Emergence of SARS-CoV-2 Variants in the World: How Could This Happen?
Abstract
:1. Introduction
2. Classification of SARS-CoV-2 Variants
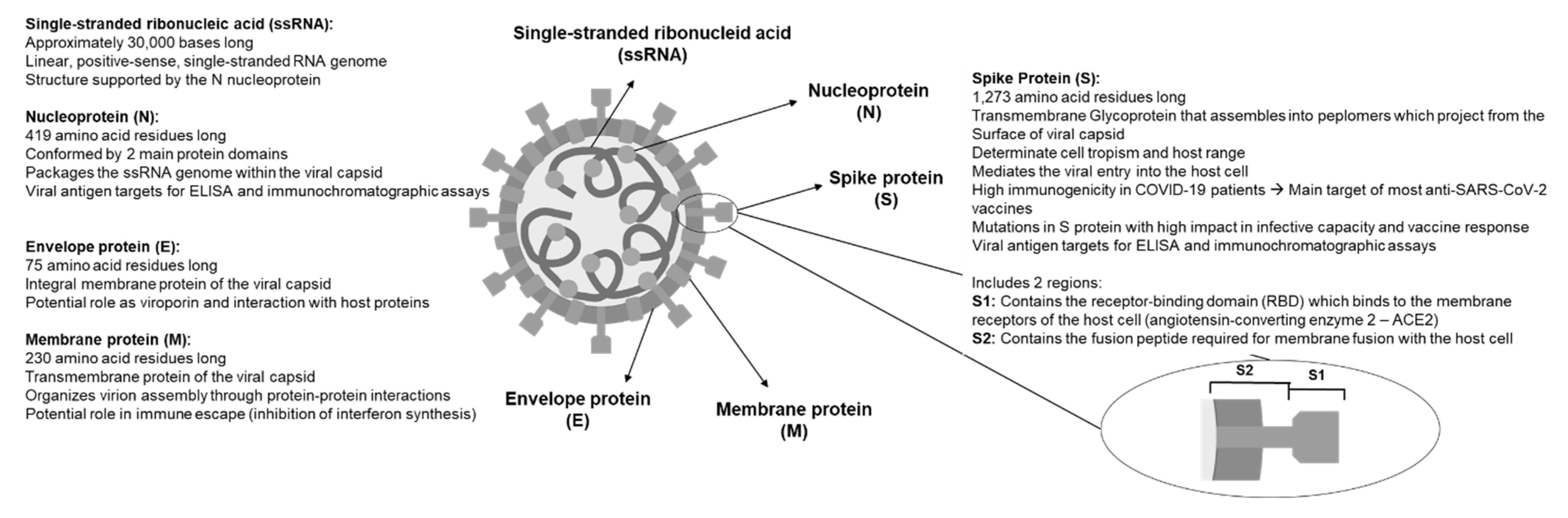
3. Virological Characteristics of SARS-CoV-2
4. Relevant Characteristics of the SARS-CoV-2 Genome
5. Genetic Changes in the SARS-CoV-2 Variants of Interest (VOIs) and Variants of Concern (VOCs)
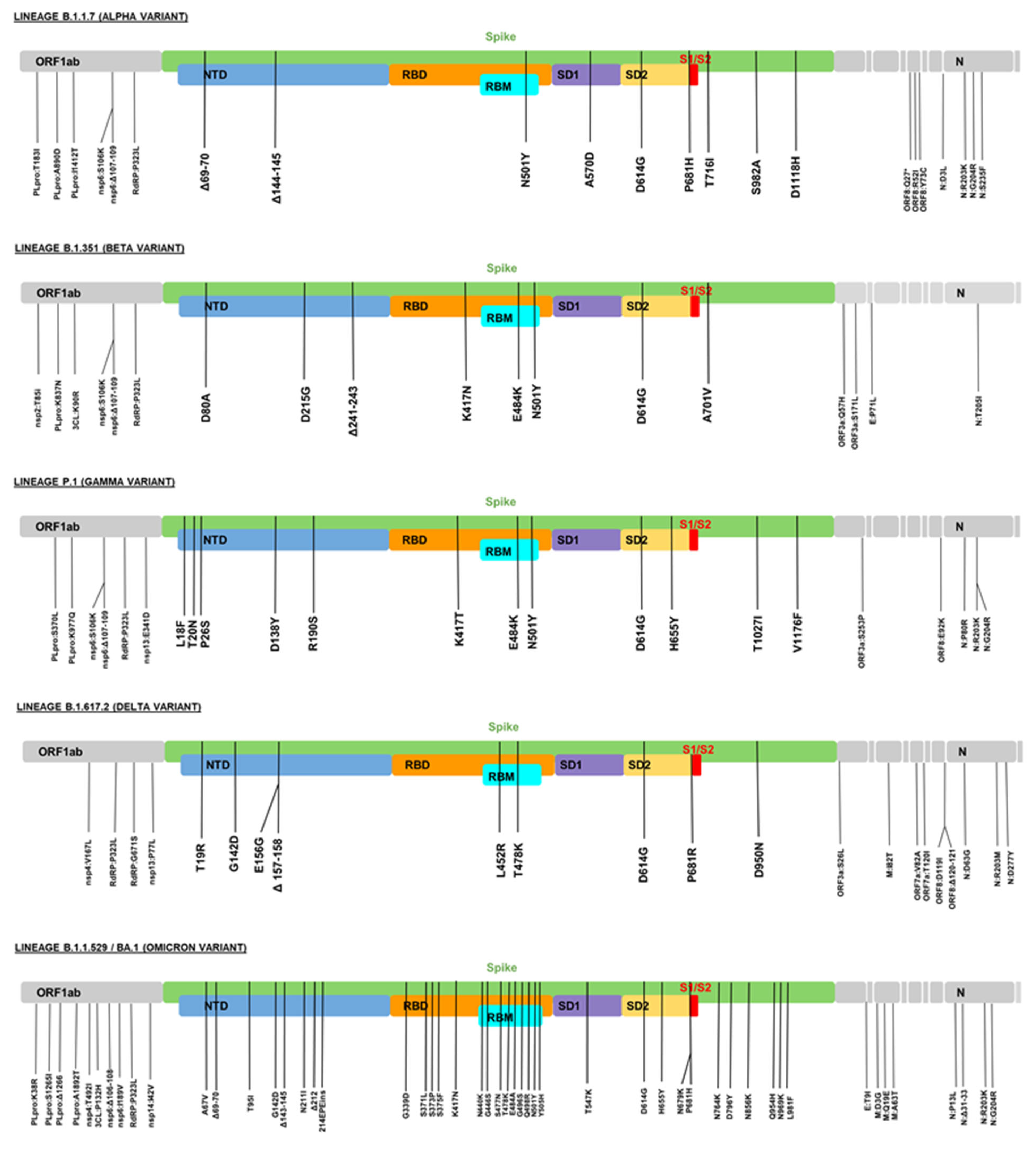
5.1. Lineage B.1.1.7 (Alpha Variant)
5.2. Lineage B.1.351 (Beta Variant)
5.3. Lineage P.1 (Gamma Variant)
5.4. Lineage B.1.617.2 (Delta Variant)
5.5. Lineage B.1.1.529/BA.1 (Omicron Variant)
6. Predisposing Geographical, Environmental, and Genetic Conditions for the Development of SARS-CoV-2 Variants
7. Influence of Social Determinants on the Development of Variants of SARS-CoV-2
7.1. Social Inequality
7.2. Poor Access to Health Systems—Vaccination Effects
7.3. Health Education
7.4. Social Capital
7.5. Overcrowding
7.6. Labor and Economic Conditions
8. Strategies to Control Potential Variants of Concern of SARS-CoV-2
8.1. Genomic Surveillance of SARS-CoV-2
8.2. International Anti-SARS-CoV-2 Vaccination Campaign
8.3. Use of Face Masks
8.4. Identification of Infected Patients and Containment Strategies
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- COVID-19 m–p—John Hopkins Coronavirus Resource Center. Jhu.Edu. Available online: https://coronavirus.jhu.edu/map.html (accessed on 31 December 2021).
