Deficiency of TTYH1 Expression Reduces the Migration and Invasion of U2OS Human Osteosarcoma Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Construction of Expression Vector and siRNA-Mediated TTYH1 Knockdown
2.3. Reverse Transcription Polymerase Chain Reaction (RT-PCR) and Real-Time Quantitative RT-PCR (qRT-PCR)
2.4. Immunoblotting
2.5. Immunofluorescence
2.6. Cell Viability Assay
2.7. Cell Invasion Assay
2.8. Cell Migration Assay
2.9. Statistical Analysis
3. Results
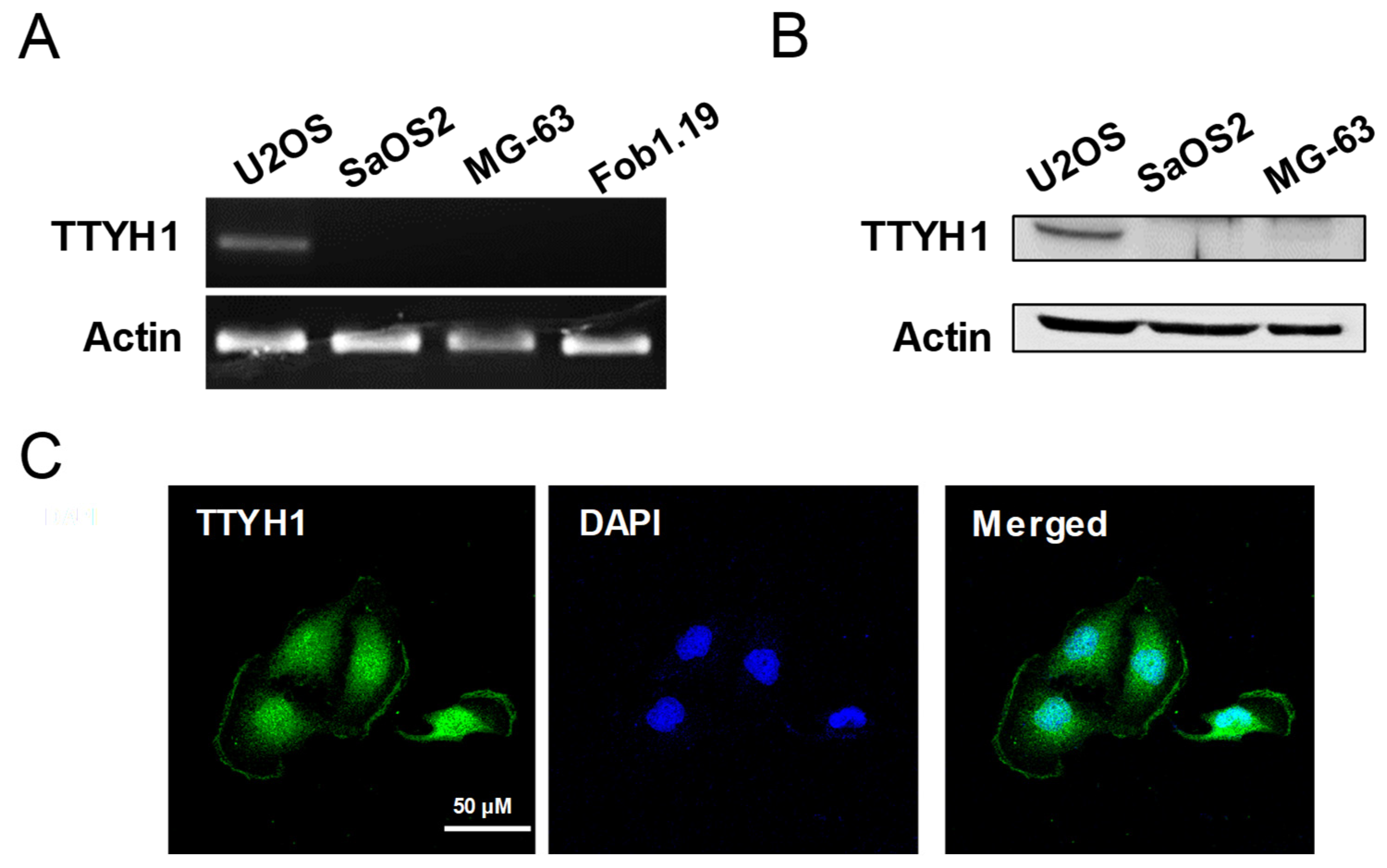
3.1. TTYH1 Was Endogenously Expressed in U2OS Cells
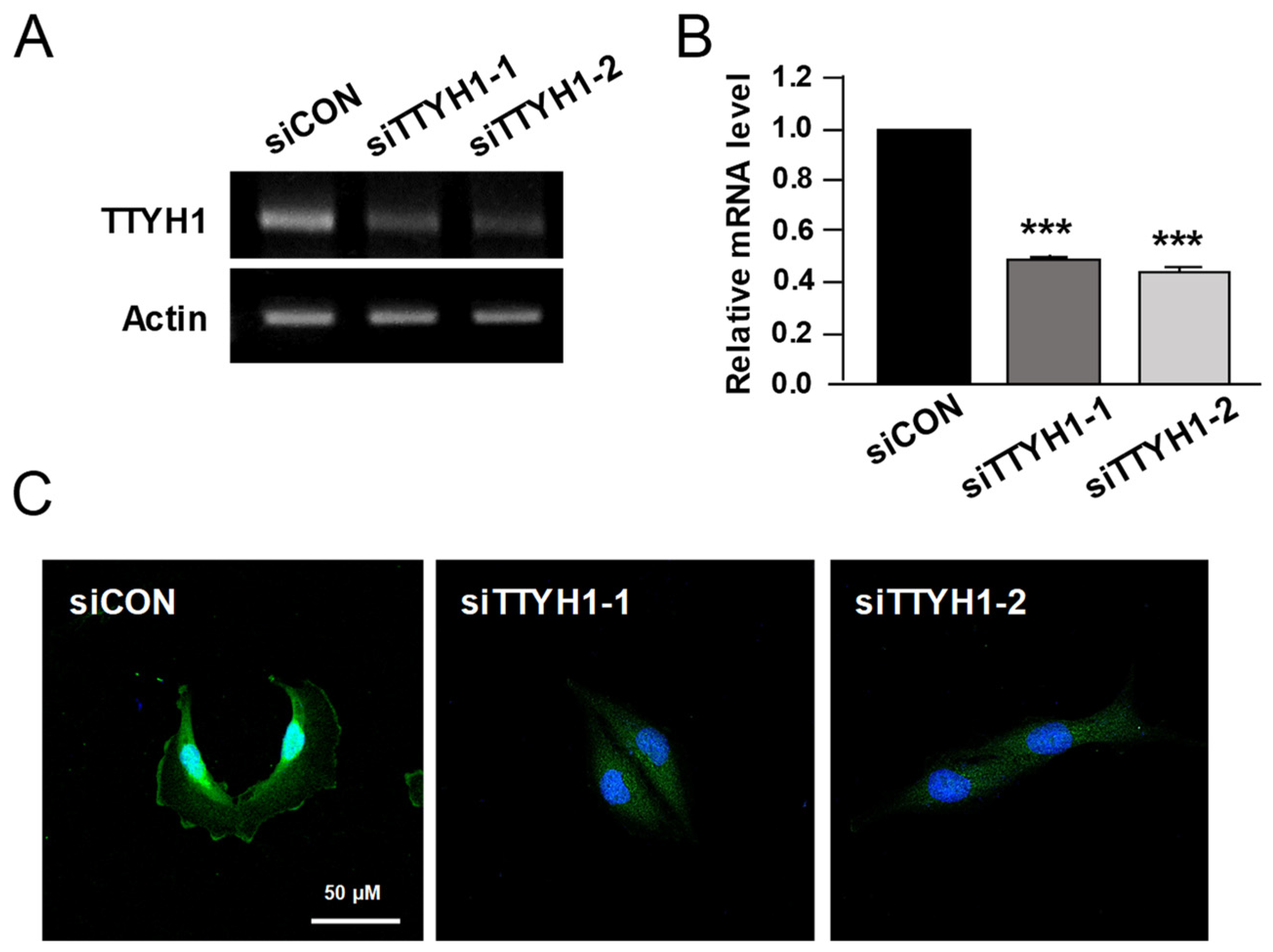
3.2. Silencing of TTYH1 Gene Expression Inhibited the Migration and Invasion of U2OS Cells
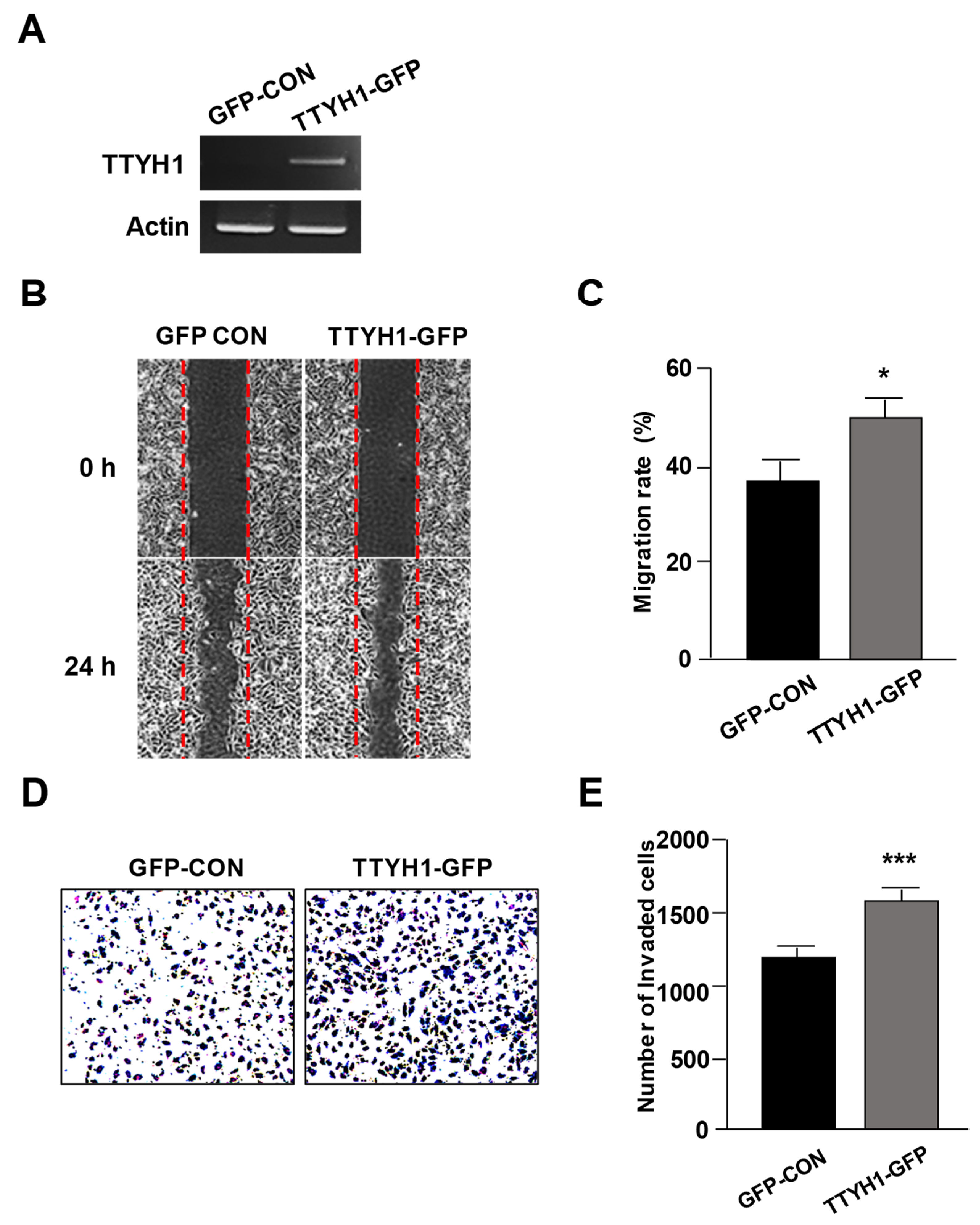
3.3. Overexpression of TTYH1 Promoted the Cell Migration and Invasion Abilities of a TTYH1 Non-Expressed Osteosarcoma Cell, SaOS2 Cell
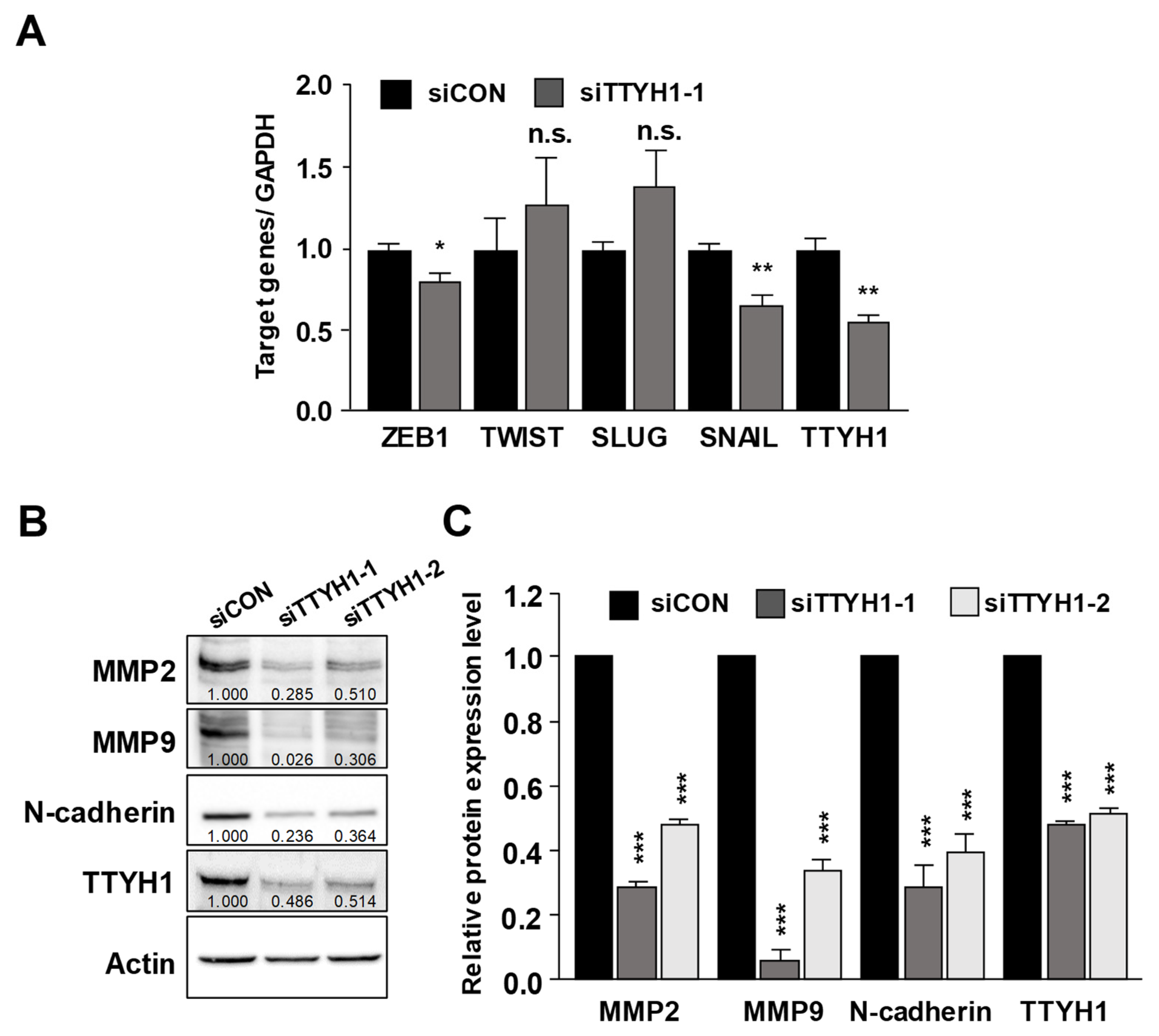
3.4. Silencing of the TTYH1 Gene Interfered with the Expression of Epithelial–Mesenchymal (EMT)-Related Factors
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Taran, S.J.; Taran, R.; Malipatil, N.B. Pediatric Osteosarcoma: An Updated Review. Indian J. Med. Paediatr. Oncol. 2017, 38, 33–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, E.; DeSantis, C.; Robbins, A.; Kohler, B.; Jemal, A. Childhood and adolescent cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 83–103. [Google Scholar] [CrossRef] [PubMed]
