Pigment Epithelia of the Eye: Cell-Type Conversion in Regeneration and Disease
Abstract
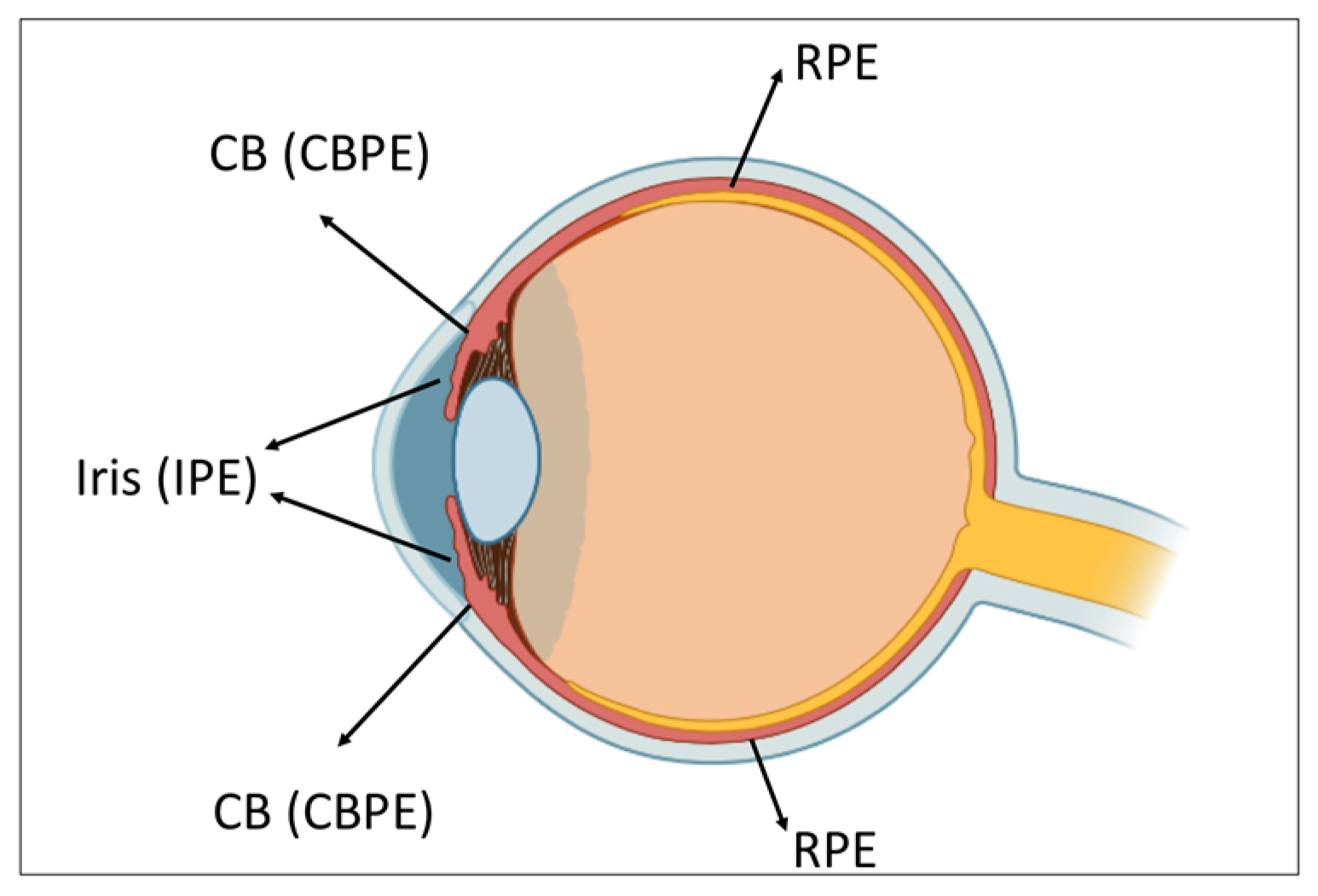
1. Introduction
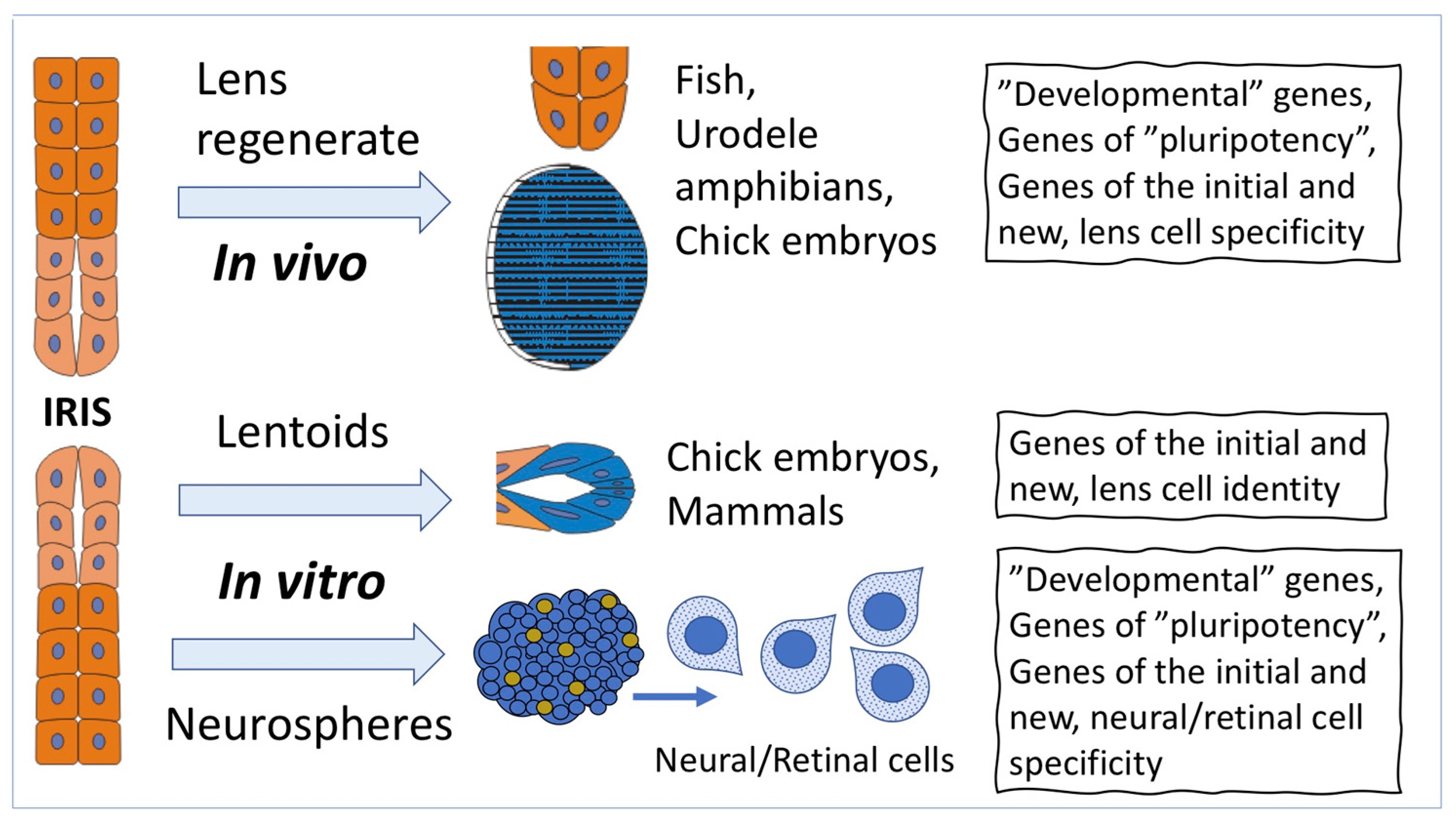
2. Cell-Type Conversion of Iris Pigment Epithelial (IPE) Cells
2.1. Cell-Type Conversion of IPE Cells In Vivo
2.2. Cell-Type Conversion of IPE Cells In Vitro
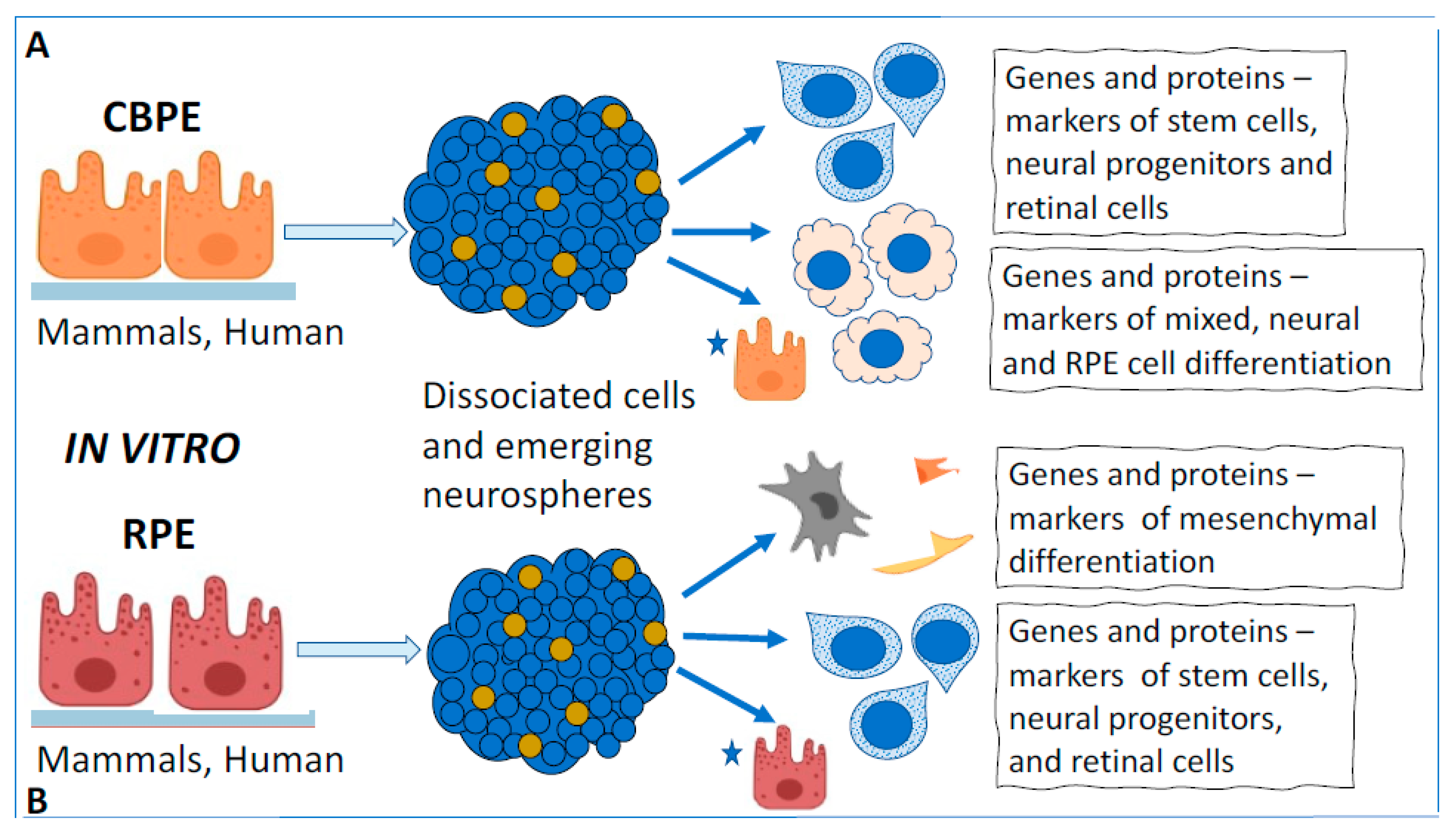
3. Cell-Type Conversion of Ciliary Body Epithelial Cells
3.1. Cell-Type Conversion of CB Cells In Vivo
3.2. Cell-Type Conversion of CB Cells In Vitro
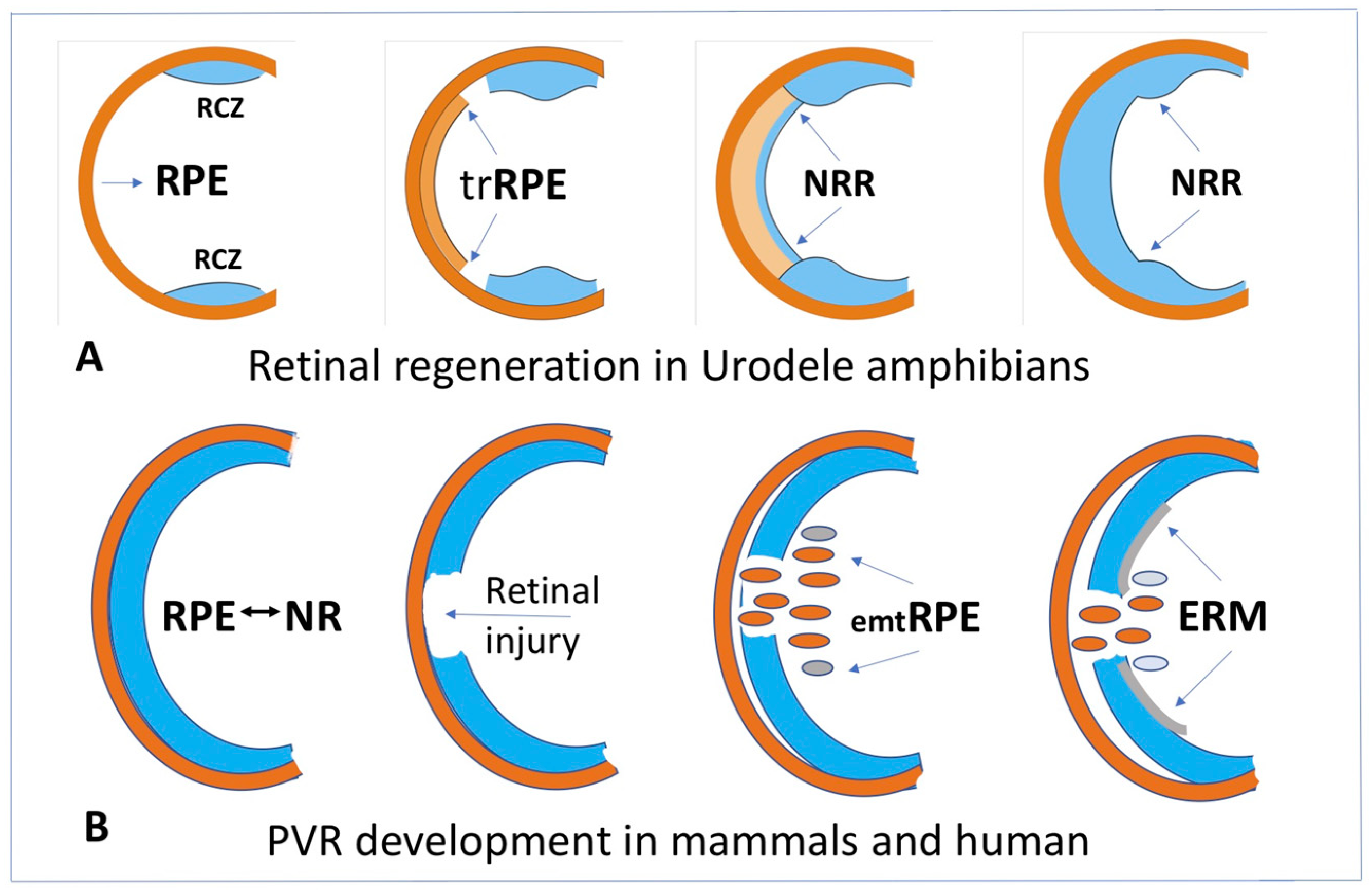
4. Cell-Type Conversion of the Retinal Pigment Epithelial (RPE) Cells
4.1. RPE Cell-Type Conversion as a Basis of NR Regeneration in Amphibians and Birds
4.2. RPE Cell-Type Conversion In Vitro
4.3. RPE Cell-Type Conversion In Vivo as a Basis of Retinal Diseases in Mammals and Humans
5. Overall Discussion
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Selman, K.; Kafatos, F.C. Transdifferentiation in the labial gland of silk moths: Is DNA required for cellular metamorphosis? Cell Diff. 1974, 3, 81–94. [Google Scholar] [CrossRef]
- Yamada, T. Control mechanisms in cell-type conversion in newt lens regeneration. In Monographs in Developmental Biology; Wolsky, A., Ed.; S. Karger: Basel, Switzerland, 1977; Volume 13, pp. 1–126. [Google Scholar]
- Okada, T.S. Transdifferentiation; Clarendon Press: Oxford, UK, 1991; pp. 1–238. [Google Scholar]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Tapscott, S.J.; Davis, R.L.; Thayer, M.J.; Cheng, P.F.; Weintraub, H.; Lassar, A.B. MyoD1: A nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 1988, 242, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, H.; Tapscott, S.J.; Davis, R.L.; Thayer, M.J.; Adam, M.A.; Lassar, A.B.; Miller, A.D. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. USA 1989, 86, 5434–5438. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Yan, R.-T.; Li, X.; Wang, S.-Z. Reprogramming RPE to differentiate towards retinal neurons with Sox2. Stem Cells 2009, 27, 1376–1387. [Google Scholar] [CrossRef]
- Yan, R.-T.; Wang, S.-Z. Differential induction of gene expression by basic fibroblast growth factor and neuroD in cultured retinal pigment epithelial cells. Vis. Neurosci. 2000, 17, 157–164. [Google Scholar] [CrossRef]
- Burke, J.M. Epithelial phenotype and the RPE: Is the answer blowing in the Wnt? Prog. Retin. Eye Res. 2008, 27, 579–595. [Google Scholar] [CrossRef]
- Chen, H.-C.; Zhu, Y.-T.; Chen, S.-Y.; Tseng, S.C.G. Wnt Signaling Induces Epithelial-Mesenchymal Transition with Proliferation in ARPE-19 Cells upon Loss of Contact Inhibition. Lab. Investig. 2012, 92, 676–687. [Google Scholar] [CrossRef]
- Xiao, W.; Chen, X.; Liu, X.; Luo, L.; Ye, S.; Liu, Y. Trichostatin A, a histone deacetylase inhibitor, suppresses proliferation and epithelial-mesenchymal transition in retinal pigment epithelium cells. Cell Mol. Med. 2014, 18, 646–655. [Google Scholar] [CrossRef]
- Royall, L.N.; Lea, D.; Matsushita, T.; Takeda, T.-A.; Taketani, S.; Araki, M. A novel culture method reveals unique neural stem/progenitors in mature porcine iris tissues that differentiate into neuronal and rod photoreceptor-like cells. Brain Res. 2017, 167515, 51–60. [Google Scholar] [CrossRef]
- Shen, C.-N.; Burke, Z.D.; Tosh, D. Transdifferentiation, metaplasia and tissue regeneration. Organogenesis 2004, 1, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Szibor, M.; Poling, J.; Warnecke, H.; Kubin, T.; Braun, T. Remodeling and dedifferentiation of adult cardiomyocytes during disease and regeneration. Cell Mol. Life Sci. 2014, 71, 1907–1916. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, C.H. Dedifferentiation: Inspiration for devising engineering strategies for regenerative medicine. NPJ Regen. Med. 2020, 5, 14. [Google Scholar] [CrossRef]
- Grigoryan, E.N.; Markitantova, Y.V. Cellular and molecular preconditions for retinal pigment epithelium (RPE) natural reprogramming during retinal regeneration in Urodela. Biomedicines 2016, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Grigoryan, E.N.; Markitantova, Y.V. Molecular Strategies for Transdifferentiation of Retinal Pigment Epithelial Cells in Amphibians and Mammals In Vivo. Russ. J. Dev. Biol. 2021, 52, 220–243. [Google Scholar] [CrossRef]
- Morescalchi, F.; Duse, S.; Gambicorti, E.; Romano, M.R.; Costagliola, C.; Semeraro, F. Proliferative Vitreoretinopathy after Eye Injuries: An Overexpression of Growth Factors and Cytokines Leading to a Retinal Keloid. Mediat. Inflamm. 2013, 2013, 269787. [Google Scholar] [CrossRef]
- Idrees, S.; Sridhar, J.; Kuriyan, A.E. Proliferative Vitreoretinopathy: A Review. Int. Ophthalmol. Clin. 2019, 59, 221–240. [Google Scholar] [CrossRef]
- Zou, H.; Shan, C.; Ma, L.; Liu, J.; Yang, N.; Zhao, J. Polarity and epithelial-mesenchymal transition of retinal pigment epithelial cells in proliferative vitreoretinopathy. PeerJ 2020, 8, e10136. [Google Scholar] [CrossRef]
- Abu El-Asrar, A.M.; Midena, E.; Al-Shabrawey, M.; Mohammad, G. New Developments in the Pathophysiology and Management of Diabetic Retinopathy. J. Diabetes Res. 2013, 2013, 424258. [Google Scholar] [CrossRef]
- Lopez, P.F.; Yan, Q.; Kohen, L.; Rao, N.A.; Spee, C.; Black, J.; Oganesian, A. Retinal pigment epithelial wound healing in vivo. Arch. Ophthalmol. 1995, 113, 1437–1446. [Google Scholar] [CrossRef]
- Yu, H.; Vu, T.H.K.; Cho, K.-S.; Guo, C.; Chen, D.F. Mobilizing endogenous stem cells for retinal repair. Transl. Res. 2014, 163, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Karl, M.O.; Reh, T.A. Regenerative medicine for retinal diseases: Activating the endogenous repair mechanisms. Trends Mol. Med. 2010, 16, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Wohl, S.G.; Schmeer, C.W.; Isenmann, S. Neurogenic potential of stem/progenitor-like cells in the adult mammalian eye. Prog. Retin. Eye Res. 2012, 31, 213–242. [Google Scholar] [CrossRef]
- Aladdad, A.M.; Kador, K.E. Adult Stem Cells, Tools for Repairing the Retina. Curr. Ophthalmol. Rep. 2019, 7, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Grigoryan, E.N. Endogenous cell sources for eye retina regeneration in vertebrate animals and human. Russ. J. Dev. Biol. 2018, 49, 314–326. [Google Scholar] [CrossRef]
- Grigoryan, E.N. Potential Endogenous Cell Sources for Retinal Regeneration in Vertebrates and Humans: Progenitor Traits and Specialization. Biomedicines 2020, 8, 208. [Google Scholar] [CrossRef]
- Pan, C.K.; Heilweil, G.; Lanza, R.; Schwartz, S.D. Embryonic stem cells as a treatment for macular degeneration. Expert Opin. Biol. Ther. 2013, 13, 1125–1133. [Google Scholar] [CrossRef]
- Singh, M.S.; Park, S.S.; Albini, T.A.; Canto-Soler, M.V.; Klassen, H.; MacLaren, R.E.; Takahashi, M.; Nagiel, A.; Schwartz, S.D.; Bharti, K. Retinal Stem Cell Transplantation: Balancing Safety and Potential. Prog. Retin. Eye Res. 2020, 75, 100779. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, Z.; Gu, P. Stem/progenitor cell-based transplantation for retinal degeneration: A review of clinical trials. Cell Death Dis. 2020, 11, 793. [Google Scholar] [CrossRef]
- Land, M.F.; Fernald, R.D. The evolution of eyes. Annu. Rev. Neurosci. 1992, 15, P1–P29. [Google Scholar] [CrossRef]
- Davis-Silberman, N.; Ashery-Padan, R. Iris development in vertebrates: Genetic and molecular considerations. Brain Res. 2008, 1192, 17–28. [Google Scholar] [CrossRef]
- Reyer, R.W. Regeneration of the lens in the amphibian eye. Q. Rev. Biol. 1954, 29, 1–46. [Google Scholar] [CrossRef]