- Sharma, A.; Ahmad Farouk, I.; Lal, S.K. COVID-19: A Review on the Novel Coronavirus Disease Evolution, Transmission, Detection, Control and Prevention. Viruses 2021, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Tzou, P.L.; Nouhin, J.; Gupta, R.K.; de Oliveira, T.; Kosakovsky Pond, S.L.; Fera, D.; Shafer, R.W. The Biological and Clinical Significance of Emerging SARS-CoV-2 Variants. Nat. Rev. Genet. 2021, 22, 757–773. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.; Govindarajulu, M.; Parise, R.S.; Neel, L.; Shankar, T.; Patel, S.; Lowery, P.; Smith, F.; Dhanasekaran, M.; Moore, T. Emerging SARS-CoV-2 Variants: A Review of its Mutations, its Implications and Vaccine Efficacy. Vaccines 2021, 9, 1195. [Google Scholar] [CrossRef] [PubMed]
- Penner, R.C. Mutagenic Distinction between the Receptor-Binding and Fusion Subunits of the SARS-CoV-2 Spike Glycoprotein and Its Upshot. Vaccines 2021, 9, 1509. [Google Scholar] [CrossRef]
- Thomas, E.; Delabat, S.; Carattini, Y.L.; Andrews, D.M. SARS-CoV-2 and Variant Diagnostic Testing Approaches in the United States. Viruses 2021, 13, 2492. [Google Scholar] [CrossRef]
- Karim, S.S.A.; Karim, Q.A. Omicron SARS-CoV-2 Variant: A New Chapter in the COVID-19 Pandemic. Lancet 2021, 398, 2126–2128. [Google Scholar] [CrossRef]
- Shanmugaraj, B.; Malla, A.; Khorattanakulchai, N.; Phoolcharoen, W. SARS-CoV-2 Omicron Variant: Could it be Another Threat? J. Med. Virol. 2021. [Google Scholar] [CrossRef]
- World Health Organization. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 31 December 2021).
- Parums, D. Editorial: Revised World Health Organization (WHO) Terminology for Variants of Concern and Variants of Interest of SARS-CoV-2. Med. Sci. Monit. 2021, 27, e933622. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Worp, N.; Nieuwenhuijse, D.F.; Sikkema, R.S.; Haagmans, B.; Fouchier, R.A.M.; Koopmans, M. The next Phase of SARS-CoV-2 Surveillance: Real-Time Molecular Epidemiology. Nat. Med. 2021, 27, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-Y.; Zhao, R.; Gao, L.-J.; Gao, X.-F.; Wang, D.-P.; Cao, J.-M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, D. What You Need to Know about the Novel Coronavirus. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). In Encyclopedia of Virology; Bamford, D.H., Zuckerman, M., Eds.; Elsevier: San Diego, CA, USA, 2021; pp. 428–440. [Google Scholar]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xie, W.; Wang, Y.; Xiong, Y.; Chen, S.; Han, J.; Wu, Q. A Comparative Overview of COVID-19, MERS and SARS: Review Article. Int. J. Surg. 2020, 81, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Raskin, S. Genetics of COVID-19. J. Pediatr. 2021, 97, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Stanford Coronavirus Antiviral & Resistance Database (CoVDB). Available online: https://covdb.stanford.edu/page/mutation-viewer/ (accessed on 28 December 2021).
- Ye, Q.; West, A.M.V.; Silletti, S.; Corbett, K.D. Architecture and Self-Assembly of the SARS-CoV-2 Nucleocapsid Protein. Protein Sci. 2020, 29, 1890–1901. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhuang, M.-W.; Han, L.; Zhang, J.; Nan, M.-L.; Zhan, P.; Kang, D.; Liu, X.; Gao, C.; Wang, P.-H. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Membrane (M) Protein Inhibits Type I and III Interferon Production by Targeting RIG-I/MDA-5 Signaling. Signal Transduct. Target. Ther. 2020, 5, 299. [Google Scholar] [CrossRef] [PubMed]
- Mandala, V.S.; McKay, M.J.; Shcherbakov, A.A.; Dregni, A.J.; Kolocouris, A.; Hong, M. Structure and Drug Binding of the SARS-CoV-2 Envelope Protein Transmembrane Domain in Lipid Bilayers. Nat. Struct. Mol. Biol. 2020, 27, 1202–1208. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Yang, R.; Lee, I.; Zhang, W.; Sun, J.; Wang, W.; Meng, X. Characterization of the SARS-CoV-2 E Protein: Sequence, Structure, Viroporin, and Inhibitors. Protein Sci. 2021, 30, 1114–1130. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; He, G.; Yin, Q.; Zeng, L.; Ye, X.; Shi, Y.; Xu, W. Molecular Biology of the SARs-CoV-2 Spike Protein: A Review of Current Knowledge. J. Med. Virol. 2021, 93, 5729–5741. [Google Scholar] [CrossRef] [PubMed]
- Kyriakidis, N.C.; López-Cortés, A.; González, E.V.; Grimaldos, A.B.; Prado, E.O. SARS-CoV-2 Vaccines Strategies: A Comprehensive Review of Phase 3 Candidates. NPJ Vaccines 2021, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, K.L.; Best, E.; Crawford, N.W.; Giles, M.; Koirala, A.; Macartney, K.; Russell, F.; Teh, B.W.; Wen, S.C. Progress and Pitfalls in the Quest for Effective SARS-CoV-2 (COVID-19) Vaccines. Front. Immunol. 2020, 11, 579250. [Google Scholar] [CrossRef] [PubMed]
- Dejnirattisai, W.; Zhou, D.; Ginn, H.M.; Duyvesteyn, H.M.E.; Supasa, P.; Case, J.B.; Zhao, Y.; Walter, T.S.; Mentzer, A.J.; Liu, C.; et al. The Antigenic Anatomy of SARS-CoV-2 Receptor Binding Domain. Cell 2021, 184, 2183–2200.e22. [Google Scholar] [CrossRef]
- Khare, S.; Azevedo, M.; Parajuli, P.; Gokulan, K. Conformational Changes of the Receptor Binding Domain of SARS-CoV-2 Spike Protein and Prediction of a B-Cell Antigenic Epitope Using Structural Data. Front. Artif. Intell. 2021, 4, 630955. [Google Scholar] [CrossRef] [PubMed]
- De Santos, I.A.; Grosche, V.R.; Bergamini, F.R.G.; Sabino-Silva, R.; Jardim, A.C.G. Antivirals against Coronaviruses: Candidate Drugs for SARS-CoV-2 Treatment? Front. Microbiol. 2020, 11, 1818. [Google Scholar] [CrossRef]
- GISAID–Initiative. Available online: https://www.gisaid.org/ (accessed on 31 December 2021).
