HIV-1 Integrase Inhibitory Effects of Major Compounds Present in CareVid™: An Anti-HIV Multi-Herbal Remedy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. In Vitro HIV-1 Integrase
2.3. Docking Methodology
3. Results
3.1. Compounds
3.2. HIV-1 Integrase Catalytic Activity Inhibition
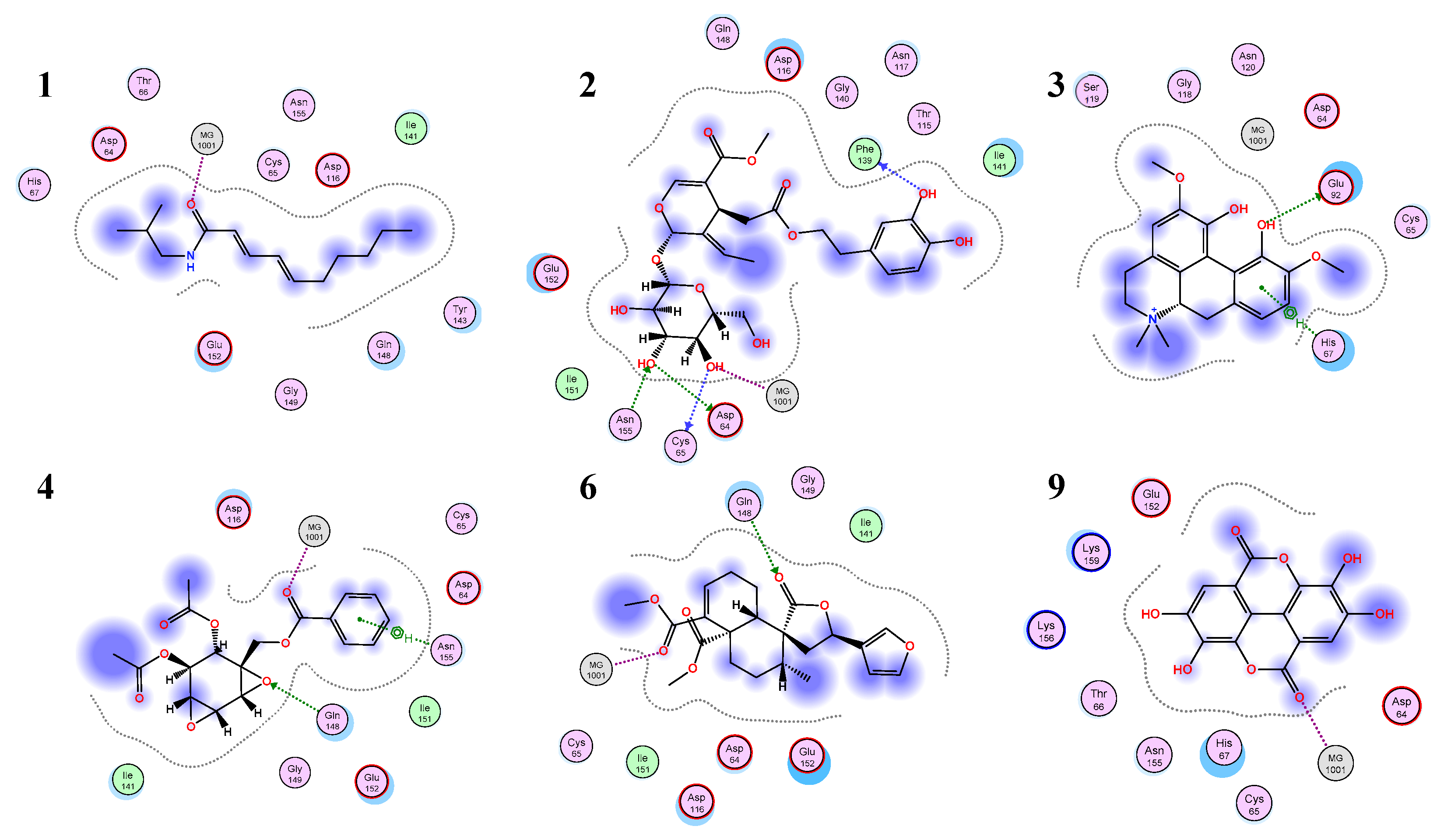
3.3. Molecular Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rotich, W.; Sadgrove, N.J.; Mas-Claret, E.; Padilla-González, G.F.; Guantai, A.; Langat, M.K. HIV-1 Reverse Transcriptase Inhibition by Major Compounds in a Kenyan Multi-Herbal Composition (CareVidTM): In Vitro and In Silico Contrast. Pharmaceuticals 2021, 14, 1009. [Google Scholar] [CrossRef]
- Chepkwony, P.C.; Medina, M.; Medina, M. Compounds and Compositions for Treating Infection. U.S. Patent No. 20120190734A1, 26 June 2012. [Google Scholar]
- Medina, M.; Chepkwony, P.C.; Medina, M. Medicinal and Herbal Compositions and Uses Thereof. U.S. Patent No. 8404284B2, 23 December 2010. [Google Scholar]
- Rotich, W. Botanical aspects, chemical overview, and pharmacological activities of 14 plants used to formulate a Kenyan multi-herbal composition (CareVid). Sci. Africam. 2022. submited. [Google Scholar]
- Nutan, M.M.; Goelb, T.; Das, T.; Malik, S.; Suri, S.; Singh Rawat, A.K.; Srivastava, S.K.; Tuli, R.; Malhotra, S.; Gupta, S.K. Ellagic acid & gallic acid from Lagerstroemia speciosa L. inhibit HIV-1 infection through inhibition of HIV-1 protease & reverse transcriptase activity. Indian J. Med. Res. 2013, 137, 540. [Google Scholar]
- Craigie, R. The molecular biology of HIV integrase. Future Virol. 2012, 7, 679–686. [Google Scholar] [CrossRef] [Green Version]
- Au, T.K.; Lam, T.L.; Ng, T.B.; Fong, W.P.; Wan, D.C.C. A comparison of HIV-1 integrase inhibition by aqueous and methanol extracts of Chinese medicinal herbs. Life Sci. 2001, 68, 1687–1694. [Google Scholar] [CrossRef]
- Thomas, M.; Brady, L. HIV integrase: A target for AIDS therapeutics. Trends Biotechnol. 1997, 15, 167–172. [Google Scholar] [CrossRef]
- Scarsi, K.K.; Havens, J.P.; Podany, A.T.; Avedissian, S.N.; Fletcher, C.V. HIV-1 Integrase Inhibitors: A Comparative Review of Efficacy and Safety. Drugs 2020, 80, 1649. [Google Scholar] [CrossRef] [PubMed]
- Kolakowska, A.; Maresca, A.F.; Collins, I.J.; Cailhol, J. Update on Adverse Effects of HIV Integrase Inhibitors. Curr. Treat. Options Infect. Dis. 2019, 11, 372–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaniad, P.; Sudsai, T.; Septama, A.W.; Chukaew, A.; Tewtrakul, S. Evaluation of Anti-HIV-1 Integrase and Anti-Inflammatory Activities of Compounds from Betula alnoides Buch-Ham. Adv. Pharmacol. Sci. 2019, 2019, 2573965. [Google Scholar] [CrossRef] [Green Version]