- Stone, L.S. An investigation recording all salamanders which can and cannot regenerate a lens from the dorsal iris. J. Exp. Zool. 1967, 164, 87–103. [Google Scholar] [CrossRef]
- Tsonis, P.A. Regeneration in vertebrates. Dev. Biol. 2000, 221, 273–284. [Google Scholar] [CrossRef]
- Tsonis, P.A.; Madhavan, M.; Tancous, E.E.; del Rio-Tsonis, K. A newt’s eye view of lens regeneration. Int. J. Dev. Biol. 2004, 48, 975–980. [Google Scholar] [CrossRef]
- Vergara, M.N.; Tsissios, G.; del Rio-Tsonis, K. Lens regeneration: A historical perspective. Int. J. Dev. Biol. 2018, 62, 351–361. [Google Scholar] [CrossRef]
- Sato, T. Uber die Linsen Regeneration bei Cobiditien Fishen. I. Misgurnus anguilicaudatus (Contor). Embriologia 1961, 6, 251–290. [Google Scholar]
- Van Deth, J.H.M.G. Induction et regeneration du cristallin chez l’embryon de la poule. Acta Neerl. Morphol. 1940, 3, 151–169. [Google Scholar]
- Genis-Galvez, J.M.G. The result of the total and partial removal of the lens primordium in the chick embryo. Contribution to the study of lens regeneration. An. Dessarollo 1962, 10, 249–267. [Google Scholar]
- Coulombre, J.L.; Coulombre, A.J. Regeneration of neural retina from the pigmented epithelium in the chick embryo. Dev. Biol. 1965, 12, 79–92. [Google Scholar] [CrossRef]
- Shekhawat, D.S.; Jangir, O.P.; Prakash, A.; Pawan, S. Lens regeneration in mice under the influence of vitamin A. J. Biosci. 2001, 26, 571–576. [Google Scholar] [CrossRef]
- Eguchi, G. Electron microscopic studies on lens regeneration. Embryologia 1964, 8, 247–287. [Google Scholar] [CrossRef]
- Eguchi, G. Cellular and molecular background of Wolffian lens regeneration. Cell Differ. Dev. 1988, 25, 147–158. [Google Scholar] [CrossRef]
- Roddy, M.; Fox, T.P.; McFadden, J.P.; Nakamura, K.; Del Rio-Tsonis, K.; Tsonis, P.A. A comparative proteomic analysis during urodele lens regeneration. Biochem. Biophys. Res. Commun. 2008, 377, 275–279. [Google Scholar] [CrossRef][Green Version]
- Tsonis, P.A.; del Rio-Tsonis, K. Lens and retina regeneration: Transdifferentiation, stem cells and clinical applications. Exp. Eye Res. 2004, 78, 161–172. [Google Scholar] [CrossRef]
- Henry, J.J.; Tsonis, P.A. Molecular and cellular aspects of amphibian lens regeneration. Prog. Retin. Eye Res. 2010, 29, 543–555. [Google Scholar] [CrossRef]
- Yang, Y.; Zalik, S.E. The cells of the dorsal iris involved in lens regeneration are myoepithelial cells whose cytoskeleton changes during cell-type conversion. Anat. Embryol. 1994, 189, 475–487. [Google Scholar] [CrossRef]
- Imokawa, Y.; Simon, A.; Brockes, J.P. A critical role for thrombin in vertebrate lens regeneration. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2004, 359, 765–776. [Google Scholar] [CrossRef]
- Godwin, J.W.; Liem, K.F.; Brockes, J.P. Tissue factor expression in newt iris coincides with thrombin activation and lens regeneration. Mech. Dev. 2010, 127, 321–328. [Google Scholar] [CrossRef]
- Del Rio-Tsonis, K.; Trombley, M.T.; McMahon, G.; Tsonis, P.A. Regulation of lens regeneration by fibroblast growth factor receptor 1. Dev. Dyn. 1998, 213, 140–146. [Google Scholar] [CrossRef]
- Makarev, E.; Call, M.K.; Grogg, M.W.; Atkinson, D.L.; Milash, B.; Odelberg, S.J.; Tsonis, P.A. Gene expression signatures in the newt irises during lens regeneration. FEBS Lett. 2007, 581, 1865–1870. [Google Scholar] [CrossRef]
- Sousounis, K.; Looso, M.; Maki, N.; Ivester, C.J.; Braun, T.; Tsonis, P.A. Transcriptome analysis of newt lens regeneration reveals distinct gradients in gene expression patterns. PLoS ONE 2013, 8, e61445. [Google Scholar] [CrossRef]
- Grogg, M.W.; Call, M.K.; Tsonis, P.A. Signaling during lens regeneration. Semin. Cell Dev. Biol. 2006, 17, 753–758. [Google Scholar] [CrossRef][Green Version]
- Hayashi, T.; Mizuno, N.; Kondoh, H. Determinative roles of FGF and Wnt signals in iris-derived lens regeneration in newt eye. Dev. Growth Differ. 2008, 50, 279–287. [Google Scholar] [CrossRef]
- Del Rio-Tsonis, K.; Tsonis, P.A. Eye regeneration at the molecular age. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2003, 226, 211–224. [Google Scholar] [CrossRef]
- Mitashov, V.I.; Grigoryan, E.N. Radiographic study of proliferation and specific proteins’ synthesis in iris cells during eye regeneration in the newt. Ontogenez 1980, 11, 160–167. [Google Scholar]
- Maki, N.; Suetsugu-Maki, R.; Tarui, H.; Agata, K.; del Rio-Tsonis, K.; Tsonis, P.A. Expression of stem cell pluripotency factors during regeneration in newts. Dev. Dyn. 2009, 238, 1613–1616. [Google Scholar] [CrossRef]
- Maki, N.; Suetsugu-Maki, R.; Sano, S.; Nakamura, K.; Nishimura, O.; Tarui, H.; del Rio-Tsonis, K.; Ohsumi, K.; Agata, K.; Tsonis, P.A. Oocyte-type linker histone B4 is required for transdifferentiation of somatic cells in vivo. FASEB J. 2010, 24, 3462–3467. [Google Scholar] [CrossRef]
- Maki, N.; Takechi, K.; Sano, S.; Tarui, H.; Sasai, Y.; Agata, K. Rapid accumulation of nucleostemin in nucleolus during newt regeneration. Dev. Dyn. 2007, 236, 941–950. [Google Scholar] [CrossRef]
- Tsai, R.Y.L.; McKay, R.D.G. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002, 16, 2991–3003. [Google Scholar] [CrossRef]
- Eguchi, G.; Eguchi, Y.; Nakamura, K.; Yadav, M.C.; Millan, J.L.; Tsonis, P.A. Regenerative capacity in newts is not altered by repeated regeneration and ageing. Nat. Commun. 2011, 2, 384. [Google Scholar] [CrossRef]
- Okamoto, M. Simultaneous demonstration of lens regeneration from dorsal iris and tumor production from ventral iris in the same newt eye after carcinogen administration. Differentiation 1997, 61, 285–292. [Google Scholar] [CrossRef]
- Grigoryan, E.N.; Radugina, E.A. Behavior of stem-like cells, precursors for tissue regeneration in Urodela, under conditions of microgravity. Stem Cells Dev. 2019, 28, 423–437. [Google Scholar] [CrossRef]
- Clayton, R.M.; Jeanny, J.-C.; Bower, D.J.; Errington, L.H. The presence of extralenticular crystallins and its relationship with transdifferentiation to lens. Curr. Top. Dev. Biol. 1986, 20, 137–151. [Google Scholar] [CrossRef]
- Juric-Lekic, G.; Svajger, A. Lentoid formation in ectopic grafts of lentectomized eyes of rat fetuses. Cell Differ. Dev. 1989, 27, 225–232. [Google Scholar] [CrossRef]
- Yang, C.; Yang, Y.; Brennan, L.; Bouhassira, E.E.; Kantorow, M.; Cvekl, A. Efficient generation of lens progenitor cells and lentoid bodies from human embryonic stem cells in chemically defined conditions. FASEB J. 2010, 24, 3274–3283. [Google Scholar] [CrossRef]
- Ali, M.; Kabir, F.; Thomson, J.J.; Ma, Y.; Qiu, C.; Delannoy, M.; Khan, S.Y.; Riazuddin, S.A. Comparative transcriptome analysis of hESC- and iPSC-derived lentoid bodies. Sci. Rep. 2019, 9, 18552. [Google Scholar] [CrossRef]
- Thumann, G. Development and Cellular Functions of the Iris Pigment Epithelium. Surv. Ophthalmol. 2001, 45, 345–354. [Google Scholar] [CrossRef]
- Sun, G.; Asami, M.; Ohta, H.; Kosaka, J.; Kosaka, M. Retinal stem/progenitior properties of iris pigment epithelial cells. Dev. Biol. 2006, 289, 243–252. [Google Scholar] [CrossRef]
- Matsushita, T.; Steinfeld, J.; Fujihara, A.; Urayama, S.; Taketani, S. Regulation of neuronal and photoreceptor cell differentiation by Wnt signaling from iris-derived stem/progenitor cells of the chick in flat vs. matrigel-embedding cultures. Brain Res. 2019, 1704, 207–218. [Google Scholar] [CrossRef]