- Rahimi, A.; Mirzazadeh, A.; Tavakolpour, S. Genetics and Genomics of SARS-CoV-2: A Review of the Literature with the Special Focus on Genetic Diversity and SARS-CoV-2 Genome Detection. Genomics 2021, 113, 1221–1232. [Google Scholar] [CrossRef]
- Jaimes, J.A.; André, N.M.; Chappie, J.S.; Millet, J.K.; Whittaker, G.R. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J. Mol. Biol. 2020, 432, 3309–3325. [Google Scholar] [CrossRef]
- Brant, A.C.; Tian, W.; Majerciak, V.; Yang, W.; Zheng, Z.-M. SARS-CoV-2: From Its Discovery to Genome Structure, Transcription, and Replication. Cell Biosci. 2021, 11, 136. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zeng, F.; Rao, J.; Yuan, S.; Ji, M.; Lei, X.; Xiao, X.; Li, Z.; Li, X.; Du, W.; et al. Comparison of Clinical Features and Outcomes of Medically Attended COVID-19 and Influenza Patients in a Defined Population in the 2020 Respiratory Virus Season. Front. Public Health 2021, 9, 587425. [Google Scholar] [CrossRef] [PubMed]
- Morales, A.C.; Rice, A.M.; Ho, A.T.; Mordstein, C.; Mühlhausen, S.; Watson, S.; Cano, L.; Young, B.; Kudla, G.; Hurst, L.D. Causes and Consequences of Purifying Selection on SARS-CoV-2. Genome Biol. Evol. 2021, 13, evab196. [Google Scholar] [CrossRef] [PubMed]
- Malone, B.; Urakova, N.; Snijder, E.J.; Campbell, E.A. Structures and Functions of Coronavirus Replication-Transcription Complexes and Their Relevance for SARS-CoV-2 Drug Design. Nat. Rev. Mol. Cell Biol. 2022, 23, 21–39. [Google Scholar] [CrossRef]
- Justo Arevalo, S.; Zapata Sifuentes, D.; Huallpa, J.C.; Landa Bianchi, G.; Castillo Chávez, A.; Garavito-Salini Casas, R.; Uribe Calampa, C.S.; Uceda-Campos, G.; Pineda Chavarría, R. Dynamics of SARS-CoV-2 Mutations Reveals Regional-Specificity and Similar Trends of N501 and High-Frequency Mutation N501Y in Different Levels of Control Measures. Sci. Rep. 2021, 11, 17755. [Google Scholar] [CrossRef]
- Yusof, W.; Irekeola, A.A.; Wada, Y.; Engku Abd Rahman, E.N.S.; Ahmed, N.; Musa, N.; Khalid, M.F.; Rahman, Z.A.; Hassan, R.; Yusof, N.Y.; et al. A Global Mutational Profile of SARS-CoV-2: A Systematic Review and Meta-Analysis of 368,316 COVID-19 Patients. Life 2021, 11, 1224. [Google Scholar] [CrossRef]
- Li, X.; Giorgi, E.E.; Marichannegowda, M.H.; Foley, B.; Xiao, C.; Kong, X.-P.; Chen, Y.; Gnanakaran, S.; Korber, B.; Gao, F. Emergence of SARS-CoV-2 through Recombination and Strong Purifying Selection. Sci. Adv. 2020, 6, eabb9153. [Google Scholar] [CrossRef]
- Haddad, D.; John, S.E.; Mohammad, A.; Hammad, M.M.; Hebbar, P.; Channanath, A.; Nizam, R.; Al-Qabandi, S.; Al Madhoun, A.; Alshukry, A.; et al. SARS-CoV-2: Possible Recombination and Emergence of Potentially More Virulent Strains. PLoS ONE 2021, 16, e0251368. [Google Scholar] [CrossRef]
- Walker, A.S.; Vihta, K.-D.; Gethings, O.; Pritchard, E.; Jones, J.; House, T.; Bell, I.; Bell, J.I.; Newton, J.N.; Farrar, J.; et al. Tracking the Emergence of SARS-CoV-2 Alpha Variant in the United Kingdom. N. Engl. J. Med. 2021, 385, 2582–2585. [Google Scholar] [CrossRef]
- B.1.1.7 Lineage Report. Alaa Abdel Latif, Julia L. Mullen, Manar Alkuzweny, Ginger Tsueng, Marco Cano, Emily Haag, Jerry Zhou, Mark Zeller, Emory Hufbauer, Nate Matteson, Chunlei Wu, Kristian G. Andersen, Andrew I. Su, Karthik Gangavarapu, Laura D. Hughes, and the Center for Viral Systems Biology. Outbreak.Info. Available online: https://outbreak.info/situation-reports?pango=B.1.1.7 (accessed on 31 December 2021).