- Durfee, R.A.; Mohammed, M.; Luu, H.H. Review of Osteosarcoma and Current Management. Rheumatol. Ther. 2016, 3, 221–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peretti, M.; Angelini, M.; Savalli, N.; Florio, T.; Yuspa, S.H.; Mazzanti, M. Chloride channels in cancer: Focus on chloride intracellular channel 1 and 4 (CLIC1 AND CLIC4) proteins in tumor development and as novel therapeutic targets. Biochim. Biophys. Acta 2015, 1848, 2523–2531. [Google Scholar] [CrossRef] [Green Version]
- Jentsch, T.J.; Stein, V.; Weinreich, F.; Zdebik, A.A. Molecular structure and physiological function of chloride channels. Physiol. Rev. 2002, 82, 503–568. [Google Scholar] [CrossRef]
- Prevarskaya, N.; Skryma, R.; Shuba, Y. Ion channels and the hallmarks of cancer. Trends Mol. Med. 2010, 16, 107–121. [Google Scholar] [CrossRef]
- Gururaja Rao, S.; Patel, N.J.; Singh, H. Intracellular Chloride Channels: Novel Biomarkers in Diseases. Front. Physiol. 2020, 11, 96. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.S.; Lee, J.K.; Bae, Y.; Lee, B.S.; Kim, E.; Cho, C.H.; Ryoo, K.; Yoo, J.; Kim, C.H.; Yi, G.S.; et al. Suppression of 14-3-3gamma-mediated surface expression of ANO1 inhibits cancer progression of glioblastoma cells. Sci. Rep. 2016, 6, 26413. [Google Scholar] [CrossRef] [Green Version]
- Britschgi, A.; Bill, A.; Brinkhaus, H.; Rothwell, C.; Clay, I.; Duss, S.; Rebhan, M.; Raman, P.; Guy, C.T.; Wetzel, K.; et al. Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc. Natl. Acad. Sci. USA 2013, 110, E1026–E1034. [Google Scholar] [CrossRef] [Green Version]