- Asami, M.; Sun, G.; Yamaguchi, M.; Kosaka, M. Multipotent cells from mammalian iris pigment epithelium. Dev. Biol. 2007, 304, 433–446. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haruta, M.; Kosaka, M.; Kanegae, Y.; Saito, I.; Inoue, T.; Kageyama, R.; Nishida, A.; Honda, Y.; Takahashi, M. Induction of photoreceptor-specific phenotypes in adult mammalian iris tissue. Nat. Neurosci. 2001, 4, 1163–1164. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T. Photoreceptors derived from adult iris tissue: Prospects for retinal transplantation. Semin. Ophthalmol. 2005, 20, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Frøen, R.C.; Johnsen, E.O.; Petrovski, G.; Berényi, E.; Facskó, A.; Berta, A.; Nicolaissen, B.; Moe, M.C. Pigment epithelial cells isolated from human peripheral iridectomies have limited properties of retinal stem cells. Acta Ophthalmol. 2011, 89, e635–e644. [Google Scholar] [CrossRef] [PubMed]
- Seko, Y.; Azuma, N.; Kaneda, M.; Nakatani, K.; Miyagawa, Y.; Noshiro, Y.; Kurokawa, R.; Okano, H.; Umezawa, A. Derivation of human differential photoreceptor-like cells from the iris by defined combinations of CRX, RX and NEUROD. PLoS ONE 2012, 7, e35611. [Google Scholar] [CrossRef]
- Bennis, A.; ten Brink, J.B.; Moerland, P.D.; Heine, V.M.; Bergen, A.A. Comparative gene expression study and pathway analysis of the human iris and the retinal pigment epithelium. PLoS ONE 2017, 12, e0182983. [Google Scholar] [CrossRef]
- Yamamoto, N.; Hiramatsu, N.; Ohkuma, M.; Hatsusaka, N.; Takeda, S.; Nagai, N.; Miyachi, E.; Kondo, M.; Imaizumi, K.; Horiguchi, M.; et al. Novel Technique for Retinal Nerve Cell Regeneration with Electrophysiological Functions Using Human Iris-Derived iPS Cells. Cells 2021, 10, 743. [Google Scholar] [CrossRef]
- Abe, T.; Yoshida, M.; Youshioka, Y.; Wakusawa, R.; Tokita-Ishikawa, Y.; Seto, H.; Tamai, M.; Nishida, K. Iris pigment epithelial cell transplantation for degenerative retinal diseases. Prog. Retin. Res. 2007, 26, 302–321. [Google Scholar] [CrossRef]
- Yosukawa, T.; Hirano, Y.; Kato, A.; Ohashi, Y.; Ogura, Y. Experimental transplantation of autologous iris pigment epithelial cell sheets to treat chorioretinal atrophy and tests for clinical applications. Cytotherapy 2021, 23, 135. [Google Scholar] [CrossRef]
- Ramos, M.F.; Baker, J.; Atzpodien, E.-A.; Bach, U.; Brassard, J.; Cartwright, J.; Farman, C.; Fishman, C.; Jacobsen, M.; Junker-Walker, U.; et al. Nonproliferative and Proliferative Lesions of the Ratand Mouse Special Sense Organs (Ocular [eye and glands], Olfactory and Otic). J. Toxicol. Pathol. 2018, 31 (Suppl. 3), 97S–214S. [Google Scholar] [CrossRef]
- Taradach, C.; Greaves, P. Spontaneous eye lesions in laboratory animals: Incidence in relation to age. Crit. Rev. Toxicol. 1984, 12, 121–147. [Google Scholar] [CrossRef] [PubMed]
- Wormstone, I.M. The human capsular bag model of posterior capsule opacification. Eye 2020, 34, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.A.; Fraile-Martínez, O.; García-Honduvilla, N.; Coca, S.; Álvarez-Mon, M.; Buján, J.; Teus, M.A. Update on uveal melanoma: Translational research from biology to clinical practice (Review). Int. J. Oncol. 2020, 57, 1262–1279. [Google Scholar] [CrossRef] [PubMed]
- Georgalas, I.; Petrou, M.D.; Papaconstantonou, D.; Brouzas, D.; Koutsandrea, C.; Kanakis, M. Iris cysts: A comprehensive review on diagnosis and treatment. Surv. Ophthalmol. 2018, 63, 347–364. [Google Scholar] [CrossRef]
- Fernández-Nogales, M.; Murcia-Belmonte, V.; Chen, H.Y.; Herrera, E. The peripheral eye: A neurogenic area with potential to treat retinal pathologies? Prog. Retin. Eye Res. 2019, 68, 110–123. [Google Scholar] [CrossRef]
- Miles, A.; Tropepe, V. Retinal Stem Cell ‘Retirement Plans’: Growth, Regulation and Species Adaptations in the Retinal Ciliary Marginal Zone. Int. J. Mol. Sci. 2021, 22, 6528. [Google Scholar] [CrossRef]
- Napier, H.R.; Kidson, S.H. Molecular events in early development of the ciliary body: A question of folding. Exp. Eye Res. 2007, 84, 615–625. [Google Scholar] [CrossRef]
- McDougal, D.H.; Gamlin, P.D. Autonomic control of the eye. Comp. Physiol. 2015, 5, 439–473. [Google Scholar] [CrossRef]
- Delamere, N.A. Ciliary Body and Ciliary Epithelium. Adv. Organ Biol. 2005, 10, 127–148. [Google Scholar] [CrossRef]
- Coca-Prados, M.; Escribano, J. New perspectives in aqueous humor secretion and in glaucoma: The ciliary body as a multifunctional neuroendocrine gland. Prog. Retin. Eye Res. 2007, 26, 239–262. [Google Scholar] [CrossRef]
- Goel, R.; Murthy, K.R.; Srikanth, S.M.; Pinto, S.M.; Bhattacharjee, M.; Kelkar, D.S.; Madugundu, A.K.; Dey, G.; Mohan, S.S.; Krishna, V.; et al. Characterizing the normal proteome of human ciliary body. Clin. Proteom. 2013, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Janssen, S.F.; Gorgels, T.G.M.F.; Bossers, K.; ten Brink, J.B.; Essing, A.H.W.; Nagtegaal, M.; van der Spek, P.; Jansonius, N.M.; Bergen, A.A.B. Gene Expression and Functional Annotation of the Human Ciliary Body Epithelia. PLoS ONE 2012, 7, e44973. [Google Scholar] [CrossRef] [PubMed]
- Coles, B.L.K.; van der Kooy, D. P-Cadherin is necessary for retinal stem cell behavior in vitro, but not in vivo. Stem Cell Res. 2017, 21, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, P.E.; Emsley, J.G.; Myers, T.; Clarke, D.B. Proliferation and expression of progenitor and mature retinal phenotypes in the adult mammalian ciliary body after retinal ganglion cell injury. Investig. Ophthalmol. Vis. Sci. 2007, 48, 5266–5275. [Google Scholar] [CrossRef][Green Version]
- Del Debbio, C.B.; Santos, M.F.; Yan, C.Y.; Ahmad, I.; Hamassaki, D.E. Rho GTPases control ciliary epithelium cells proliferation and progenitor profile induction in vivo. Investig. Ophthalmol. Vis. Sci. 2014, 55, 2631–2641. [Google Scholar] [CrossRef]
- Coles, B.L.; Horsford, D.J.; McInnes, R.R.; van der Kooy, D. Loss of retinal progenitor cells leads to an increase in the retinal stem cell population in vivo. Eur. J. Neurosci. 2006, 23, 75–82. [Google Scholar] [CrossRef]
- Ahmad, I.; Tang, L.; Pham, H. Identification of neural progenitors in the adult mammalian eye. Biochem. Biophys. Res. Commun. 2000, 270, 517–521. [Google Scholar] [CrossRef]
- Das, A.V.; James, J.; Rahnenfuhrer, J.; Thoreson, W.B.; Bhattacharya, S.; Zhao, X.; Ahmad, I. Retinal properties and potential of the adult mammalian ciliary epithelium stem cells. Vis. Res. 2005, 45, 1653–1666. [Google Scholar] [CrossRef]
- Lord-Grignon, J.; Abdouh, M.; Bernier, G. Identification of genes expressed in retinal progenitor/stem cell colonies isolated from the ocular ciliary body of adult mice. Gene Expr. Patterns 2006, 6, 992–999. [Google Scholar] [CrossRef]
- MacNeil, A.; Pearson, R.A.; MacLaren, R.E.; Smith, A.J.; Sowden, J.C.; Ali, R.R. Comparative analysis of progenitor cells isolated from the iris, pars plana, and ciliary body of the adult porcine eye. Stem Cells 2007, 25, 2430–2438. [Google Scholar] [CrossRef]
- Martinez-Navarrete, G.C.; Angulo, A.; Martin-Nieto, J.; Cuenca, N. Gradual morphogenesis of retinal neurons in the peripheral retinal margin of adult monkeys and humans. J. Comp. Neurol. 2008, 511, 557–580. [Google Scholar] [CrossRef]
- Tropepe, V.; Coles, B.L.; Chiasson, B.J.; Horsford, D.J.; Elia, A.J.; McInnes, R.R.; van der Kooy, D. Retinal stem cells in the adult mammalian eye. Science 2000, 287, 2032–2036. [Google Scholar] [CrossRef]
- Coles, B.L.; Angenieux, B.; Inoue, T.; del Rio-Tsonis, K.; Spence, J.R.; McInnes, R.R.; Arsenijevic, Y.; van der Kooy, D. Facile isolation and the characterization of human retinal stem cells. Proc. Natl. Acad. Sci. USA 2004, 101, 15772–15777. [Google Scholar] [CrossRef]
- Kohno, R.; Ikeda, Y.; Yonemitsu, Y.; Hisatomi, T.; Yamaguchi, M.; Miyazaki, M.; Takeshita, H.; Ishibashi, T.; Sueishi, K. Sphere formation of ocular epithelial cells in the ciliary body is a reprogramming system for neural differentiation. Brain Res. 2006, 1093, 54–70. [Google Scholar] [CrossRef]
