Relaxed Substrate Specificity in Qβ Replicase through Long-Term In Vitro Evolution
Abstract
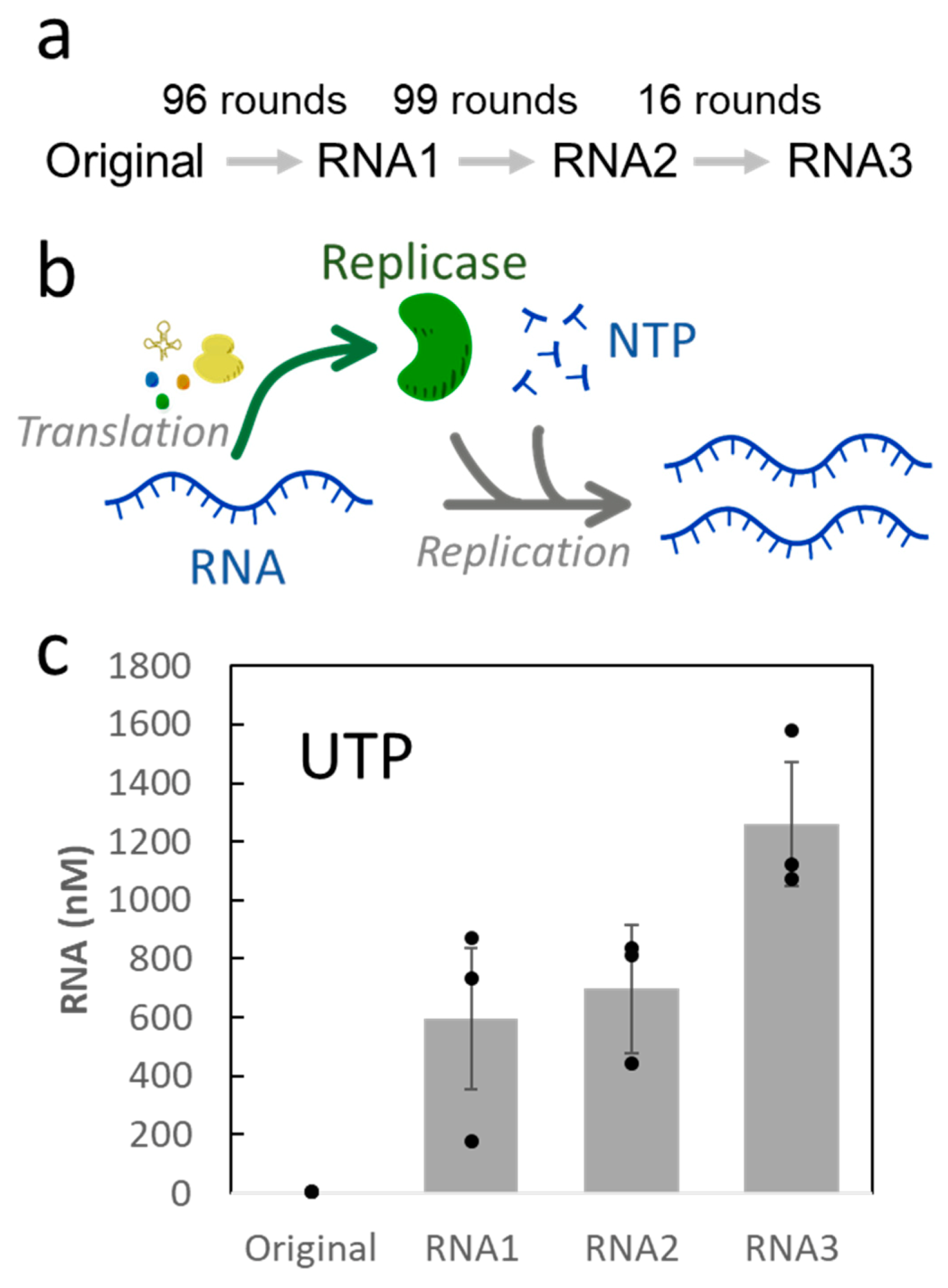
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. RNA Preparation
4.2. RNA Replication and Measurement
4.3. Labeled RNA Recovery
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lazcano, A.; Guerrero, R.; Margulis, L.; Oró, J. The evolutionary transition from RNA to DNA in early cells. J. Mol. Evol. 1988, 27, 283–290. [Google Scholar] [CrossRef]
- Poole, A.M.; Jeffares, D.C.; Penny, D. The path from the RNA world. J. Mol. Evol. 1998, 46, 1–17. [Google Scholar] [CrossRef]
- Forterre, P. The two ages of the RNA world, and the transition to the DNA world: A story of viruses and cells. Biochimie 2005, 87, 793–803. [Google Scholar] [CrossRef]
- Lazcano, A.; Fastag, J.; Gariglio, P.; Ramírez, C.; Oró, J. On the early evolution of RNA polymerase. J. Mol. Evol. 1988, 27, 365–376. [Google Scholar] [CrossRef]
- Cojocaru, R.; Unrau, P.J. Transitioning to DNA genomes in an RNA world. Elife 2017, 6, e32330. [Google Scholar] [CrossRef] [Green Version]
- Gavette, J.V.; Stoop, M.; Hud, N.V.; Krishnamurthy, R. RNA-DNA Chimeras in the Context of an RNA World Transition to an RNA/DNA World. Angew. Chem. 2016, 128, 13398–13403. [Google Scholar] [CrossRef]
- Leu, K.; Obermayer, B.; Rajamani, S.; Gerland, U.; Chen, I.A. The prebiotic evolutionary advantage of transferring genetic information from RNA to DNA. Nucleic Acids Res. 2011, 39, 8135–8147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, N.; Hogeweg, P.; Koonin, E.V. On the Origin of DNA Genomes: Evolution of the Division of Labor between Template and Catalyst in Model Replicator Systems. PLoS Comput. Biol. 2011, 7, e10020240. Available online: https://pubmed.ncbi.nlm.nih.gov/21455287/ (accessed on 27 November 2021). [CrossRef] [PubMed] [Green Version]
- Takahashi, S.; Okura, H.; Sugimoto, N. Bisubstrate Function of RNA Polymerases Triggered by Molecular Crowding Conditions. Biochemistry 2019, 58, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Krupovic, M.; Ishino, S.; Ishino, Y. The replication machinery of LUCA: Common origin of DNA replication and transcription. BMC Biol. 2020, 18, 61. [Google Scholar] [CrossRef]
- Traut, T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994, 140, 1–22. [Google Scholar] [CrossRef]
- Jensen, R.A. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 1976, 30, 409–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, D.R.; Peterson, R.L.; Spiegelman, S. An Extracellular Darwinian Experiment with a Self-Duplicating Nucleic Acid Molecule. Proc. Natl. Acad. Sci. USA 1967, 58, 217–224. [Google Scholar] [CrossRef] [Green Version]
- Kita, H.; Matsuura, T.; Sunami, T.; Hosoda, K.; Ichihashi, N.; Tsukada, K.; Urabe, I.; Yomo, T. Replication of genetic information with self-encoded replicase in liposomes. ChemBioChem 2008, 9, 2403–2410. [Google Scholar] [CrossRef]