- Shiwarski, D.J.; Shao, C.; Bill, A.; Kim, J.; Xiao, D.; Bertrand, C.A.; Seethala, R.S.; Sano, D.; Myers, J.N.; Ha, P.; et al. To “grow” or “go”: TMEM16A expression as a switch between tumor growth and metastasis in SCCHN. Clin. Cancer Res. 2014, 20, 4673–4688. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Cao, Q.H.; Lu, D.J.; Luo, B.; Lu, X.F.; Luo, R.C.; Wang, X.G. TMEM16A overexpression contributes to tumor invasion and poor prognosis of human gastric cancer through TGF-beta signaling. Oncotarget 2015, 6, 11585–11599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crottes, D.; Jan, L.Y. The multifaceted role of TMEM16A in cancer. Cell Calcium 2019, 82, 102050. [Google Scholar] [CrossRef] [PubMed]
- Campbell, H.D.; Schimansky, T.; Claudianos, C.; Ozsarac, N.; Kasprzak, A.B.; Cotsell, J.N.; Young, I.G.; de Couet, H.G.; Miklos, G.L. The Drosophila melanogaster flightless-I gene involved in gastrulation and muscle degeneration encodes gelsolin-like and leucine-rich repeat domains and is conserved in Caenorhabditis elegans and humans. Proc. Natl. Acad. Sci. USA 1993, 90, 11386–11390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, H.D.; Kamei, M.; Claudianos, C.; Woollatt, E.; Sutherland, G.R.; Suzuki, Y.; Hida, M.; Sugano, S.; Young, I.G. Human and mouse homologues of the Drosophila melanogaster tweety (tty) gene: A novel gene family encoding predicted transmembrane proteins. Genomics 2000, 68, 89–92. [Google Scholar] [CrossRef]