- Bertolotti, E.; Neri, A.; Camparini, M.; Macaluso, C.; Marigo, V. Stem cells as source for retinal pigment epithelium transplantation. Prog. Retin. Eye Res. 2014, 42, 130–144. [Google Scholar] [CrossRef]
- Guduric-Fuchs, J.; Chen, W.; Price, H.; Archer, D.B.; Cogliati, T. RPE and neuronal differentiation of allotransplantated porcine ciliary epithelium-derived cells. Mol. Vis. 2011, 17, 2580. [Google Scholar]
- Jasty, S.; Srinivasan, P.; Pasricha, G.; Chatterjee, N.; Subramanian, K. Gene expression profiles and retinal potential of stem/progenitor cells derived from human iris and ciliary pigment epithelium. Stem Cell Rev. 2012, 8, 1163–1177. [Google Scholar] [CrossRef]
- Cicero, S.A.; Johnson, D.; Reyntjens, S.; Frase, S.; Connell, S.; Chow, L.M.; Baker, S.J.; Sorrentino, B.P.; Dyer, M.A. Cells previously identified as retinal stem cells are pigmented ciliary epithelial cells. Proc. Natl. Acad. Sci. USA 2009, 106, 6685–6690. [Google Scholar] [CrossRef]
- Gualdoni, S.; Baron, M.; Lakowski, J.; Decembrini, S.; Smith, A.J.; Pearson, R.A.; Ali, R.R.; Sowden, J.C. Adult ciliary epithelial cells, previously identified as retinal stem cells with potential for retinal repair, fail to differentiate into new rod photoreceptors. Stem Cells 2010, 28, 1048–1059. [Google Scholar] [CrossRef]
- Coles, B.L.; van der Kooy, D. Isolation of retinal stem cells from the mouse eye. J. Vis. Exp. 2010, 43, 2209. [Google Scholar] [CrossRef]
- Frøen, R.; Johnsen, E.O.; Nicolaissen, B.; Facskó, A.; Petrovski, G.; Moe, M.C. Does the adult human ciliary body epithelium contain “true” retinal stem cells? Biomed. Res. Int. 2013, 2013, 531579. [Google Scholar] [CrossRef]
- Ahmad, I.; Das, A.V.; James, J.; Bhattacharya, S.; Zhao, X. Neural stem cells in the mammalian eye: Types and regulation. Semin. Cell Dev. Biol. 2004, 15, 53–62. [Google Scholar] [CrossRef]
- Das, A.V.; James, J.; Zhao, X.; Rahnenführer, J.; Ahmad, I. Identification of c-Kit receptor as a regulator of adult neural stem cells in the mammalian eye: Interactions with Notch signaling. Dev. Biol. 2004, 273, 87–105. [Google Scholar] [CrossRef][Green Version]
- Pang, J.; Le, L.; Zhou, Y.; Tu, R.; Hou, Q.; Tsuchiya, D.; Thomas, N.; Wang, Y.; Yu, Z.; Alexander, R.; et al. NOTCH Signaling Controls Ciliary Body Morphogenesis and Secretion by Directly Regulating Nectin Protein Expression. Cell Rep. 2021, 34, 108603. [Google Scholar] [CrossRef]
- Jasty, S.; Krishnakumar, S. Profiling of DNA and histone methylation reveals epigenetic-based regulation of gene expression during retinal differentiation of stem/progenitor cells isolated from the ciliary pigment epithelium of human cadaveric eyes. Brain Res. 2016, 165115, 1–10. [Google Scholar] [CrossRef]
- Oliver, V.F.; van Bysterveldt, K.A.; Merbs, S.L. Epigenetics in ocular medicine. In Medical Epigenetics; Elsevier Inc.: Amsterdam, The Netherlands, 2016; Volume 22, pp. 391–412. [Google Scholar]
- Aldiri, I.; Xu, B.; Wang, L.; Chen, X.; Hiler, D.; Griffiths, L.; Valentine, M.; Shirinifard, A.; Thiagarajan, S.; Sablauer, A.; et al. The Dynamic Epigenetic Landscape of the Retina During Development, Reprogramming, and Tumorigenesis. Neuron 2017, 94, 550–568. [Google Scholar] [CrossRef]
- Chen, E.; Bohm, K.; Rosenblatt, M.; Kang, K. Epigenetic regulation of anterior segment diseases and potential therapeutics. Ocul. Surf. 2020, 18, 383–395. [Google Scholar] [CrossRef]
- Kumar, J.B.; Proia, A.D.; Sharma, S. Primary adenocarcinoma of pigmented ciliary epithelium in a phthisical eye. Surv. Ophthalmol. 2016, 61, 502–505. [Google Scholar] [CrossRef]
- Li, Y.; Shi, J.; Yang, J.; Ge, S.; Zhang, J.; Jia, R.; Fan, X. Uveal melanoma: Progress in molecular biology and therapeutics. Ther. Adv. Med. Oncol. 2020, 12, 1758835920965852. [Google Scholar] [CrossRef]
- Fallico, M.; Raciti, G.; Longo, A.; Reibaldi, M.; Bonfiglio, V.; Russo, A.; Caltabiano, R.; Gattuso, G.; Falzone, L.; Avitabile, T. Current molecular and clinical insights into uveal melanoma (Review). Int. J. Oncol. 2021, 58, 10. [Google Scholar] [CrossRef]
- Hägglund, A.C.; Jones, I.; Carlsson, L. A novel mouse model of anterior segment dysgenesis (ASD): Conditional deletion of Tsc1 disrupts ciliary body and iris development. Dis. Model. Mech. 2017, 10, 245–257. [Google Scholar] [CrossRef]
- Ni, A.; Wu, M.J.; Nakanishi, Y.; Chavala, S.H. Facile and efficient reprogramming of ciliary body epithelial cells into induced pluripotent stem cells. Stem Cells Dev. 2013, 22, 2543–2550. [Google Scholar] [CrossRef]
- Sparrow, J.R.; Hicks, D.; Hamel, C.P. The Retinal Pigment Epithelium in Health and Disease. Curr. Mol. Med. 2010, 10, 802–823. [Google Scholar] [CrossRef]
- Koster, C.; Wever, K.E.; Wagstaff, P.E.; van den Hurk, K.T.; Hooijmans, C.R.; Bergen, A.A. A Systematic Review on Transplantation Studies of the Retinal Pigment Epithelium in Animal Models. Int. J. Mol. Sci. 2020, 21, 2719. [Google Scholar] [CrossRef]
- Caceres, P.S.; Rodriguez-Boulan, E. Retinal Pigment Epithelium Polarity in Health and Blinding Diseases. Curr. Opin. Cell Biol. 2020, 62, 37–45. [Google Scholar] [CrossRef]
- Markitantova, Y.; Simirskii, V. Inherited Eye Diseases with Retinal Manifestations through the Eyes of Homeobox Genes. Int. J. Mol. Sci. 2020, 21, 1602. [Google Scholar] [CrossRef]
- Strauss, O. The Retinal Pigment Epithelium in Visual Function. Physiol. Rev. 2005, 85, 845–881. [Google Scholar] [CrossRef]
- Fuhrmann, S.; Zou, C.J.; Levine, E.M. Retinal pigment epithelium development, plasticity, and tissue homeostasis. Exp. Eye Res. 2014, 123, 141–150. [Google Scholar] [CrossRef]
- Hasegawa, M. Restitution of the eye after removal of the retina and lens in the newt Triturus pyrrhogaster. Embryologia 1958, 4, 1–32. [Google Scholar] [CrossRef]
- Keefe, J.R. An analysis of urodelean retinal regeneration. I–IV. J. Exp. Zool. 1973, 184, 185–257. [Google Scholar] [CrossRef]
- Mitashov, V.I. Mechanisms of retina regeneration in vertebrates. Int. J. Dev. Biol. 1996, 40, 833–844. [Google Scholar]
- Mitashov, V.I. Retinal regeneration in amphibians. Int. J. Dev. Biol. 1997, 41, 893–905. [Google Scholar]
- Chiba, C.; Mitashov, V.I. Cellular and molecular events in the adult newt retinal regeneration. In Strategies for Retinal Tissue Repair and Regeneration in Vertebrates: From Fish to Human; Chiba, C., Ed.; Research Signpost: Kerala, India, 2007; pp. 15–33. [Google Scholar]
- Yasumuro, H.; Sakurai, K.; Toyama, F.; Maruo, F.; Chiba, C. Implications of a Multi-Step Trigger of Retinal Regeneration in the Adult Newt. Biomedicines 2017, 5, 25. [Google Scholar] [CrossRef]
- Markitantova, Y.V.; Makar’ev, E.O.; Smirnova, Y.A.; Zinov’eva, R.D.; Mitashov, V.I. Analysis of the expression pattern of regulatory genes pax6, prox1, and six3 during regeneration of eye structures in the newt. Biol. Bull. 2004, 31, 428–436. [Google Scholar] [CrossRef]
- Markitantova, Y.V.; Avdonin, P.P.; Grigoryan, E.N.; Zinovieva, R.D. Identification of the pitx1 embryogenesis regulatory gene in a regenerating newt retina. Dokl. Biol. Sci. 2010, 435, 421–424. [Google Scholar] [CrossRef]
- Avdonin, P.P.; Markitantova, Y.V.; Zinovieva, R.D.; Mitashov, V.I. Expression of regulatory genes Pax6, Otx2, Six3, and FGF2 during newt retina regeneration. Biol. Bull. 2008, 35, 355. [Google Scholar] [CrossRef]
- Avdonin, P.P.; Grigoryan, E.N.; Markitantova, Y.V. Transcriptional factor Pitx2: Localization during newt retina regeneration. Biol. Bull. 2010, 37, 231–235. [Google Scholar] [CrossRef]
