Structural and Functional Characterization of Camelus dromedarius Glutathione Transferase M1-1
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Expression and Purification
2.2.2. Assay of the Enzyme Activity, Protein and Kinetic Analysis
2.3. Inhibition Analysis
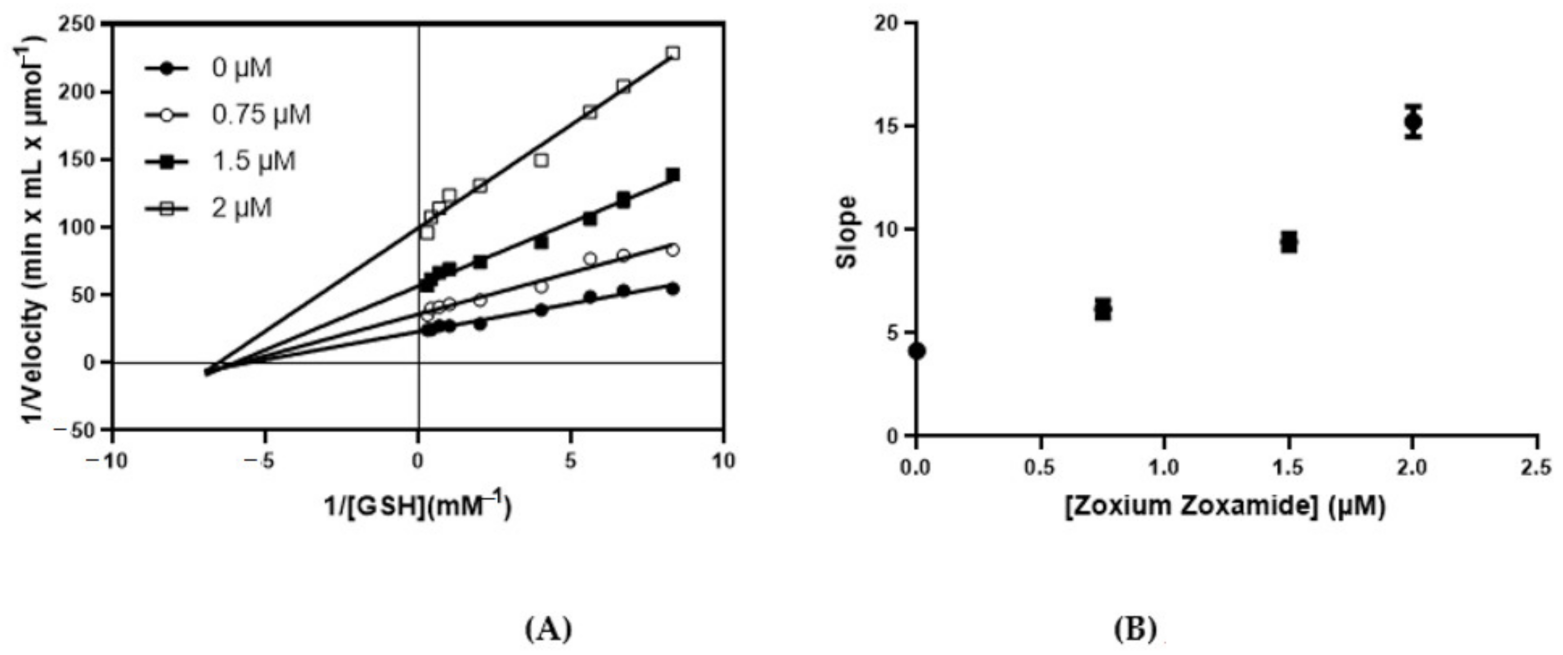
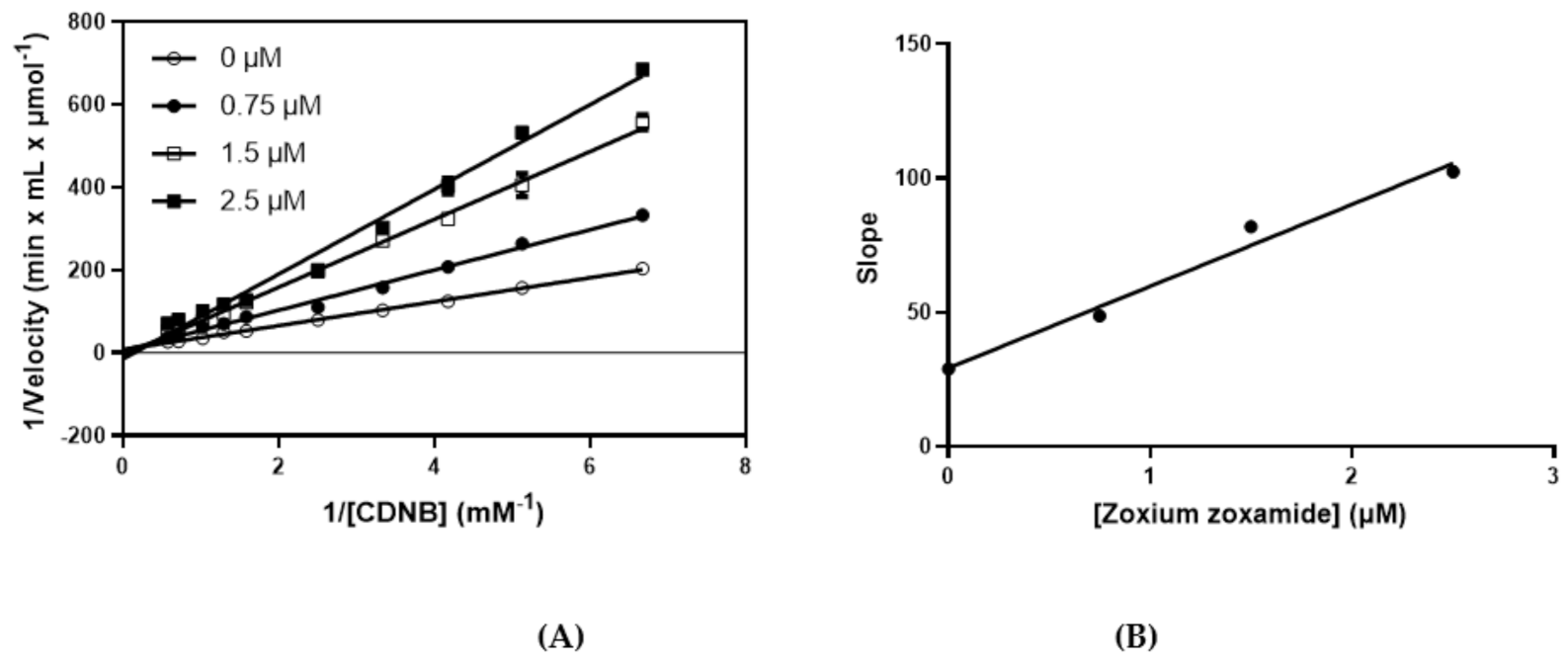
2.4. Kinetic Inhibition Studies
2.5. Thermal Stability
2.6. Crystallization
2.7. Structure Solution and Refinement