- Jung, E.; Osswald, M.; Blaes, J.; Wiestler, B.; Sahm, F.; Schmenger, T.; Solecki, G.; Deumelandt, K.; Kurz, F.T.; Xie, R.; et al. Tweety-Homolog 1 Drives Brain Colonization of Gliomas. J. Neurosci. 2017, 37, 6837–6850. [Google Scholar] [CrossRef]
- Bae, Y.; Kim, A.; Cho, C.H.; Kim, D.; Jung, H.G.; Kim, S.S.; Yoo, J.; Park, J.Y.; Hwang, E.M. TTYH1 and TTYH2 Serve as LRRC8A-Independent Volume-Regulated Anion Channels in Cancer Cells. Cells 2019, 8, 562. [Google Scholar] [CrossRef] [Green Version]
- Moon, D.K.; Bae, Y.J.; Jeong, G.R.; Cho, C.H.; Hwang, S.C. Upregulated TTYH2 expression is critical for the invasion and migration of U2OS human osteosarcoma cell lines. Biochem. Biophys. Res. Commun. 2019, 516, 521–525. [Google Scholar] [CrossRef]
- Nalamalapu, R.R.; Yue, M.; Stone, A.R.; Murphy, S.; Saha, M.S. The tweety Gene Family: From Embryo to Disease. Front. Mol. Neurosci. 2021, 14, 672511. [Google Scholar] [CrossRef]
- Schmalhofer, O.; Brabletz, S.; Brabletz, T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009, 28, 151–166. [Google Scholar] [CrossRef]
- Kim, J.; Han, D.; Byun, S.H.; Kwon, M.; Cho, J.Y.; Pleasure, S.J.; Yoon, K. Ttyh1 regulates embryonic neural stem cell properties by enhancing the Notch signaling pathway. EMBO Rep. 2018, 19, e45472. [Google Scholar] [CrossRef]
- Jentsch, T.J. VRACs and other ion channels and transporters in the regulation of cell volume and beyond. Nat. Rev. Mol. Cell Biol. 2016, 17, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.K.; Sorensen, B.H.; Sauter, D.P.; Lambert, I.H. Role of volume-regulated and calcium-activated anion channels in cell volume