- Tewtrakul, S.; Subhadhirasakul, S.; Kummee, S. Anti-HIV-1 integrase activity of medicinal plants used as self medication by AIDS patients. Songklanakarin J. Sci. Technol. 2006, 28, 785–790. [Google Scholar]
- Serna-Arbeláez, M.S.; Florez-Sampedro, L.; Orozco, L.P.; Ramírez, K.; Galeano, E.; Zapata, W. Natural Products with Inhibitory Activity against Human Immunodeficiency Virus Type 1. Adv. Virol. 2021, 2021, 5552088. [Google Scholar] [CrossRef] [PubMed]
- Ee, G.C.L.; Lim, C.M.; Rahmani, M.; Shaari, K.; Bong, C.F.J. Pellitorine, a Potential Anti-Cancer Lead Compound against HL60 and MCT-7 Cell Lines and Microbial Transformation of Piperine from Piper Nigrum. Molecules 2010, 15, 2398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omar, S.H. Oleuropein in Olive and its Pharmacological Effects. Sci. Pharm. 2010, 78, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, T.; Kuang, T.; Du, H.; Li, Q.; Feng, T.; Zhang, Y.; Fan, G. Magnoflorine: A review of its pharmacology, pharmacokinetics and toxicity. Pharmacol. Res. 2020, 152, 104632. [Google Scholar] [CrossRef]
- Kupchan, S.M.; Hemingway, R.J.; Coggon, P.; McPhail, A.T.; Sim, G.A. Crotepoxide, a novel cyclohexane diepoxide tumor inhibitor from Croton macrostachys. J. Am. Chem. Soc. 1968, 90, 2982–2983. [Google Scholar] [CrossRef] [PubMed]
- Maroyi, A. Ethnopharmacological Uses, Phytochemistry, and Pharmacological Properties of Croton macrostachyus Hochst. Ex Delile: A Comprehensive Review. Evid.-Based Complement. Altern. Med. 2017, 2017, 1694671. [Google Scholar] [CrossRef] [Green Version]
- Evtyugin, D.D.; Magina, S.; Evtuguin, D.V. Recent Advances in the Production and Applications of Ellagic Acid and Its Derivatives. A Review. Molecules 2020, 25, 2745. [Google Scholar] [CrossRef]
- Selma, M.V.; Beltrán, D.; Luna, M.C.; Romo-Vaquero, M.; García-Villalba, R.; Mira, A.; Espín, J.C.; Tomás-Barberán, F.A. Isolation of human intestinal bacteria capable of producing the bioactive metabolite isourolithin a from ellagic acid. Front. Microbiol. 2017, 8, 1521. [Google Scholar] [CrossRef]
- Pflieger, A.; Teguo, P.W.; Papastamoulis, Y.; Chaignepain, S.; Subra, F.; Munir, S.; Delelis, O.; Lesbats, P.; Calmels, C.; Andreola, M.L.; et al. Natural Stilbenoids Isolated from Grapevine Exhibiting Inhibitory Effects against HIV-1 Integrase and Eukaryote MOS1 Transposase In Vitro Activities. PLoS ONE 2013, 8, e81184. [Google Scholar] [CrossRef]