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Chakraborty, C.; Saha, A.; Sharma, A.R.; Bhattacharya, M.; Lee, S.-S.; Agoramoorthy, G. D614G Mutation Eventuates in All VOI and VOC in SARS-CoV-2: Is It Part of the Positive Selection Pioneered by Darwin? Mol. Ther. Nucleic Acids 2021, 26, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated Transmissibility and Impact of SARS-CoV-2 Lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef] [PubMed]
- Volz, E.; Mishra, S.; Chand, M.; Barrett, J.C.; Johnson, R.; Geidelberg, L.; Hinsley, W.R.; Laydon, D.J.; Dabrera, G.; O’Toole, Á.; et al. Assessing Transmissibility of SARS-CoV-2 Lineage B.1.1.7 in England. Nature 2021, 593, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, M.U.G.; Hill, V.; Ruis, C.; Dellicour, S.; Bajaj, S.; McCrone, J.T.; Baele, G.; Parag, K.V.; Battle, A.L.; Gutierrez, B.; et al. Spatiotemporal Invasion Dynamics of SARS-CoV-2 Lineage B.1.1.7 Emergence. Science 2021, 373, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Jarvis, C.I.; CMMID COVID-19 Working Group; Edmunds, W.J.; Jewell, N.P.; Diaz-Ordaz, K.; Keogh, R.H. Increased Mortality in Community-Tested Cases of SARS-CoV-2 Lineage B.1.1.7. Nature 2021, 593, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Challen, R.; Brooks-Pollock, E.; Read, J.M.; Dyson, L.; Tsaneva-Atanasova, K.; Danon, L. Risk of Mortality in Patients Infected with SARS-CoV-2 Variant of Concern 202012/1: Matched Cohort Study. BMJ 2021, 372, n579. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.S.; Sudre, C.H.; May, A.; Antonelli, M.; Murray, B.; Varsavsky, T.; Kläser, K.; Canas, L.S.; Molteni, E.; Modat, M.; et al. Changes in Symptomatology, Reinfection, and Transmissibility Associated with the SARS-CoV-2 Variant B.1.1.7: An Ecological Study. Lancet Public Health 2021, 6, e335–e345. [Google Scholar] [CrossRef]
- Charmet, T.; Schaeffer, L.; Grant, R.; Galmiche, S.; Chény, O.; Von Platen, C.; Maurizot, A.; Rogoff, A.; Omar, F.; David, C.; et al. Impact of Original, B.1.1.7, and B.1.351/P.1 SARS-CoV-2 Lineages on Vaccine Effectiveness of Two Doses of COVID-19 MRNA Vaccines: Results from a Nationwide Case-Control Study in France. Lancet Reg. Health Eur. 2021, 8, 100171. [Google Scholar] [CrossRef]
- Cevik, M.; Grubaugh, N.D.; Iwasaki, A.; Openshaw, P. COVID-19 Vaccines: Keeping Pace with SARS-CoV-2 Variants. Cell 2021, 184, 5077–5081. [Google Scholar] [CrossRef]
- Choi, J.Y.; Smith, D.M. SARS-CoV-2 Variants of Concern. Yonsei Med. J. 2021, 62, 961–968. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 Variant of Concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef] [PubMed]
- B.1.351 Lineage Report. Alaa Abdel Latif, Julia L. Mullen, Manar Alkuzweny, Ginger Tsueng, Marco Cano, Emily Haag, Jerry Zhou, Mark Zeller, Emory Hufbauer, Nate Matteson, Chunlei Wu, Kristian G. Andersen, Andrew I. Su, Karthik Gangavarapu, Laura D. Hughes, and the Center for Viral Systems Biology. Outbreak.Info. Available online: https://outbreak.info/situation-reports?pango=B.1.351 (accessed on 31 December 2021).
- Khan, A.; Zia, T.; Suleman, M.; Khan, T.; Ali, S.S.; Abbasi, A.A.; Mohammad, A.; Wei, D.-Q. Higher Infectivity of the SARS-CoV-2 New Variants Is Associated with K417N/T, E484K, and N501Y Mutants: An Insight from Structural Data. J. Cell. Physiol. 2021, 236, 7045–7057. [Google Scholar] [CrossRef] [PubMed]
- Funk, T.; Pharris, A.; Spiteri, G.; Bundle, N.; Melidou, A.; Carr, M.; Gonzalez, G.; Garcia-Leon, A.; Crispie, F.; O’Connor, L.; et al. Characteristics of SARS-CoV-2 Variants of Concern B.1.1.7, B.1.351 or P.1: Data from Seven EU/EEA Countries, Weeks 38/2020 to 10/2021. Eurosurveillance 2021, 26, 2100348. [Google Scholar] [CrossRef] [PubMed]
- Nyberg, T.; Twohig, K.A.; Harris, R.J.; Seaman, S.R.; Flannagan, J.; Allen, H.; Charlett, A.; De Angelis, D.; Dabrera, G.; Presanis, A.M. Risk of Hospital Admission for Patients with SARS-CoV-2 Variant B.1.1.7: Cohort Analysis. BMJ 2021, 373, n1412. [Google Scholar] [CrossRef]
- Mahase, E. COVID-19: Novavax Vaccine Efficacy Is 86% against UK Variant and 60% against South African Variant. BMJ 2021, 372, 296. [Google Scholar] [CrossRef]
- Naveca, F.G.; Nascimento, V.; de Souza, V.C.; Corado, A.; de Nascimento, L.F.; Silva, G.; Costa, Á.; Duarte, D.; Pessoa, K.; Mejía, M.; et al. COVID-19 in Amazonas, Brazil, Was Driven by the Persistence of Endemic Lineages and P.1 Emergence. Nat. Med. 2021, 27, 1230–1238. [Google Scholar] [CrossRef]
- P.1 Lineage Report. Alaa Abdel Latif, Julia L. Mullen, Manar Alkuzweny, Ginger Tsueng, Marco Cano, Emily Haag, Jerry Zhou, Mark Zeller, Emory Hufbauer, Nate Matteson, Chunlei Wu, Kristian G. Andersen, Andrew I. Su, Karthik Gangavarapu, Laura D. Hughes, and the Center for Viral Systems Biology. Outbreak.Info. Available online: https://outbreak.info/situation-reports?pango=P.1 (accessed on 31 December 2021).
- Sabino, E.C.; Buss, L.F.; Carvalho, M.P.S.; Prete, C.A., Jr.; Crispim, M.A.E.; Fraiji, N.A.; Pereira, R.H.M.; Parag, K.V.; da Silva Peixoto, P.; Kraemer, M.U.G.; et al. Resurgence of COVID-19 in Manaus, Brazil, despite High Seroprevalence. Lancet 2021, 397, 452–455. [Google Scholar] [CrossRef]
- Freitas, A.R.R.; Beckedorff, O.A.; de Cavalcanti, G.L.P.; Siqueira, A.M.; de Castro, D.B.; da Costa, C.F.; Lemos, D.R.Q.; Barros, E.N.C. The Emergence of Novel SARS-CoV-2 Variant P.1 in Amazonas (Brazil) Was Temporally Associated with a Change in the Age and Sex Profile of COVID-19 Mortality: A Population Based Ecological Study. Lancet Reg. Health Am. 2021, 1, 100021. [Google Scholar] [CrossRef]
- Taylor, L. COVID-19: Brazil’s Spiralling Crisis is Increasingly Affecting Young People. BMJ 2021, 373, n879. [Google Scholar] [CrossRef]
- The Catastrophic Brazilian Response to COVID-19 may Amount to a Crime against Humanity. Available online: https://blogs.bmj.com/bmj/2021/04/05/the-catastrophic-brazilian-response-to-COVID-19-may-amount-to-a-crime-against-humanity/ (accessed on 29 December 2021).