- Sakami, S.; Hisatomi, O.; Sakakibara, S.; Liu, J. Downregulation of Otx2 in the dedifferentiated RPE cells of regenerating newt retina. Dev. Brain Res. 2005, 155, 49–59. [Google Scholar] [CrossRef]
- Markitantova, Y.V.; Avdonin, P.P.; Grigoryan, E.N. Nucleostemin expression in the process of reprogramming of pigment epithelium cells in situ during retinal regeneration in an adult newt. Tsitologiya 2014, 56, 671–672. [Google Scholar]
- Markitantova, Y.V.; Avdonin, P.P.; Grigoryan, E.N. Identification of the gene encoding nucleostemin in the eye tissues of Pleurodeles waltl. Biol. Bull. 2015, 42, 379–386. [Google Scholar] [CrossRef]
- Markitantova, Y.V.; Novikova, Y.P.; Poplinskaya, V.A.; Grigoryan, E.N. Expression of FGF2 and nucleostemin in models of retinal regeneration in the newt under conditions of 3D organotypic culture in vitro. EC Ophthalmol. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Grigoryan, E.N.; Bazhin, A.V.; Krasnov, M.S.; Philippov, P.P. Study of calcium-binding protein recoverin expression in normal, surviving and regenerating retina of the newt Pleurodeles waltl. Klet. Tekhnol. Biol. Med. 2009, N3, 169–173. [Google Scholar]
- Islam, M.R.; Nakamura, K.; Casco-Robles, M.M.; Kunahong, A.; Inami, W.; Toyama, F.; Maruo, F.; Chiba, C. The newt reprograms mature RPE cells into a unique multipotent state for retinal regeneration. Sci. Rep. 2014, 4, 6043. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, J.; Chiba, C. Immunohistochemical analysis of Musashi-1 expression during retinal regeneration of adult newt. Neurosci. Lett. 2009, 450, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Casco-Robles, M.M.; Islam, M.R.; Inami, W.; Tanaka, H.V.; Kunahong, A.; Yasumuro, H.; Hanzawa, S.; Casco-Robles, R.M.; Toyama, F.; Maruo, F.; et al. Turning the fate of reprogramming cells from retinal disorder to regeneration by Pax6 in newts. Sci. Rep. 2016, 6, 33761. [Google Scholar] [CrossRef] [PubMed]
- Grigoryan, E.N. Shared triggering mechanisms of retinal regeneration in lower vertebrates and retinal rescue in higher ones. In Tissue Regeneration—From Basic Biology to Clinical Application; Davies, J., Ed.; In Tech: Rijeka, Croatia, 2012; pp. 145–164. [Google Scholar]
- Pastor, J.C.; Rojas, J.; Pastor-Idoate, S.; di Lauro, S.; Gonzalez-Buendia, L.; Delgado-Tirado, S. Proliferative vitreoretinopathy: A new concept of disease pathogenesis and practical consequences. Prog. Retin. Eye Res. 2016, 51, 125–155. [Google Scholar] [CrossRef]
- Kaneko, Y.; Hirota, K.; Matsumoto, G.; Hanyu, Y. Expression pattern of a newt Notch homologue in regenerating newt retina. Brain Res. Dev. Brain Res. 2001, 31, 53–62. [Google Scholar] [CrossRef]
- Nakamura, K.; Chiba, C. Evidence for Notch signaling involvement in retinal regeneration of adult newt. Brain Res. 2007, 1136, 28–42. [Google Scholar] [CrossRef]
- Mercer, S.E.; Cheng, C.-H.; Atkinson, D.L.; Krcmery, J.; Guzman, C.E.; Kent, D.T.; Zukor, K.; Marx, K.A.; Odelberg, S.J.; Simon, H.-J. Multi-tissue microarray analysis identifies a molecular signature of regeneration. PLoS ONE 2012, 7, e52375. [Google Scholar] [CrossRef]
- Grigoryan, E.N. Molecular factors of the maintenance and activation of the juvenile phenotype of cellular sources for eye tissue regeneration. Biochemistry 2018, 83, 1627–1642. [Google Scholar] [CrossRef]
- Sherpa, T.; Lankford, T.; McGinn, T.E.; Hunter, S.S.; Frey, R.A.; Sun, C.; Ryan, M.; Robison, B.D.; Stenkamp, D.L. Retinal regeneration is facilitated by the presence of surviving neurons. Dev. Neurobiol. 2014, 74, 851–876. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.R.; Aycinena, J.-C.; del Rio-Tsonis, K. FGF-Hedgehog Interdependence During Retina Regeneration. Dev. Dyn. 2007, 236, 1161–1174. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Kidd, A.R., 3rd; Thomas, J.L.; Poss, K.D.; Hyde, D.R.; Raymond, P.A.; Thummel, R. FGF signaling regulates rod photoreceptor cell maintenance and regeneration zebrafish. Exp. Eye Res. 2011, 93, 726–734. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fukui, L.; Henry, J.J. FGF signaling is required for lens regeneration in Xenopus laevis. Biol. Bull. 2011, 221, 137–145. [Google Scholar] [CrossRef]
- Susaki, K.; Chiba, C. Pigment MEK mediates in vitro neural transdifferentiation of the adult newt retinal pigment epithelium cells: Is FGF2 an induction factor? Cell Res. 2007, 20, 364–379. [Google Scholar] [CrossRef]
- Markitantova, Y.V.; Avdonin, P.P.; Grigoryan, E.N. FGF2 signaling pathway components in tissues of the posterior eye sector in the adult newt Pleurodeles waltl. Biol. Bull. 2014, 41, 297–305. [Google Scholar] [CrossRef]
- Mitsuda, S.; Yoshii, C.; Ikegami, Y.; Araki, M. Tissue interaction between the retinal pigment epithelium and the choroid triggers retinal regeneration of the newt Cynops pyrrhogaster. Dev. Biol. 2005, 280, 122–132. [Google Scholar] [CrossRef]
- Araki, M. Regeneration of the amphibian retina: Role of tissue interaction and related signaling molecules on RPE transdifferentiation. Dev. Growth Differ. 2007, 49, 109–120. [Google Scholar] [CrossRef]
- Nikolaev, A.A. Epigenetic features of pigment epithelium reprogramming during retinal regeneration after photo-induced detachment in Pleurodeles waltl newt. In Bachelor’s Final Qualification Work; Moscow State University: Moscow, Russia, 2018. [Google Scholar]
- Dvoriantchikova, D.; Seemungal, R.J.; Ivanov, D. The epigenetic basis for the impaired ability of adult murine retinal pigment epithelium cells to regenerate retinal tissue. Sci. Rep. 2019, 9, 3860. [Google Scholar] [CrossRef]
- Grigoryan, E.N. Study of natural long-life juvenility and tissue regeneration in caudate amphibians and potential application of resulting data in biomedicine. J. Dev. Biol. 2021, 9, 2. [Google Scholar] [CrossRef]
- Yoshii, C.; Ueda, Y.; Okamoto, M.; Araki, M. Neural retinal regeneration in the anuran amphibian Xenopus laevis post-metamorphosis: Transdifferentiation of retinal pigmented epithelium regenerates the neural retina. Dev. Biol. 2007, 303, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Vergara, M.N.; del Rio-Tsonis, K. Retinal regeneration in the Xenopus laevis tadpole: A new model system. Mol. Vis. 2009, 15, 1000–1013. [Google Scholar] [PubMed]
- Sakaguchi, D.S.; Janick, L.M.; Reh, T.A. Basic Fibroblast Growth Factor (FGF-2) induced transdifferentiation of retinal pigment epithelium: Generation of retinal neurons and glia. Dev. Dyn. 1997, 209, 387–398. [Google Scholar] [CrossRef]
- Kuriyama, F.; Ueda, Y.; Araki, M. Complete reconstruction of the retinal laminar structure from a cultured retinal pigment epithelium is triggered by altered tissue interaction and promoted by overlaid extracellular matrices. Dev. Neurobiol. 2009, 69, 950–958. [Google Scholar] [CrossRef] [PubMed]
- Naitoh, H.; Suganuma, Y.; Ueda, Y.; Sato, T.; Hiramuki, Y.; Fujisawa-Sehara, A.; Taketani, S.; Araki, M. Upregulation of matrix metalloproteinase triggers transdifferentiation of retinal pigmented epithelial cells in Xenopus laevis: A Link between inflammatory response and regeneration. Dev. Neurobiol. 2017, 77, 1086–1100. [Google Scholar] [CrossRef]
- Arresta, E.; Bernardini, S.; Bernardini, E.; Filoni, S.; Cannata, S.M. Pigmented epithelium to retinal transdifferentiation and Pax6 expression in larval Xenopus laevis. J. Exp. Zool. A Comp. Exp. Biol. 2005, 303, 958–967. [Google Scholar] [CrossRef]
- Nabeshima, A.; Nishibayashi, C.; Ueda, Y.; Ogino, H.; Araki, M. Loss of cell-extracellular matrix interaction triggers retinal regeneration accompanied by Rax and Pax6 activation. Genesis 2013, 51, 410–419. [Google Scholar] [CrossRef]