homeostasis, cancer and drug resistance. Channels 2015, 9, 380–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsey, B.A.; Markel, J.E.; Kleinerman, E.S. Osteosarcoma Overview. Rheumatol. Ther. 2017, 4, 25–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- PosthumaDeBoer, J.; Witlox, M.A.; Kaspers, G.J.; van Royen, B.J. Molecular alterations as target for therapy in metastatic osteosarcoma: A review of literature. Clin. Exp. Metastasis 2011, 28, 493–503. [Google Scholar] [CrossRef] [Green Version]
- Krakhmal, N.V.; Zavyalova, M.V.; Denisov, E.V.; Vtorushin, S.V.; Perelmuter, V.M. Cancer Invasion: Patterns and Mechanisms. Acta Nat. 2015, 7, 17–28. [Google Scholar] [CrossRef] [Green Version]
- Sannino, G.; Marchetto, A.; Kirchner, T.; Grunewald, T.G.P. Epithelial-to-Mesenchymal and Mesenchymal-to-Epithelial Transition in Mesenchymal Tumors: A Paradox in Sarcomas? Cancer Res. 2017, 77, 4556–4561. [Google Scholar] [CrossRef] [Green Version]
- Demirkan, B. The Roles of Epithelial-to-Mesenchymal Transition (EMT) and Mesenchymal-to-Epithelial Transition (MET) in Breast Cancer Bone Metastasis: Potential Targets for Prevention and Treatment. J. Clin. Med. 2013, 2, 264–282. [Google Scholar] [CrossRef] [Green Version]
- Niinaka, Y.; Harada, K.; Fujimuro, M.; Oda, M.; Haga, A.; Hosoki, M.; Uzawa, N.; Arai, N.; Yamaguchi, S.; Yamashiro, M.; et al. Silencing of autocrine motility factor induces mesenchymal-to-epithelial transition and suppression of osteosarcoma pulmonary metastasis. Cancer Res. 2010, 70, 9483–9493. [Google Scholar] [CrossRef] [Green Version]