- Genome Sequencing by INSACOG Shows Variants of Concern and a Novel Variant in India. Available online: https://pib.gov.in/PressReleaseIframePage.aspx?PRID=1707177 (accessed on 29 December 2021).
- B.1.617.2 Lineage Report. Alaa Abdel Latif, Julia L. Mullen, Manar Alkuzweny, Ginger Tsueng, Marco Cano, Emily Haag, Jerry Zhou, Mark Zeller, Emory Hufbauer, Nate Matteson, Chunlei Wu, Kristian G. Andersen, Andrew I. Su, Karthik Gangavarapu, Laura D. Hughes, and the Center for Viral Systems Biology. Outbreak.Info. Available online: https://outbreak.info/situation-reports?pango=B.1.617.2 (accessed on 31 December 2021).
- Chen, J.; Wang, R.; Wang, M.; Wei, G.-W. Mutations Strengthened SARS-CoV-2 Infectivity. J. Mol. Biol. 2020, 432, 5212–5226. [Google Scholar] [CrossRef]
- Teng, S.; Sobitan, A.; Rhoades, R.; Liu, D.; Tang, Q. Systemic Effects of Missense Mutations on SARS-CoV-2 Spike Glycoprotein Stability and Receptor-Binding Affinity. Brief. Bioinform. 2021, 22, 1239–1253. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.; Vusirikala, A.; Flannagan, J.; Twohig, K.A.; Zaidi, A.; Chudasama, D.; Lamagni, T.; Groves, N.; Turner, C.; Rawlinson, C.; et al. Household Transmission of COVID-19 Cases Associated with SARS-CoV-2 Delta Variant (B.1.617.2): National Case-Control Study. Lancet Reg. Health Eur. 2022, 12, 100252. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, K.; Mannell, M.; Naqvi, O.; Matson, D.; Stone, J. SARS-CoV-2 B.1.617.2 (Delta) Variant COVID-19 Outbreak Associated with a Gymnastics Facili–y—Oklahoma, April-May 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1004–1007. [Google Scholar] [CrossRef] [PubMed]
- Australian Government Department of Health. Available online: https://www.health.gov.au/health-alerts/covid-19/symptoms-and-variants/delta (accessed on 29 December 2021).
- Ong, S.W.X.; Chiew, C.J.; Ang, L.W.; Mak, T.-M.; Cui, L.; Toh, M.P.H.S.; Lim, Y.D.; Lee, P.H.; Lee, T.H.; Chia, P.Y.; et al. Clinical and Virological Features of SARS-CoV-2 Variants of Concern: A Retrospective Cohort Study Comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta). Clin. Infect. Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Fisman, D.N.; Tuite, A.R. Evaluation of the Relative Virulence of Novel SARS-CoV-2 Variants: A Retrospective Cohort Study in Ontario, Canada. CMAJ 2021, 193, E1619–E1625. [Google Scholar] [CrossRef]
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital Admission and Emergency Care Attendance Risk for SARS-CoV-2 Delta (B.1.617.2) Compared with Alpha (B.1.1.7) Variants of Concern: A Cohort Study. Lancet Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef]
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of COVID-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef]
- Department of Health, Government of South Africa. COVID-19. Available online: https://sacoronavirus.co.za/ (accessed on 2 December 2021).
- BA.1 Lineage Report. Alaa Abdel Latif, Julia L. Mullen, Manar Alkuzweny, Ginger Tsueng, Marco Cano, Emily Haag, Jerry Zhou, Mark Zeller, Emory Hufbauer, Nate Matteson, Chunlei Wu, Kristian G. Andersen, Andrew I. Su, Karthik Gangavarapu, Laura D. Hughes, and the Center for Viral Systems Biology. Outbreak.Info. Available online: https://outbreak.info/situation-reports?pango=BA.1 (accessed on 31 December 2021).
- Torjesen, I. COVID-19: Omicron May Be More Transmissible than Other Variants and Partly Resistant to Existing Vaccines, Scientists Fear. BMJ 2021, 375, n2943. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradník, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.E.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. Omicron-B.1.1.529 Leads to Widespread Escape from Neutralizing Antibody Responses. bioRxiv 2021, 471045. [Google Scholar] [CrossRef]
- Meng, B.; Ferreira, I.A.T.M.; Abdullahi, A.; Saito, A.; Kimura, I.; Yamasoba, D.; Kemp, S.A.; Goonawardane, N.; Papa, G.; Fatihi, S.; et al. SARS-CoV-2 Omicron Spike Mediated Immune Escape, Infectivity and Cell-Cell Fusion. bioRxiv 2021, 473248. [Google Scholar] [CrossRef]
- Callaway, E.; Ledford, H. How Bad Is Omicron? What Scientists Know so Far. Nature 2021, 600, 197–199. [Google Scholar] [CrossRef]
- CDC. Omicron Variant: What You Need to Know. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/omicron-variant.html (accessed on 28 December 2021).
- Burki, T.K. Omicron Variant and Booster COVID-19 Vaccines. Lancet Respir. Med. 2021. [Google Scholar] [CrossRef]
- Pfizer and BioNTech Provide Update on Omicron Variant. Available online: https://www.pfizer.com/news/press-release/press-release-detafizerzer-and-biontech-provide-update-omicron-variant (accessed on 30 December 2021).
- Vaxzevria Significantly Boosted Antibody Levels against Omicron. Available online: https://www.astrazeneca.com/content/astraz/media-centre/press-releases/2021/vaxzevria-significantly-boosted-antibody-levels-against-omicron.html (accessed on 30 December 2021).
- Pulliam, J.R.C.; van Schalkwyk, C.; Govender, N.; von Gottberg, A.; Cohen, C.; Groome, M.J.; Dushoff, J.; Mlisana, K.; Moultrie, H. Increased Risk of SARS-CoV-2 Reinfection Associated with Emergence of the Omicron Variant in South Africa. bioRxiv 2021. [Google Scholar] [CrossRef]
- SARS-CoV-2 and Omicron: The Need to Optimise Genome Surveillance and Tracing. BMJ 2021, 375, 3133. [CrossRef]
- BBC News. COVID: US Reports Record Infections as Europe’s Omicron Cases Also Soar. BBC News, 29 December 2021. [Google Scholar]
- Helmore, E. US Sets New Record for Daily COVID Cases as Omicron Spreads across Country. The Guardian, 30 December 2021. [Google Scholar]
- Pan American Health Organization. COVID-19 Genomic Surveillance Regional Network. Available online: https://www.paho.org/en/topics/influenza-and-other-respiratory-viruses/covid-19-genomic-surveillance-regional-network (accessed on 23 January 2021).