- El-Hodiri, H.M.; Martinez-De Luna, R.I.; Kelly, L.E. The retinal homeobox (Rx) gene is necessary for retinal regeneration in Xenopus laevis tadpoles. Dev. Biol. 2010, 344, 423. [Google Scholar] [CrossRef][Green Version]
- Martinez-De Luna, R.I.; Kelly, L.E.; El-Hodiri, H.M. The Retinal Homeobox (Rx) gene is necessary for retinal regeneration. Dev. Biol. 2011, 353, 10–18. [Google Scholar] [CrossRef][Green Version]
- Tseng, A.-S. Seeing the future: Using Xenopus to understand eye regeneration. Genesis 2017, 55, e23003. [Google Scholar] [CrossRef]
- Kha, C.X.; Son, P.H.; Son, P.H.; Tseng, K.A.-S. A model for investigating developmental eye repair in Xenopus laevis. Exp. Eye Res. 2018, 169, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Ochi, H. Regeneration enhancers: A clue to reactivation of developmental genes. Dev. Growth Differ. 2020, 62, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Park, C.M.; Hollenberg, M.J. Growth factor-induced retinal regeneration in vivo. Int. Rev. Cytol. 1993, 146, 49–74. [Google Scholar] [CrossRef] [PubMed]
- Haynes, T.; Luz-Madrigal, A.; Reis, E.S.; Echeverri Ruiz, N.P.; Grajales-Esquivel, E.; Tzekou, A.; Tsonis, P.A.; Lambris, J.D.; del Rio-Tsonis, K. Complement anaphylatoxin C3a is a potent inducer of embryonic chick retina regeneration. Nat. Commun. 2013, 4, 2312. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Luz-Madrigal, A.; Haynes, T.; Zavada, J.; Burke, A.K.; del Rio-Tsonis, K. β-Catenin Inactivation Is a Pre-Requisite for Chick Retina Regeneration. PLoS ONE 2014, 9, e101748. [Google Scholar] [CrossRef]
- Spence, J.R.; Madhavan, M.; Aycinena, J.-C.; del Rio-Tsonis, K. Retina regeneration in the chick embryo is not induced by spontaneous Mitf downregulation but requires FGF/FGFR/MEK/Erk dependent upregulation of Pax6. Mol. Vis. 2007, 13, 57–65. [Google Scholar]
- Tangeman, J.R.; Luz-Madrigal, A.; Sreeskandarajan, S.; Grajales-Esquivel, E.; Liu, L.; Liang, C.; Tsonis, P.A.; del Rio-Tsonis, K. Transcriptome Profiling of Embryonic Retinal Pigment Epithelium Reprogramming. Genes 2021, 12, 840. [Google Scholar] [CrossRef]
- Luz-Madrigal, A.; Grajales-Esquivel, E.; McCorkle, A.; DiLorenzo, A.M.; Barbosa-Sabanero, K.; Tsonis, P.A.; del Rio-Tsonis, K. Reprogramming of the chick retinal pigmented epithelium after retinal injury. BMC Biol. 2014, 12, 28. [Google Scholar] [CrossRef]
- Spence, J.R.; Madhavan, M.; Ewing, J.D.; Jones, D.K.; Lehman, B.M.; del Rio-Tsonis, K. The hedgehog pathway is a modulator of retina regeneration. Development 2004, 131, 4607–4621. [Google Scholar] [CrossRef][Green Version]
- Steinfeld, J.; Steinfeld, I.; Bausch, A.; Coronato, N.; Hampel, M.-L.; Depner, H.; Layer, P.G.; Vogel-Höpker, A. BMP-induced reprogramming of the neural retina into retinal pigment epithelium requires Wnt signalling. Biol. Open. 2017, 6, 979–992. [Google Scholar] [CrossRef]
- Lee, I.; Rasoul, B.A.; Holub, A.S.; Lejeune, A.; Enke, R.A.; Timp, W. Whole genome DNA methylation sequencing of the chicken retina, cornea and brain. Sci. Data 2017, 4, 170148. [Google Scholar] [CrossRef] [PubMed]
- Luz-Madrigal, A.; Grajales-Esquivel, E.; Tangeman, J.; Kosse, S.; Liu, L.; Wang, K.; Fausey, A.; Liang, C.; Tsonis, P.A.; del Rio-Tsonis, K. DNA demethylation is a driver for chick retina regeneration. Epigenetics 2020, 15, 998–1019. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, M.; Bogdahn, U.; Aigner, L. Adult retinal pigment epithelium cells express neural progenitor properties and the neuronal precursor protein doublecortin. Brain Res. 2005, 1040, 98–111. [Google Scholar] [CrossRef]
- Kuznetsova, A.V.; Rzhanova, L.A.; Aleksandrova, M.A. Small non-coding RNA in regulation of differentiation of retinal pigment epithelium. Russ. J. Dev. Biol. 2021, 52, 305–314. [Google Scholar] [CrossRef]
- Milyushina, L.A.; Verdiev, B.I.; Kuznetsova, A.V.; Aleksandrova, M.A. Expression of multipotent and retinal markers in pigment epithelium of adult human in vitro. Bull. Exp. Biol. Med. 2012, 153, 157. [Google Scholar] [CrossRef]
- Amemiya, K.; Haruta, M.; Takahashi, M.; Kosaka, M.; Eguchi, G. Adult human retinal pigment epithelial cells capable of differentiating into neurons. Biochem. Biophys. Res. Commun. 2004, 316, 1–5. [Google Scholar] [CrossRef]
- Li, S.; Zhang, H.; Wang, A.; Liu, Y.; Liu, H.; Yue, F.; Abulaiti, X.; Zhang, C.; Li, L. Differentiation of adult human retinal pigment epithelial cells into dopaminergic-like cells in vitro and in the recipient monkey brain. Mol. Med. 2019, 25, 9. [Google Scholar] [CrossRef]
- Sakami, S.; Etter, P.; Reh, T.A. Activin signaling limits the competence for retinal regeneration from the pigmented epithelium. Mech. Dev. 2008, 125, 106–116. [Google Scholar] [CrossRef]
- Chen, F.; Liu, X.; Chen, Y.; Liu, J.Y.; Lu, H.; Wang, W.; Lu, X.; Dean, K.C.; Gao, L.G.; Kaplan, H.J.; et al. Sphere-induced reprogramming of RPE cells into dual-potential RPE stem-like cells. EBioMedicine 2020, 52, 102618. [Google Scholar] [CrossRef]
- Milyushina, L.A.; Kuznetsova, A.V.; Grigoryan, E.N.; Aleksandrova, M.A. Phenotypic plasticity of retinal pigment epithelium cells of adult human eye in vitro. Klet. Tekhnol. Biol. Med. 2011, 2, 71–76. [Google Scholar]
- Kuznetsova, A.V.; Kurinov, A.M.; Aleksandrova, M.A. Cell models to study regulation of cell transformation in pathologies of retinal pigment epithelium. J. Ophthalmol. 2014, 2014, 801787. [Google Scholar] [CrossRef] [PubMed]
- Shafei, E.V.; Kurinov, A.M.; Kuznetsova, A.V.; Aleksandrova, M.A. Reprogramming of human retinal pigment epithelial cells under the effect of bFGF in vitro. Bull. Exp. Biol. Med. 2017, 163, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Bok, D. A cell culture medium that supports the differentiation of human retinal pigment epithelium into functionally polarized monolayers. Mol. Vis. 2001, 7, 14–19. [Google Scholar] [PubMed]
- Blenkinsop, T.A.; Salero, E.; Stern, J.H.; Temple, S. The culture and maintenance of functional retinal pigment epithelial monolayers from adult human eye. Methods Mol. Biol. 2013, 945, 45–55. [Google Scholar] [CrossRef]
- Blenkinsop, T.A.; Saini, J.S.; Maminishkis, A.; Bharti, K.; Wan, Q.; Banzon, T.; Lotfi, M.; Davis, J.; Singh, D.; Rizzolo, L.J.; et al. Human Adult Retinal Pigment Epithelial Stem Cell-Derived RPE Monolayers Exhibit Key Physiological Characteristics of Native Tissue. Investig. Ophthalmol. Vis. Sci. 2015, 56, 7085–7099. [Google Scholar] [CrossRef]
- Samuel, W.; Jaworski, C.; Postnikova, O.A.; Kutty, R.K.; Duncan, T.; Tan, L.X.; Poliakov, E.; Lakkaraju, A.; Redmond, T.M. Appropriately differentiated ARPE-19 cells regain phenotype and gene expression profiles similar to those of native RPE cells. Mol. Vis. 2017, 23, 60–89. [Google Scholar]
- Salero, E.; Blenkinsop, T.A.; Corneo, B.; Harris, A.; Rabin, D.; Stern, J.H.; Temple, S. Adult human RPE can be activated into a multipotent stem cell that produces mesenchymal derivatives. Cell Stem Cell 2012, 10, 88–95. [Google Scholar] [CrossRef]
- Saini, J.S.; Temple, S.; Stern, J.H. Human Retinal Pigment Epithelium Stem Cell (RPESC). Adv. Exp. Med. Biol. 2016, 854, 557–562. [Google Scholar] [CrossRef]
- Burke, J.M.; Hjelmeland, L.M. Mosaicism of the retinal pigment epithelium: Seeing the small picture. Mol. Interv. 2005, 5, 241–249. [Google Scholar] [CrossRef]
- Hjelmeland, L.M.; Fujikawa, A.; Oltjen, S.L.; Smit-McBride, Z.; Braunschweig, D. Quantification of retinal pigment epithelial phenotypic variation using laser scanning cytometry. Mol. Vis. 2010, 16, 1108–1121. [Google Scholar]