- Sanna, C.; Marengo, A.; Acquadro, S.; Caredda, A.; Lai, R.; Corona, A.; Tramontano, E.; Rubiolo, P.; Esposito, F. In Vitro Anti-HIV-1 Reverse Transcriptase and Integrase Properties of Punica granatum L. Leaves, Bark, and Peel Extracts and Their Main Compounds. Plants 2021, 10, 2124. [Google Scholar] [CrossRef] [PubMed]
- Promsong, A.; Chuenchitra, T.; Saipin, K.; Tewtrakul, S.; Panichayupakaranant, P.; Satthakarn, S.; Nittayananta, W. Ellagic acid inhibits HIV-1 infection in vitro: Potential role as a novel microbicide. Oral Dis. 2018, 24, 249–252. [Google Scholar] [CrossRef]
- Nobsathian, S.; Tuchinda, P.; Soorukram, D.; Pohmakotr, M.; Reutrakul, V.; Yoosook, C.; Kasisit, J.; Napaswad, C. A new conjugated amide-dimer from the aerial parts of Piper submultinerve. Nat. Prod. Res. 2012, 26, 1824–1830. [Google Scholar] [CrossRef] [PubMed]
- Lee-Huang, S.; Huang, P.L.; Zhang, D.; Lee, J.W.; Bao, J.; Sun, Y.; Chang, Y.T.; Zhang, J.; Huang, P.L. Discovery of small-molecule HIV-1 fusion and integrase inhibitors oleuropein and hydroxytyrosol: Part I. Integrase inhibition. Biochem. Biophys. Res. Commun. 2007, 354, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Hădărugă, D.I. Natural and synthetic HIV-1 non-nucleoside reverse transcriptase inhibitors: A theoretical approach. J. Agroaliment. Process. Technol. 2011, 17, 360–370. [Google Scholar]
- Hongthong, S.; Kuhakarn, C.; Jaipetch, T.; Prabpai, S.; Kongsaeree, P.; Piyachaturawat, P.; Jariyawat, S.; Suksen, K.; Limthongkul, J.; Panthong, A.; et al. Polyoxygenated cyclohexene derivatives isolated from Dasymaschalon sootepense and their biological activities. Fitoterapia 2015, 106, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Hung, Y.C.; Chang, F.R.; Cosentino, M.; Wang, H.K.; Lee, K.H. Identification of ent-16β,17-Dihydroxykauran-19-oic Acid as an Anti-HIV Principle and Isolation of the New Diterpenoids Annosquamosins A and B from Annona squamosa. J. Nat. Prod. 1996, 59, 635–637. [Google Scholar] [CrossRef]
- Neamati, N.; Lin, Z.; Karki, R.G.; Orr, A.; Cowansage, K.; Strumberg, D.; Pais, G.C.G.; Voigt, J.H.; Nicklaus, M.C.; Winslow, H.E.; et al. Metal-Dependent Inhibition of HIV-1 Integrase. J. Med. Chem. 2002, 45, 5661–5670. [Google Scholar] [CrossRef]
- McColl, D.J.; Chen, X. Strand transfer inhibitors of HIV-1 integrase: Bringing IN a new era of antiretroviral therapy. Antiviral Res. 2010, 85, 101–118. [Google Scholar] [CrossRef]
- Goldgur, Y.; Craigie, R.; Cohen, G.H.; Fujiwara, T.; Yoshinaga, T.; Fujishita, T.; Sugimoto, H.; Endo, T.; Murai, H.; Davies, D.R. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: A platform for antiviral drug design. Proc. Natl. Acad. Sci. USA 1999, 96, 13040–13043. [Google Scholar] [CrossRef] [Green Version]