- Duffy, S. Why Are RNA Virus Mutation Rates so Damn High? PLoS Biol. 2018, 16, e3000003. [Google Scholar] [CrossRef] [Green Version]
- Mandary, M.B.; Masomian, M.; Poh, C.L. Impact of RNA Virus Evolution on Quasispecies Formation and Virulence. Int. J. Mol. Sci. 2019, 20, 4657. [Google Scholar] [CrossRef] [Green Version]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef]
- Javanian, M.; Barary, M.; Ghebrehewet, S.; Koppolu, V.; Vasigala, V.; Ebrahimpour, S. A Brief Review of Influenza Virus Infection. J. Med. Virol. 2021, 93, 4638–4646. [Google Scholar] [CrossRef]
- Nypaver, C.; Dehlinger, C.; Carter, C. Influenza and Influenza Vaccine: A Review. J. Midwifery Women’s Health 2021, 66, 45–53. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. GISAID: Global Initiative on Sharing All Influenza Data—From Vision to Reality. Eurosurveillance 2017, 22, 13. [Google Scholar] [CrossRef] [Green Version]
- Justo Arevalo, S.; Zapata Sifuentes, D.; Huallpa, C.J.; Landa Bianchi, G.; Castillo Chávez, A.; Garavito-Salini Casas, R.; Uceda-Campos, G.; Pineda Chavarria, R. Global Geographic and Temporal Analysis of SARS-CoV-2 Haplotypes Normalized by COVID-19 Cases during the Pandemic. Front. Microbiol. 2021, 12, 612432. [Google Scholar] [CrossRef] [PubMed]
- Koshy, J. The Story of how a Mystifying Novel Coronavirus Variant, Delta, has India and the Globe in Its Grip. Available online: https://www.thehindu.com/sci-tech/health/the-story-of-how-a-mystifying-novel-coronavirus-variant-delta-has-india-and-the-globe-in-its-grip/article36772942.ece (accessed on 30 December 2021).
- Yang, W.; Shaman, J. SARS-CoV-2 Transmission Dynamics in South Africa and Epidemiological Characteristics of the Omicron Variant. bioRxiv 2021, 21268073. [Google Scholar] [CrossRef]
- Castillo, A.E.; Parra, B.; Tapia, P.; Lagos, J.; Arata, L.; Acevedo, A.; Andrade, W.; Leal, G.; Tambley, C.; Bustos, P.; et al. Geographical Distribution of Genetic Variants and Lineages of SARS-CoV-2 in Chile. Front. Public Health 2020, 8, 562615. [Google Scholar] [CrossRef] [PubMed]
- Lechien, J.R.; Saussez, S. Importance of Epidemiological Factors in the Evaluation of Transmissibility and Clinical Severity of SARS-CoV-2 Variants. Lancet Infect. Dis. 2022, 22, 2–3. [Google Scholar] [CrossRef]
- Salleh, M.Z.; Derrick, J.P.; Deris, Z.Z. Structural Evaluation of the Spike Glycoprotein Variants on SARS-CoV-2 Transmission and Immune Evasion. Int. J. Mol. Sci. 2021, 22, 7425. [Google Scholar] [CrossRef]
- Muth, D.; Corman, V.M.; Roth, H.; Binger, T.; Dijkman, R.; Gottula, L.T.; Gloza-Rausch, F.; Balboni, A.; Battilani, M.; Rihtarič, D.; et al. Attenuation of Replication by a 29 Nucleotide Deletion in SARS-Coronavirus Acquired during the Early Stages of Human-to-Human Transmission. Sci. Rep. 2018, 8, 15177. [Google Scholar] [CrossRef]
- Callaway, E. The Coronavirus Is Mutati–g—Does It Matter? Nature 2020, 585, 174–177. [Google Scholar] [CrossRef]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.T.M.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta Variant Replication and Immune Evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced Sensitivity of SARS-CoV-2 Variant Delta to Antibody Neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef]
- Edara, V.-V.; Pinsky, B.A.; Suthar, M.S.; Lai, L.; Davis-Gardner, M.E.; Floyd, K.; Flowers, M.W.; Wrammert, J.; Hussaini, L.; Ciric, C.R.; et al. Infection and Vaccine-Induced Neutralizing-Antibody Responses to the SARS-CoV-2 B.1.617 Variants. N. Engl. J. Med. 2021, 385, 664–666. [Google Scholar] [CrossRef]
- Gorczynski, R.M.; Lindley, R.A.; Steele, E.J.; Wickramasinghe, N.C. Nature of Acquired Immune Responses, Epitope Specificity and Resultant Protection from SARS-CoV-2. J. Pers. Med. 2021, 11, 1253. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.-C.; Lu, J.; Xu, Q.-F.; Guo, Q.; Xu, D.-Z.; Sun, Q.-W.; Yang, H.; Zhao, G.-M.; Jiang, Q.-W. Influence of Meteorological Factors and Air Pollution on the Outbreak of Severe Acute Respiratory Syndrome. Public Health 2007, 121, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Audi, A.; AlIbrahim, M.; Kaddoura, M.; Hijazi, G.; Yassine, H.M.; Zaraket, H. Seasonality of Respiratory Viral Infections: Will COVID-19 Follow Suit? Front. Public Health 2020, 8, 567184. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhou, J.; Yao, J.; Zhang, X.; Li, L.; Xu, X.; He, X.; Wang, B.; Fu, S.; Niu, T.; et al. Impact of Meteorological Factors on the COVID-19 Transmission: A Multi-City Study in China. Sci. Total Environ. 2020, 726, 138513. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, H.; Khandait, H.; Narlawar, U.W.; Rathod, P.; Mamtani, M. Independent Association of Meteorological Characteristics with Initial Spread of COVID-19 in India. Sci. Total Environ. 2021, 764, 142801. [Google Scholar] [CrossRef] [PubMed]
- Brandt, E.B.; Mersha, T.B. Environmental Determinants of Coronavirus Disease 2019 (COVID-19). Curr. Allergy Asthma Rep. 2021, 21, 15. [Google Scholar] [CrossRef]
- Pan American Health Organization. Determinantes Sociales de la Salud. Available online: https://www.paho.org/es/temas/determinantes-sociales-salud (accessed on 30 December 2021).
- World Health Organization. Social determinants of Health. Available online: https://www.who.int/health-topics/social-determinants-of-health (accessed on 30 December 2021).