- Smith, B.N.; Bhowmick, N.A. Role of EMT in Metastasis and Therapy Resistance. J. Clin. Med. 2016, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Garg, M. Epithelial-mesenchymal transition–activating transcription factors–multifunctional regulators in cancer. World J. Stem Cells 2013, 5, 188–195. [Google Scholar] [CrossRef]
- Gonzalez, D.M.; Medici, D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 2014, 7, re8. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Wang, R.; Zhao, C.; Wen, J.; Dong, H.; Wang, S.; Li, Y.; Zhao, Y.; Li, J.; Yang, Y.; et al. Notch signaling regulates osteosarcoma proliferation and migration through Erk phosphorylation. Tissue Cell 2019, 59, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Xia, K.; Gao, T.; Chen, J.; Zhang, Z.; Sun, X.; Simões, B.M.; Eyre, R.; Fan, Z.; Guo, W.; et al. The Notch Pathway Promotes Osteosarcoma Progression through Activation of Ephrin Reverse Signaling. Mol. Cancer Res. 2019, 17, 2383–2394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.N.; Cao, X.L.; Fang, Z.; Zhang, Y.F.; Han, W.J.; Yue, K.Y.; Cao, Y.; Zheng, M.H.; Wang, L.L.; Han, H. Deficiency of Ttyh1 downstream to Notch signaling results in precocious differentiation of neural stem cells. Biochem. Biophys. Res. Commun. 2019, 514, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Sukalskaia, A.; Straub, M.S.; Deneka, D.; Sawicka, M.; Dutzler, R. Cryo-EM Structures of the TTYH Family Reveal a Novel Architecture for Lipid Interactions. Nat. Commun. 2021, 12, 4893. [Google Scholar] [CrossRef]