- Jaskolski, M.; Alexandratos, J.N.; Bujacz, G.; Wlodawer, A. Piecing together the structure of retroviral integrase, an important target in AIDS therapy. FEBS J. 2009, 276, 2926. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Treatment | % Activity rel. to Control | % Inhibition |
|---|---|---|
| HIV Integrase (no inhibition) | 100 | 0 |
| Pellitorine (1) | 81.0 ± 9.1 * | 19.0 |
| Oleuropein (2) | 95.3 ± 4.2 | 4.7 |
| Magnoflorine (3) | 88.0 ± 7.5 | 12.0 |
| Crotepoxide (4) | 91.0 ± 5.0 | 9.0 |
| Ent-kaurane-16β,17-diol (5) | 86.6 ± 2.1 | 13.4 |
| Crotocorylifuran (6) | 90.9 ± 5.0 | 9.1 |
| Lupeol (7) | 81.5 ± 1.2 * | 18.5 |
| Betulin (8) | 83.2 ± 13.6 | 16.8 |
| Ellagic acid (9) | 78.9 ± 5.7 * | 21.1 |
| Urolithin a (10) | 88.7 ± 5.4 | 11.3 |
| Urolithin b (11) | 90.1 ± 3.8 | 9.9 |
| Free Energy of Binding (ΔG) kcal/mol | ||
|---|---|---|
| Compound | Rigid Receptor | Induced Fit |
| 5CITEP | −4.42 | −4.18 |
| Pellitorine (1) | −4.34 | −4.22 |
| Oleuropein (2) | −5.67 | −5.94 |
| Magnoflorine (3) | −4.24 | −4.25 |
| Crotepoxide (4) | −4.75 | −4.96 |
| Ent-kaurane-16β,17-diol (5) | >0 | >0 |
| Crotocorylifuran (6) | −4.33 | −4.21 |
| Lupeol (7) | >0 | >0 |
| Betulin (8) | >0 | >0 |
| Ellagic acid (9) | −4.16 | −4.60 |
| Urolithin A (10) | −3.95 | −3.91 |
| Urolithin B (11) | −3.80 | −3.81 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rotich, W.; Mas-Claret, E.; Sadgrove, N.; Guantai, A.; Padilla-González, G.F.; Langat, M.K. HIV-1 Integrase Inhibitory Effects of Major Compounds Present in CareVid™: An Anti-HIV Multi-Herbal Remedy. Life 2022, 12, 417. https://doi.org/10.3390/life12030417
Rotich W, Mas-Claret E, Sadgrove N, Guantai A, Padilla-González GF, Langat MK. HIV-1 Integrase Inhibitory Effects of Major Compounds Present in CareVid™: An Anti-HIV Multi-Herbal Remedy. Life. 2022; 12(3):417. https://doi.org/10.3390/life12030417
Chicago/Turabian StyleRotich, Winnie, Eduard Mas-Claret, Nicholas Sadgrove, Anastasia Guantai, Guillermo F. Padilla-González, and Moses K. Langat. 2022. "HIV-1 Integrase Inhibitory Effects of Major Compounds Present in CareVid™: An Anti-HIV Multi-Herbal Remedy" Life 12, no. 3: 417. https://doi.org/10.3390/life12030417
APA StyleRotich, W., Mas-Claret, E., Sadgrove, N., Guantai, A., Padilla-González, G. F., & Langat, M. K. (2022). HIV-1 Integrase Inhibitory Effects of Major Compounds Present in CareVid™: An Anti-HIV Multi-Herbal Remedy. Life, 12(3), 417. https://doi.org/10.3390/life12030417