- Braveman, P.; Gottlieb, L. The Social Determinants of Health: It’s Time to Consider the Causes of the Causes. Public Health Rep. 2014, 129 (Suppl. 2), 19–31. [Google Scholar] [CrossRef] [Green Version]
- Petersen, E.; Ntoumi, F.; Hui, D.S.; Abubakar, A.; Kramer, L.D.; Obiero, C.; Tambyah, P.A.; Blumberg, L.; Yapi, R.; Al-Abri, S.; et al. Emergence of New SARS-CoV-2 Variant of Concern Omicron (B.1.1.52–)—Highlights Africa’s Research Capabilities, but Exposes Major Knowledge Gaps, Inequities of Vaccine Distribution, Inadequacies in Global COVID-19 Response and Control Efforts. Int. J. Infect. Dis. 2021, 114, 268–272. [Google Scholar] [CrossRef]
- Kim, S.J.; Bostwick, W. Social Vulnerability and Racial Inequality in COVID-19 Deaths in Chicago. Health Educ. Behav. 2020, 47, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Rae, M. Omicron: A Failure to Act with a Global Focus Will Continue the Proliferation of New Variants of COVID-19. BMJ 2021, 375, 3095. [Google Scholar] [CrossRef]
- Wise, J. COVID-19: G7 Vaccine Promises Fail to Meet Scale of Challenge, Say Critics. BMJ 2021, 373, 1520. [Google Scholar] [CrossRef]
- Omicron Is Bad but the Global Response Is Worse. Nature 2021, 600, 190. [CrossRef] [PubMed]
- Christie, B. COVID-19: Early Studies Give Hope Omicron Is Milder than Other Variants. BMJ 2021, 375, 3144. [Google Scholar] [CrossRef] [PubMed]
- Howard, J.; Huang, A.; Li, Z.; Tufekci, Z.; Zdimal, V.; van der Westhuizen, H.-M.; von Delft, A.; Price, A.; Fridman, L.; Tang, L.-H.; et al. An Evidence Review of Face Masks against COVID-19. Proc. Natl. Acad. Sci. USA 2021, 118, e2014564118. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.G.L.; LePrevost, C.E.; Harwell, E.L.; Bloss, J.E.; Cofie, L.E.; Wiggins, M.F.; Firnhaber, G.C. Coronavirus Pandemic Highlights Critical Gaps in Rural Internet Access for Migrant and Seasonal Farmworkers: A Call for Partnership with Medical Libraries. J. Med. Libr. Assoc. 2020, 108, 651–655. [Google Scholar] [CrossRef]
- Wang, Z.; Tang, K. Combating COVID-19: Health Equity Matters. Nat. Med. 2020, 26, 458. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.S. Social Capital in the Creation of Human Capital. Am. J. Sociol. 1988, 94, S95–S120. [Google Scholar] [CrossRef]
- Pitas, N.; Ehmer, C. Social Capital in the Response to COVID-19. Am. J. Health Promot. 2020, 34, 942–944. [Google Scholar] [CrossRef]
- Wong, A.S.Y.; Kohler, J.C. Social Capital and Public Health: Responding to the COVID-19 Pandemic. Global. Health 2020, 16, 88. [Google Scholar] [CrossRef]
- Patel, J.A.; Nielsen, F.B.H.; Badiani, A.A.; Assi, S.; Unadkat, V.A.; Patel, B.; Ravindrane, R.; Wardle, H. Poverty, Inequality and COVID-19: The Forgotten Vulnerable. Public Health 2020, 183, 110–111. [Google Scholar] [CrossRef]
- Juliano, C.; Castrucci, B.; Fraser, M.R. COVID-19 and Public Health: Looking Back, Moving Forward. J. Public Health Manag. Pract. 2021, 27 (Suppl. 1), S1–S4. [Google Scholar] [CrossRef] [PubMed]
- Pan American Health Organization. Recommendations for Reporting and Notification of SARS-CoV-2 Variants of Concern and Variants of Interest. Available online: https://www.paho.org/en/documents/recommendations-reporting-and-notification-sars-cov-2-variants-concern-and-variants (accessed on 31 December 2021).
- Li, J.; Lai, S.; Gao, G.F.; Shi, W. The Emergence, Genomic Diversity and Global Spread of SARS-CoV-2. Nature 2021, 600, 408–418. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation—COVID-19 Vaccine Tracker. Available online: https://covid19.trackvaccines.org/agency/who/ (accessed on 31 December 2021).
- World Population Clock: 7.9 Billion People (202–)—Worldometer. Available online: https://www.worldometers.info/world-population/ (accessed on 31 December 2021).
- John Hopkins Coronavirus Resource Center. Understanding Vaccination Progress by Country. Available online: https://coronavirus.jhu.edu/vaccines/international (accessed on 31 December 2021).
- COVID-19 Vaccine Market Dashboard. Available online: https://www.unicef.org/supply/covid-19-vaccine-market-dashboard (accessed on 31 December 2021).
- Leffler, C.T.; Ing, E.; Lykins, J.D.; Hogan, M.C.; McKeown, C.A.; Grzybowski, A. Association of Country-Wide Coronavirus Mortality with Demographics, Testing, Lockdowns, and Public Wearing of Masks. Am. J. Trop. Med. Hyg. 2020, 103, 2400–2411. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S.; Lane, H.C.; Redfield, R.R. COVID-9—Navigating the Uncharted. N. Engl. J. Med. 2020, 382, 1268–1269. [Google Scholar] [CrossRef]
- Kaimann, D.; Tanneberg, I. What Containment Strategy Leads Us through the Pandemic Crisis? An Empirical Analysis of the Measures against the COVID-19 Pandemic. PLoS ONE 2021, 16, e0253237. [Google Scholar] [CrossRef]
- CDC. Ending Isolation and Precautions for People with COVID-19: Interim Guidance. Available online: https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html (accessed on 31 December 2021).
- Tu, H.; Hu, K.; Zhang, M.; Zhuang, Y.; Song, T. Effectiveness of 14 Day Quarantine Strategy: Chinese Experience of Prevention and Control. BMJ 2021, 375, e066121. [Google Scholar] [CrossRef]
- Nussbaumer-Streit, B.; Mayr, V.; Dobrescu, A.I.; Chapman, A.; Persad, E.; Klerings, I.; Wagner, G.; Siebert, U.; Ledinger, D.; Zachariah, C.; et al. Quarantine alone or in combination with other public health measures to control COVID-19: A rapid review. Cochrane Database Syst. Rev. 2020, CD013574. [Google Scholar] [CrossRef]
- World Health Organization. What Is the ACT Accelerator. Available online: https://www.who.int/initiatives/act-accelerator/about (accessed on 31 December 2021).