- Shimizu, Y.; Inoue, A.; Tomari, Y.; Suzuki, T.; Yokogawa, T.; Nishikawa, K.; Ueda, T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001, 19, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Blumenthal, T.; Carmichael, G.G. RNA Replication: Function and Structure of QBeta-Replicase. Annu. Rev. Biochem. 1979, 48, 525–548. Available online: http://www.ncbi.nlm.nih.gov/pubmed/382992 (accessed on 27 November 2021). [CrossRef]
- Ichihashi, N.; Usui, K.; Kazuta, Y.; Sunami, T.; Matsuura, T.; Yomo, T. Darwinian evolution in a translation-coupled RNA replication system within a cell-like compartment. Nat. Commun. 2013, 4, 2494. [Google Scholar] [CrossRef] [PubMed]
- Furubayashi, T.; Ueda, K.; Bansho, Y.; Motooka, D.; Nakamura, S.; Mizuuchi, R.; Ichihashi, N. Emergence and diversification of a host-parasite RNA ecosystem through Darwinian evolution. Elife 2020, 9, e56038. [Google Scholar] [CrossRef]
- Tiwari, V.K.; Mishra, B.B.; Mishra, K.B.; Mishra, N.; Singh, A.S.; Chen, X. Cu-Catalyzed Click Reaction in Carbohydrate Chemistry. Chem. Rev. 2016, 116, 3086–3240. [Google Scholar] [CrossRef]
- Takeshita, D.; Tomita, K. Assembly of Qβ viral RNA polymerase with host translational elongation factors EF-Tu and -Ts. Proc. Natl. Acad. Sci. USA 2010, 107, 15733–15738. [Google Scholar] [CrossRef] [Green Version]
- Kidmose, R.T.; Vasiliev, N.N.; Chetverin, A.B.; Andersen, G.R.; Knudsen, C.R. Structure of the Qβ replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 10884–10889. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.A.; Suo, Z. Unlocking the Sugar ‘Steric Gate’ of DNA Polymerases. Biochemistry 2011, 50, 1135–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, C.; Arnold, J.J.; Cameron, C.E. Incorporation fidelity of the viral RNA-dependent RNA polymerase: A kinetic, thermodynamic and structural perspective. Virus Res. 2005, 107, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Poole, A.M.; Horinouchi, N.; Catchpole, R.J.; Si, D.; Hibi, M.; Tanaka, K.; Ogawa, J. The Case for an Early Biological Origin of DNA. J. Mol. Evol. 2014, 79, 204–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yukawa, K.; Mizuuchi, R.; Ichihashi, N. Relaxed Substrate Specificity in Qβ Replicase through Long-Term In Vitro Evolution. Life 2022, 12, 32. https://doi.org/10.3390/life12010032
Yukawa K, Mizuuchi R, Ichihashi N. Relaxed Substrate Specificity in Qβ Replicase through Long-Term In Vitro Evolution. Life. 2022; 12(1):32. https://doi.org/10.3390/life12010032
Chicago/Turabian StyleYukawa, Kohtoh, Ryo Mizuuchi, and Norikazu Ichihashi. 2022. "Relaxed Substrate Specificity in Qβ Replicase through Long-Term In Vitro Evolution" Life 12, no. 1: 32. https://doi.org/10.3390/life12010032
APA StyleYukawa, K., Mizuuchi, R., & Ichihashi, N. (2022). Relaxed Substrate Specificity in Qβ Replicase through Long-Term In Vitro Evolution. Life, 12(1), 32. https://doi.org/10.3390/life12010032