- Li, B.; Hoel, C.M.; Brohawn, S.G. Structures of Tweety Homolog Proteins TTYH2 and TTYH3 Reveal a Ca2+-Dependent Switch from Intra- to Intermembrane Dimerization. Nat. Commun. 2021, 12, 6913. [Google Scholar] [CrossRef]
- Stefaniuk, M.; Swiech, L.; Dzwonek, J.; Lukasiuk, K. Expression of Ttyh1, a member of the Tweety family in neurons in vitro and in vivo and its potential role in brain pathology. J. Neurochem. 2010, 115, 1183–1194. [Google Scholar] [CrossRef]
- Matthews, C.A.; Shaw, J.E.; Hooper, J.A.; Young, I.G.; Crouch, M.F.; Campbell, H.D. Expression and evolution of the mammalian brain gene Ttyh1. J. Neurochem. 2007, 100, 693–707. [Google Scholar] [CrossRef]
- Wiernasz, E.; Kaliszewska, A.; Brutkowski, W.; Bednarczyk, J.; Gorniak, M.; Kaza, B.; Lukasiuk, K. Ttyh1 protein is expressed in glia in vitro and shows elevated expression in activated astrocytes following status epilepticus. Neurochem. Res. 2014, 39, 2516–2526. [Google Scholar] [CrossRef] [Green Version]

| Gene | Primer | Sequence |
|---|---|---|
| TTYH1 | Forward | 5′-CGAGAAGCTGTGCCTCAGTT-3′ |
| Reverse | 5′-TGCAGACAGCAGGGAGAAGA-3′ | |
| Actin | Forward | 5′-CACCAACTGGGACGACAT-3′ |
| Reverse | 5′-ACAGCCTGGATAGCAACG-3′ |
| Gene | Primer | Sequence |
|---|---|---|
| TTYH1 | Forward | 5′-ACAGTGAGACCAGTGATGGG-3′ |
| Reverse | 5′-GGTCAATGGTGCTGAGTGTG-3′ | |
| SNAIL | Forward | 5′-GGACCCACACTGGCGAGAAG-3′ |
| Reverse | 5′-ATTCGGGAGAAGGTCCGAGC-3′ | |
| SLUG | Forward | 5′-TGTGACAAGGAATATGTGAGCC-3′ |