- UNICEF. Mission Possible: Getting Affordable COVID-19 Tests to Those Who Need It Most. Available online: https://www.unicef.org/supply/stories/mission-possible-getting-affordable-covid-19-tests-those-who-need-it-most (accessed on 31 December 2021).
| Variant of Interest (VOI) | Variant of Concern (VOC) | Variant Under Monitoring (VUM) | Former Monitored Variant | |
|---|---|---|---|---|
| World Health Organization working definition | 1. SARS-CoV-2 variant with genetic changes that are predicted or known to affect virus characteristics such as transmisibility, disease severity, immune escape, diagnostic and therapeutic escape. 2. Identified to cause significant community transmission or multiple COVID-19 clusters, in multiple countries with increasing relative prevalence alongside increasing number of cases over time, or other apppernt epidemiological impacts to suggest and emerging risk to global public health. | 1. SARS-CoV-2 variant that meets VOI criteria. 2. Associated with one or more of the following characteristics of global public health relevance: (a) Increase in transmisibility or detrimental change in COVID-19 epidemiology. (b) Increase in virulence or change in clinical disease presentation. (c) Decrease in effectiveness of public health and social measures or available diagnostics, vaccines, therapeutics. | SARS-CoV-2 variant with genetic changes suspected to affect virus characteristics with a potential future risk, but unclear evidence of phenotypic or epidemiological impact. | Previous VOCs/VOIs/VUMs that have been reclassified on at least one of the following criteria: (a) The variant is no longer circulating at levels of global public health significance. (b) The variant has been circulating for a long time without any significant epidemiological impact. (c) Scientific evidence demonstrate that the variant is not associated with concerning properties. |
| Designed SARS-CoV-2 variants as of 31 December 2021 (Pango lineage) | C.37 (Lambda variant) B.1.621 (Mu variant) | B.1.1.7 (Alpha variant) B.1.351 (Beta variant) P.1 (Gamma variant) B.1.617.2 (Delta variant) B.1.1.529/BA.1 (Omicron variant) | AZ.5 C.1.2 B.1.617.1 * B.1.525 * B.1.526 * B.1.630 B.1.640 | AV.1 AT.1 P.2 * P.3 * R.1 B.1.466.2 B.1.1.519 C.36.3 B.1.214.2 B.1.427/B.1.429 * B.1.1.523 B.1.619 B.1.620 |
| WHO Label | Pango Lineage | GISAID Clade | Nextstrain Clade | Origin of First Samples Detected | Date of First Documented Case | VOC Designation Date | Transmissibility | Clinical Severity | Clinical Response to Anti-SARS-CoV-2 Vaccines |
|---|---|---|---|---|---|---|---|---|---|
| Alpha | B.1.1.7 | GRY | 20I (V1) | United Kingdom | sept-20 | 18-12-2020 | Higher than previous variants (virological and epidemiological evidence) | Possibly similar incidence of severe COVID-19 and death (epidemiological studies with mixed results) | Vaccines prevent infection and adverse events |
| Beta | B.1.351 | GH/501Y.V2 | 20H (V2) | South Africa | sept-20 | 18-12-2020 | Higher than previous variants (virological and epidemiological evidence) | Possibly similar incidence of severe COVID-19 and death (epidemiological studies with mixed results) | Vaccines prevent infection and adverse events (possibly less effective in preventing infection) |
| Gamma | P.1 | GR/501Y.V3 | 20J (V3) | Brazil | nov-20 | 11-01-2021 | Higher than previous variants (virological and epidemiological evidence) | Possibly similar incidence of severe COVID-19 and death (epidemiological studies with mixed results) | Vaccines prevent infection and adverse events (possibly less effective in preventing infection) |
| Delta | B.1.617.2 | G/478K.V1 | 21A, 21I, 21J | India | oct-20 | 11-05-2021 * | Much higher than previous variants (virological and epidemiological evidence) | Increased incidence of severe COVID-19 and death (virological and epidemiological evidence) | Vaccines prevent infection and adverse events (slightly lower efficacy in preventing infection) |
| Omicron | B.1.1.529 BA.1 # | GRA | 21K, 21L, 21M | South Africa | nov-21 | 26-11-2021 ** | Extremely higher than previous variants (virological and epidemiological evidence)—higher than lineage B.1.617.2 | Preliminary data suggests that it has a lower incidence of severe COVID-19 and death—lower than lineage B.1.617.2 | Preliminary data suggest that vaccines prevent infection and adverse events (possible lower efficacy versus other VOCs) |
| Variant of Interest (VOI) | Variant of Concern (VOC) | |
|---|---|---|
| Primary actions by World Health Organization (WHO) for a potential VOI/VOC | Comparative assessment of variant characteristics and public health risks by WHO. | Comparative assessment of variant characteristics and public health risks by WHO and the Technical advisory Group on Virus Evolution. |
| If determined necessary, coordinated laboratory investigations with Member States and partners. | If determined necessary, coordinate additional laboratory investigations with Member States and partners. | |
| Review global epidemiology of VOI. | Communicate new designations and findings with Member States and public through established mechanisms. | |
| Monitor and track global spread of VOI. | Evaluate WHO guidance through established WHO mechanisms and update, if necessary. | |
| Primary actions by a Member State if a new potential VOI/VOC is identified | Inform WHO through established WHO Country or Regional Office reporting channels with supporting information about VOI-associated cases (person, place, time, clinical and other relevant characteristics). | Submit complete genome sequences and associated metadata to a publicly available database, such as GISAID. |
| Submit complete genome sequences and associated metadata to a publicly available database, such as GISAID. | Report initial cases/clusters associated with VOC infection to WHO through the International Health Regulations (IHR) mechanism. | |
| Perform field investigations to improve understanding of the potential impacts of the VOI on COVID-19 epidemiology, severity, effectiveness of public health and social measures, or other relevant characteristics. | Where capacity exists and in coordination with the international community, perform field investigations and laboratory assessments to improve understanding of the potential impacts of the VOC on COVID-19 epidemiology, severity, effectiveness of public health and social measures, diagnostic methods, immune responses, antibody neutralization, or other relevant characteristics. | |
| Perform laboratory assessments according to capacity or contact WHO for support to conduct laboratory assessments on the impact of the VOI on relevant topics. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parra-Lucares, A.; Segura, P.; Rojas, V.; Pumarino, C.; Saint-Pierre, G.; Toro, L. Emergence of SARS-CoV-2 Variants in the World: How Could This Happen? Life 2022, 12, 194. https://doi.org/10.3390/life12020194
Parra-Lucares A, Segura P, Rojas V, Pumarino C, Saint-Pierre G, Toro L. Emergence of SARS-CoV-2 Variants in the World: How Could This Happen? Life. 2022; 12(2):194. https://doi.org/10.3390/life12020194
Chicago/Turabian StyleParra-Lucares, Alfredo, Paula Segura, Verónica Rojas, Catalina Pumarino, Gustavo Saint-Pierre, and Luis Toro. 2022. "Emergence of SARS-CoV-2 Variants in the World: How Could This Happen?" Life 12, no. 2: 194. https://doi.org/10.3390/life12020194
APA StyleParra-Lucares, A., Segura, P., Rojas, V., Pumarino, C., Saint-Pierre, G., & Toro, L. (2022). Emergence of SARS-CoV-2 Variants in the World: How Could This Happen? Life, 12(2), 194. https://doi.org/10.3390/life12020194