- Boulton, M.E. Studying melanin and lipofuscin in RPE cell culture models. Exp. Eye Res. 2014, 126, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Al-Hussaini, H.; Vugler, A.; Semo, M.; Jeffery, G. Mature mammalian retinal pigment epithelium cells proliferate in vivo. Mol. Vis. 2008, 14, 1784–1791. [Google Scholar] [PubMed]
- Zhang, W.; Liu, H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Hecquet, C.; Lefevre, G.; Valtink, M.; Engelmann, K.; Mascarelli, F. Activation and role of MAP kinase-dependent pathways in retinal pig-ment epithelial cells: ERK and RPE cell proliferation. Investig. Ophthalmol. Vis. Sci. 2002, 43, 3091–3098. [Google Scholar]
- Bharti, K.; Nguyen, M.-T.T.; Skuntz, S.; Bertuzzi, S.; Arnheiter, H. The other pigment cell: Specification and development of the pigmented epithelium of the vertebrate eye. Pigment Cell Res. 2006, 19, 380–394. [Google Scholar] [CrossRef]
- Rzhanova, L.A.; Kuznetsova, A.V.; Aleksandrova, M.A. Reprogramming of differentiated mammalian and human retinal pigment epithelium: Current achievements and prospects. Russ. J. Dev. Biol. 2020, 51, 212–230. [Google Scholar] [CrossRef]
- Martinez-De Luna, R.I.; Zuber, M.E. Putting Regeneration into Regenerative Medicine. J. Ophthalmic Vis. Res. 2014, 9, 126–133. [Google Scholar]
- George, S.M.; Lu, F.; Rao, M.; Leach, L.L. Gross JM. The retinal pigment epithelium: Development, injury responses, and regenerative potential in mammalian and non-mammalian systems. Prog. Retin. Eye Res. 2021, 23, 100969. [Google Scholar] [CrossRef]
- Westenskow, P.; Piccolo, S.; Fuhrmann, S. Beta-catenin controls differentiation of the retinal pigment epithelium in the mouse optic cup by regulating Mitf and Otx2 expression. Development 2009, 136, 2505–2510. [Google Scholar] [CrossRef]
- Bumsted, K.M.; Barnstable, C.J. Dorsal retinal pigment epithelium differentiates as neural retina in the microphthalmia (mi/mi) mouse. Investig. Ophthalmol. Vis. Sci. 2000, 41, 903–908. [Google Scholar]
- Kirchhof, B.; Sorgente, N. Pathogenesis of proliferative vitreoretinopathy. Modulation of retinal pigment epithelial cell functions by vitreous and macrophages. Dev. Ophthalmol. 1989, 16, 1–53. [Google Scholar] [PubMed]
- Abe, T.; Sato, M.; Tamai, M. Dedifferentiation of the retinal pigment epithelium compared to the proliferative membranes of proliferative vitreoretinopathy. Curr. Eye Res. 1998, 17, 1103–1109. [Google Scholar] [CrossRef] [PubMed]
- Tamiya, S.; Kaplan, H.J. Role of epithelial-mesenchymal transition in proliferative vitreoretinopathy. Exp. Eye Res. 2016, 142, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, X.; Liu, X.; Huang, S.; He, C.; Chen, B.; Liu, Y. Autophagy regulates TGF-beta2-induced epithelial-mesenchymal transition in human retinal pigment epithelium cells. Mol. Med. Rep. 2018, 17, 3607–3614. [Google Scholar] [CrossRef]
- Tikhonovich, M.V.; Iojleva, E.J.; Gavrilova, S.A. The role of inflammation in the development of proliferative vitreoretinopathy. Klin. Med. 2015, 93, 14–20. [Google Scholar]
- Garweg, J.G.; Tappeiner, C.; Halberstadt, M. Pathophysiology of proliferative vitreoretinopathy in retinal detachment. Surv. Ophthalmol. 2013, 58, 321–329. [Google Scholar] [CrossRef]
- Zhou, M.; Geathers, J.S.; Grillo, S.L.; Weber, S.R.; Wang, W.; Zhao, Y.; Sundstrom, J.M. Role of epithelial-mesenchymal transition in retinal pigment epithelium dysfunction. Front. Cell Dev. Biol. 2020, 8, 501. [Google Scholar] [CrossRef]
- Han, J.W.; Lyu, J.; Park, Y.J.; Jang, S.Y.; Park, T.K. Wnt/β-catenin signaling mediates regeneration of retinal pigment epithelium after laser photocoagulation in mouse eye. Investig. Ophthalmol. Vis. Sci. 2015, 56, 8314–8324. [Google Scholar] [CrossRef]
- Kent, D.; Sheridan, C. Choroidal neovascularization: A wound healing perspective. Mol. Vis. 2003, 9, 747–755. [Google Scholar]
- Ishikawa, K.; Kannan, R.; Hinton, D.R. Molecular mechanisms of subretinal fibrosis in age-related macular degeneration. Exp. Eye Res. 2016, 142, 19–25. [Google Scholar] [CrossRef]
- Shu, D.Y.; Butcher, E.; Saint-Geniez, M. EMT and EndMT: Emerging Roles in Age-Related Macular Degeneration. Int. J. Mol. Sci. 2020, 21, 4271. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, P.G.; Moreno-Bueno, G.; Portillo, F.; Cano, A. EMT: Present and future in clinical oncology. Mol. Oncol. 2017, 11, 718–738. [Google Scholar] [CrossRef] [PubMed]
- Francou, A.; Anderson, K.V. The Epithelial-to-Mesenchymal Transition (EMT) in Development and Cancer. Annu. Rev. Cancer Biol. 2020, 4, 197–220. [Google Scholar] [CrossRef] [PubMed]
- Sisto, M.; Ribatti, D.; Lisi, S. Organ Fibrosis and Autoimmunity: The Role of Inflammation in TGFβ-Dependent EMT. Biomolecules 2021, 11, 310. [Google Scholar] [CrossRef]
- Snead, D.R.J.; James, S.; Snead, M.P. Pathological changes in the vitreoretinal junction 1: Epiretinal membrane formation. Eye 2008, 22, 1310–1317. [Google Scholar] [CrossRef]
- Philp, N.J.; Nachmias, V.T. Polarized distribution of integrin and fibronectin in retinal pigment epithelium. Investig. Ophthalmol. Vis. Sci. 1987, 28, 1275–1280. [Google Scholar]
- Huang, X.; Wei, Y.; Ma, H.; Zhang, S. Vitreous-induced cytoskeletal rearrangements via the Rac1 GTPase- dependent signaling pathway in human retinal pigment epithelial cells. Biochem. Biophys. Res. Commun. 2012, 419, 395–400. [Google Scholar] [CrossRef]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef]
- Tamiya, S.; Liu, L.; Kaplan, H.J. Epithelial-mesenchymal transition and proliferation of retinal pigment epithelial cells initiated upon loss of cell-cell contact. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2755–2763. [Google Scholar] [CrossRef]
- Maeda, M.; Johnson, K.R.; Wheelock, M.J. Cadherin switching: Essential for behavioral but not morphological changes during an epithelium-to-mesenchyme transition. J. Cell Sci. 2005, 118, 873–887. [Google Scholar] [CrossRef]
- Nieto, M.A. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu. Rev. Cell Dev. Biol. 2011, 27, 347–376. [Google Scholar] [CrossRef] [PubMed]
- Hiscott, P.; Sheridan, C.; Magee, R.M.; Grierson, I. Matrix and the retinal pigment epithelium in proliferative retinal disease. Prog. Ret. Eye Res. 1999, 18, 167–190. [Google Scholar] [CrossRef]
- Georgiadis, A.; Tschernutter, M.; Bainbridge, J.W.B.; Balaggan, K.S.; Mowat, F.; West, E.; Munro, P.M.G.; Thrasher, A.J.; Matter, K.; Balda, M.S.; et al. The tight junction associated signalling proteins ZO-1 and ZONAB regulate retinal pigment epithelium homeostasis in mice. PLoS ONE 2010, 5, e15730. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, C.; Hiscott, P.; Grierson, I. Retinal Pigment Epithelium Differentiation and Dedifferentiation—Vitreo-Retinal Surgery; Kirchhof, B., Wong, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 101–119. [Google Scholar]
- Imamichi, Y.; Menke, A. Signaling pathways involved in collagen-induced disruption of the E-cadherin complex during epithelial-mesenchymal transition. Cells Tissues Organs 2007, 185, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; O’Meara, S.J.; O’Brien, C.; Kane, R. The role of gremlin, a bmp antagonist, and epithelial-to-mesenchymal transition in proliferative vitreoretinopathy. Investig. Ophthalmol. Vis. Sci. 2007, 48, 4291–4299. [Google Scholar] [CrossRef]
- Choudhary, P.; Dodsworth, B.T.; Sidders, B.; Gutteridge, A.; Michaelides, C.; Duckworth, J.K.; Sidders, B.; Gutteridge, A.; Michaelides, C.; Duckworth, J.K.; et al. A FOXM1 dependent mesenchymal-epithelial transition in retinal pigment epithelium cells. PLoS ONE 2015, 10, e0130379. [Google Scholar] [CrossRef]