| Reverse | 5′-TGAGCCCTCAGATTTGACCTG-3′ | |
| ZEB1 | Forward | 5′-CAGCCCTGCAGTCCAAGAAC-3′ |
| Reverse | 5′-TTGTCTTTCATCCTGATTTCCATTT-3′ | |
| TWIST | Forward | 5′-CGCCCCGCTCTTCTCCTCT-3′ |
| Reverse | 5′-GACTGTCCATTTTCTCCTTCTCTG-3′ | |
| GAPDH | Forward | 5′-GTCTTCACCACCATGGAGAA-3′ |
| Reverse | 5′-GCATGGACTGTGGTCATGAG-3′ | |
| Actin | Forward | 5′-CACCAACTGGGACGACAT-3′ |
| Reverse | 5′-ACAGCCTGGATAGCAACG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.-S.; Kwon, O.; Jeong, G.-R.; Noh, J.; Kim, S.E.; Yi, G.-S.; Hwang, E.M.; Park, J.-Y. Deficiency of TTYH1 Expression Reduces the Migration and Invasion of U2OS Human Osteosarcoma Cells. Life 2022, 12, 530. https://doi.org/10.3390/life12040530
Lee Y-S, Kwon O, Jeong G-R, Noh J, Kim SE, Yi G-S, Hwang EM, Park J-Y. Deficiency of TTYH1 Expression Reduces the Migration and Invasion of U2OS Human Osteosarcoma Cells. Life. 2022; 12(4):530. https://doi.org/10.3390/life12040530
Chicago/Turabian StyleLee, Young-Sun, Osung Kwon, Geuk-Rae Jeong, Junyeol Noh, Sung Eun Kim, Gwan-Su Yi, Eun Mi Hwang, and Jae-Yong Park. 2022. "Deficiency of TTYH1 Expression Reduces the Migration and Invasion of U2OS Human Osteosarcoma Cells" Life 12, no. 4: 530. https://doi.org/10.3390/life12040530
APA StyleLee, Y.-S., Kwon, O., Jeong, G.-R., Noh, J., Kim, S. E., Yi, G.-S., Hwang, E. M., & Park, J.-Y. (2022). Deficiency of TTYH1 Expression Reduces the Migration and Invasion of U2OS Human Osteosarcoma Cells. Life, 12(4), 530. https://doi.org/10.3390/life12040530








