio | r = 0.513, p = 0.007 |
| sACE2 and CRP | r = 0.44, p = 0.024 |
| sACE2 and aPTT | r = 0.458, p = 0.021 |
| miR-4677-3p and ANC/ALC ratio | r = 0.468, p = 0.018 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elemam, N.M.; Hasswan, H.; Aljaibeji, H.; Sharif-Askari, N.S.; Halwani, R.; Taneera, J.; Sulaiman, N. Profiling Levels of Serum microRNAs and Soluble ACE2 in COVID-19 Patients. Life 2022, 12, 575. https://doi.org/10.3390/life12040575
Elemam NM, Hasswan H, Aljaibeji H, Sharif-Askari NS, Halwani R, Taneera J, Sulaiman N. Profiling Levels of Serum microRNAs and Soluble ACE2 in COVID-19 Patients. Life. 2022; 12(4):575. https://doi.org/10.3390/life12040575
Chicago/Turabian StyleElemam, Noha Mousaad, Hind Hasswan, Hayat Aljaibeji, Narjes Saheb Sharif-Askari, Rabih Halwani, Jalal Taneera, and Nabil Sulaiman. 2022. "Profiling Levels of Serum microRNAs and Soluble ACE2 in COVID-19 Patients" Life 12, no. 4: 575. https://doi.org/10.3390/life12040575
APA StyleElemam, N. M., Hasswan, H., Aljaibeji, H., Sharif-Askari, N. S., Halwani, R., Taneera, J., & Sulaiman, N. (2022). Profiling Levels of Serum microRNAs and Soluble ACE2 in COVID-19 Patients. Life, 12(4), 575. https://doi.org/10.3390/life12040575