- Shu, D.Y.; Lovicu, F.J. Myofibroblast transdifferentiation: The dark force in ocular wound healing and fibrosis. Prog. Retin. Eye Res. 2017, 60, 44–65. [Google Scholar] [CrossRef]
- Hua, X.; Liu, X.; Ansari, D.O.; Lodish, H.F. Synergistic cooperation of TFE3 and SMAD proteins in TGF-beta-induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev. 1998, 12, 3084–3095. [Google Scholar] [CrossRef]
- Kang, Y.; Massague, J. Epithelial-mesenchymal transitions: Twist in development and metastasis. Cell 2004, 118, 277–279. [Google Scholar] [CrossRef]
- Pratt, C.H.; Vadigepalli, R.; Chakravarthula, P.; Gonye, G.E.; Philp, N.J.; Grunwald, G.B. Transcriptional regulatory network analysis during epithelial-mesenchymal transformation of retinal pigment epithelium. Mol. Vis. 2008, 14, 1414–1428. [Google Scholar]
- Vaajasaari, H.; Ilmarinen, T.; Juuti-Uusitalo, K.; Rajala, K.; Onnela, N.; Narkilahti, S.; Suuronen, R.; Hyttinen, J.; Uusitalo, H.; Skottman, H. Toward the defined and xeno-free differentiation of functional human pluripotent stem cell-derived retinal pigment epithelial cells. Mol. Vis. 2011, 17, 558–575. [Google Scholar] [PubMed]
- Buchholz, D.E.; Pennington, B.O.; Croze, R.H.; Hinman, C.R.; Coffey, P.J.; Clegg, D.O. Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Transl. Med. 2013, 2, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Lynn, S.A.; Gareth, W.; Keeling, E.; Scott, J.A.; Cree, A.J.; Johnston, D.A.; Page, A.; Cuan-Urquizo, E.; Bhaskar, A.; Grossel, M.C.; et al. Ex-vivo models of the retinal pigment epithelium (RPE) in long-term culture faithfully recapitulate key structural and physio-logical features of native RPE. Tissue Cell 2017, 49, 447–460. [Google Scholar] [CrossRef]
- Benayoun, B.A.; Caburet, S.; Veitia, R.A. Forkhead transcription factors: Key players in health and disease. Trends Genet. 2011, 27, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Muller, G.A.; Quaas, M.; Fischer, M.; Han, N.; Stutchbury, B.; Sharrocks, A.D.; Engeland, K. The forkhead transcription factor FoxM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol. Cell. Biol. 2013, 33, 227–236. [Google Scholar] [CrossRef]
- Wang, I.C.; Chen, Y.J.; Hughes, D.; Petrovic, V.; Major, M.L.; Park, H.J.; Tan, Y.; Ackerson, T.; Costa, R.H. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 2005, 25, 10875–10894. [Google Scholar] [CrossRef]
- Qu, K.; Xu, X.; Liu, C.; Wu, Q.; Wei, J.; Meng, F.; Zhou, L.; Wang, Z.; Lei, L.; Liu, P. Negative regulation of transcription factor FoxM1 by p53 enhances oxaliplatin-induced senescence in hepatocellular carcinoma. Cancer Lett. 2013, 331, 105–114. [Google Scholar] [CrossRef]
- Toro, M.D.; Reibaldi, M.; Avitabile, T.; Bucolo, C.; Salomone, S.; Rejdak, R.; Nowomiejska, K.; Tripodi, S.; Posarelli, C.; Ragusa, M.; et al. MicroRNAs in the Vitreous Humor of Patients with Retinal Detachment and a Different Grading of Proliferative Vitreoretinopathy: A Pilot Study. Transl. Vis. Sci. Technol. 2020, 9, 23. [Google Scholar] [CrossRef]
- Chaudhary, R.; Scott, R.A.H.; Wallace, G.; Berry, M.; Logan, A.; Blanch, R.J. Inflammatory and Fibrogenic Factors in Proliferative Vitreoretinopathy Development. Transl. Vis. Sci. Technol. 2020, 9, 23. [Google Scholar] [CrossRef]
- Dai, Y.; Dai, C.; Sun, T. Inflammatory mediators of proliferative vitreoretinopathy: Hypothesis and review. Int. Ophthalmol. 2020, 40, 1587–1601. [Google Scholar] [CrossRef]
- Saika, S. TGF-beta pathobiology in the eye. Lab. Investig. 2006, 86, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Kita, T.; Hata, Y.; Arita, R.; Kawahara, S.; Miura, M.; Nakao, S.; Mochizuki, Y.; Enaida, N.; Goto, Y.; Shimokawa, H.; et al. Role of TGF-β in proliferative vitreoretinal diseases and ROCK as a therapeutic target. Proc. Natl. Acad. Sci. USA 2008, 105, 17504–17509. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, T.; Kayama, M.; Ryu, M.; Kunikata, H.; Watanabe, R.; Yasuda, M.; Kinugawa, J.; Vavvas, D.; Miller, J.W. Tumor Necrosis Factor-α Mediates Photoreceptor Death in a Rodent Model of Retinal Detachment. Invest. Ophthalmol. Vis. Sci. 2011, 52, 1384–1391. [Google Scholar] [CrossRef] [PubMed]
- Korthagen, N.M.; van Bilsen, K.; Swagemakers, S.M.; van de Peppel, J.; Bastiaans, J.; van der Spek, P.J.; van Hagen, P.M.; Dik, W.A. Retinal pigment epithelial cells display specific transcriptional responses upon TNF-alpha stimulation. Br. J. Ophthalmol. 2015, 99, 700–704. [Google Scholar] [CrossRef]
- Boles, N.C.; Fernandes, M.; Swigut, T.; Srinivasan, R.; Schiff, L.; Rada-Iglesias, A.; Wang, Q.; Saini, J.S.; Kiehl, T.; Stern, J.H.; et al. Epigenomic and Transcriptomic Changes During Human RPE EMT in a Stem Cell Model of Epiretinal Membrane Pathogenesis and Prevention by Nicotinamide. Stem Cell Rep. 2020, 14, 631–647. [Google Scholar] [CrossRef]
- Saika, S.; Yamanaka, O.; Okada, Y.; Tanaka, S.; Miyamoto, T.; Sumioka, T.; Kitano, A.; Shirai, K.; Ikeda, K. TGFβ in fibroproliferative diseases in the eye. Front. Biosci. 2009, 1, 376–390. [Google Scholar] [CrossRef]
- Yan, X.; Liu, Z.; Chen, Y. Regulation of TGF-β signaling by Smad7. Acta Biochim. Biophys. Sin. 2009, 41, 263–272. [Google Scholar] [CrossRef]
- Schiff, L.; Boles, N.C.; Fernandes, M.; Nachmani, B.; Gentile, R.; Blenkinsop, T.A. P38 inhibition reverses TGFb1 and TNFa-induced contraction in a model of proliferative vitreoretinopathy. Commun. Biol. 2019, 2, 162. [Google Scholar] [CrossRef]
- Saini, J.S.; Corneo, B.; Miller, J.D.; Kiehl, T.R.; Wang, Q.; Boles, N.C.; Blenkinsop, T.A.; Stern, J.H.; Temple, S. Nicotinamide Ameliorates Disease Phenotypes in a Human iPSC Model of Age-Related Macular Degeneration. Cell Stem Cell 2017, 20, 635–647. [Google Scholar] [CrossRef]
- Nassar, K.; Grisanti, S.; Tura, A.; Lüke, J.; Lüke, M.; Soliman, M.; Grisanti, S. A TGF-β receptor 1 inhibitor for prevention of proliferative vitreoretinopathy. Exp. Eye Res. 2014, 123, 72–86. [Google Scholar] [CrossRef]
- Sherpa, R.D.; Hui, S.P. An insight on established retinal injury mechanisms and prevalent retinal stem cell activation pathways in vertebrate models. Anim. Models Exp. Med. 2021, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.V.; Wang, J.; Boyd, P.; Wang, F.; Santiago, C.; Sooyeon, J.L.; Jiang, L.; Lahne, M.; Todd, L.J.; Jia, M.; et al. Gene regulatory networks controlling vertebrate retinal regeneration. Science 2020, 370, eabb8598. [Google Scholar] [CrossRef] [PubMed]
- Hamon, A.; Roger, J.E.; Yang, X.J.; Perron, M. Muller glial cell dependent regeneration of the neural retina: An overview across vertebrate model systems. Dev. Dyn. 2016, 245, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Miesfeld, J.B.; Brown, N.L. Eye organogenesis: A hierarchical view of ocular development. Curr. Top. Dev. Biol. 2019, 132, 351–393. [Google Scholar] [CrossRef] [PubMed]
- Godwin, J.W.; Rosenthal, N. Scar-free wound healing and regeneration in amphibians: Immunological influences on regenerative success. Differentiation 2014, 87, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grigoryan, E.N. Pigment Epithelia of the Eye: Cell-Type Conversion in Regeneration and Disease. Life 2022, 12, 382. https://doi.org/10.3390/life12030382
Grigoryan EN. Pigment Epithelia of the Eye: Cell-Type Conversion in Regeneration and Disease. Life. 2022; 12(3):382. https://doi.org/10.3390/life12030382
Chicago/Turabian StyleGrigoryan, Eleonora N. 2022. "Pigment Epithelia of the Eye: Cell-Type Conversion in Regeneration and Disease" Life 12, no. 3: 382. https://doi.org/10.3390/life12030382
APA StyleGrigoryan, E. N. (2022). Pigment Epithelia of the Eye: Cell-Type Conversion in Regeneration and Disease. Life, 12(3), 382. https://doi.org/10.3390/life12030382