3. Results and Discussion
3.1. Purification and Kinetic Analysis
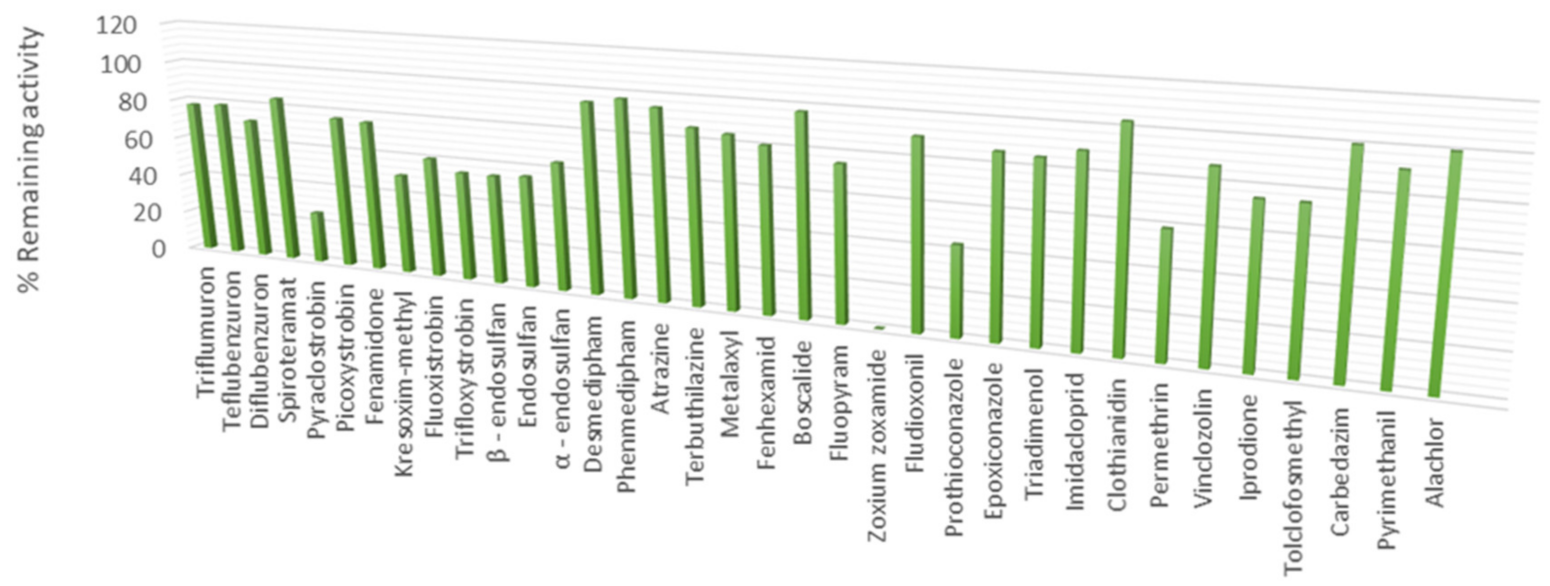
3.2. The Ability of CdGSTM1-1 to Bind Xenobiotic Compounds
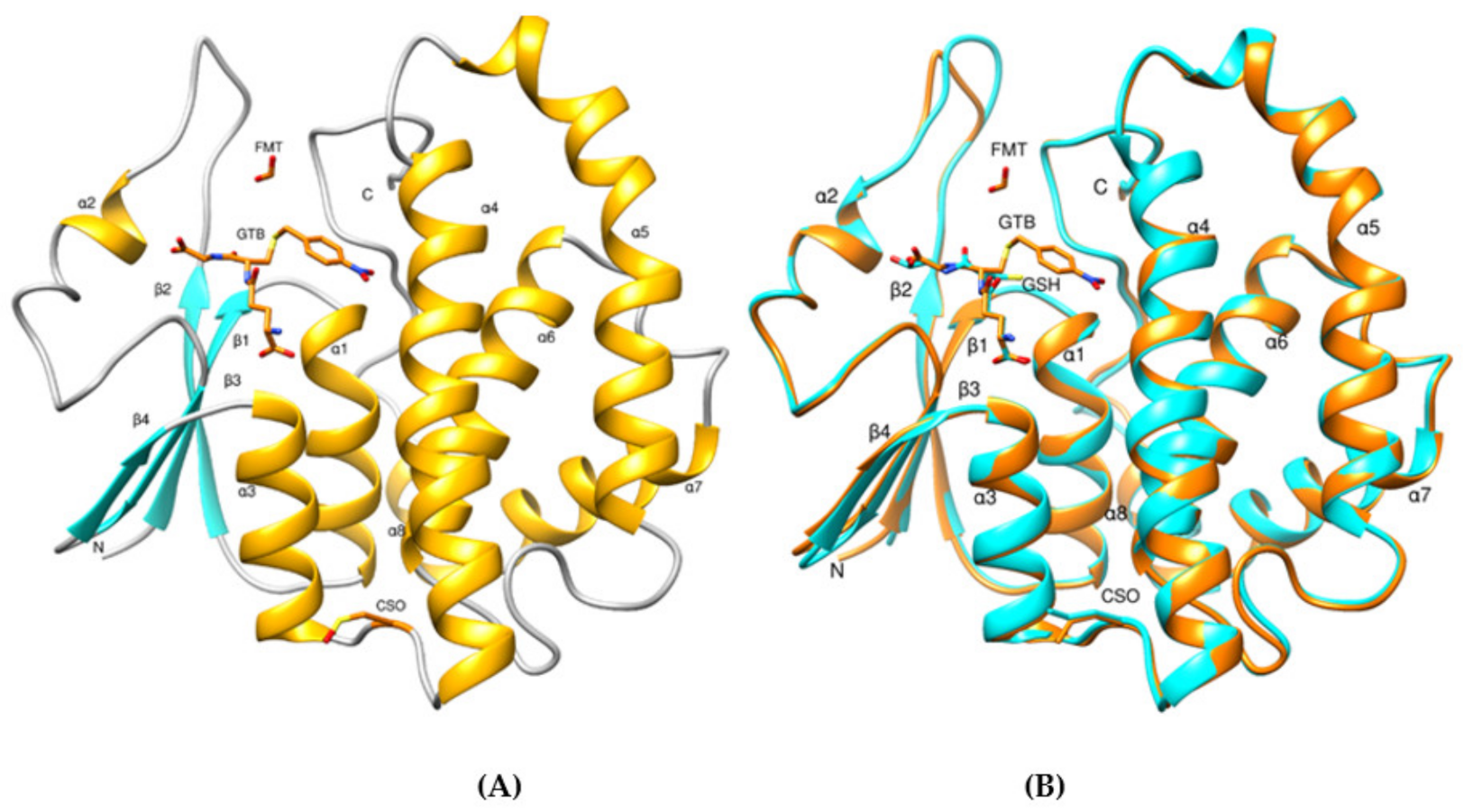
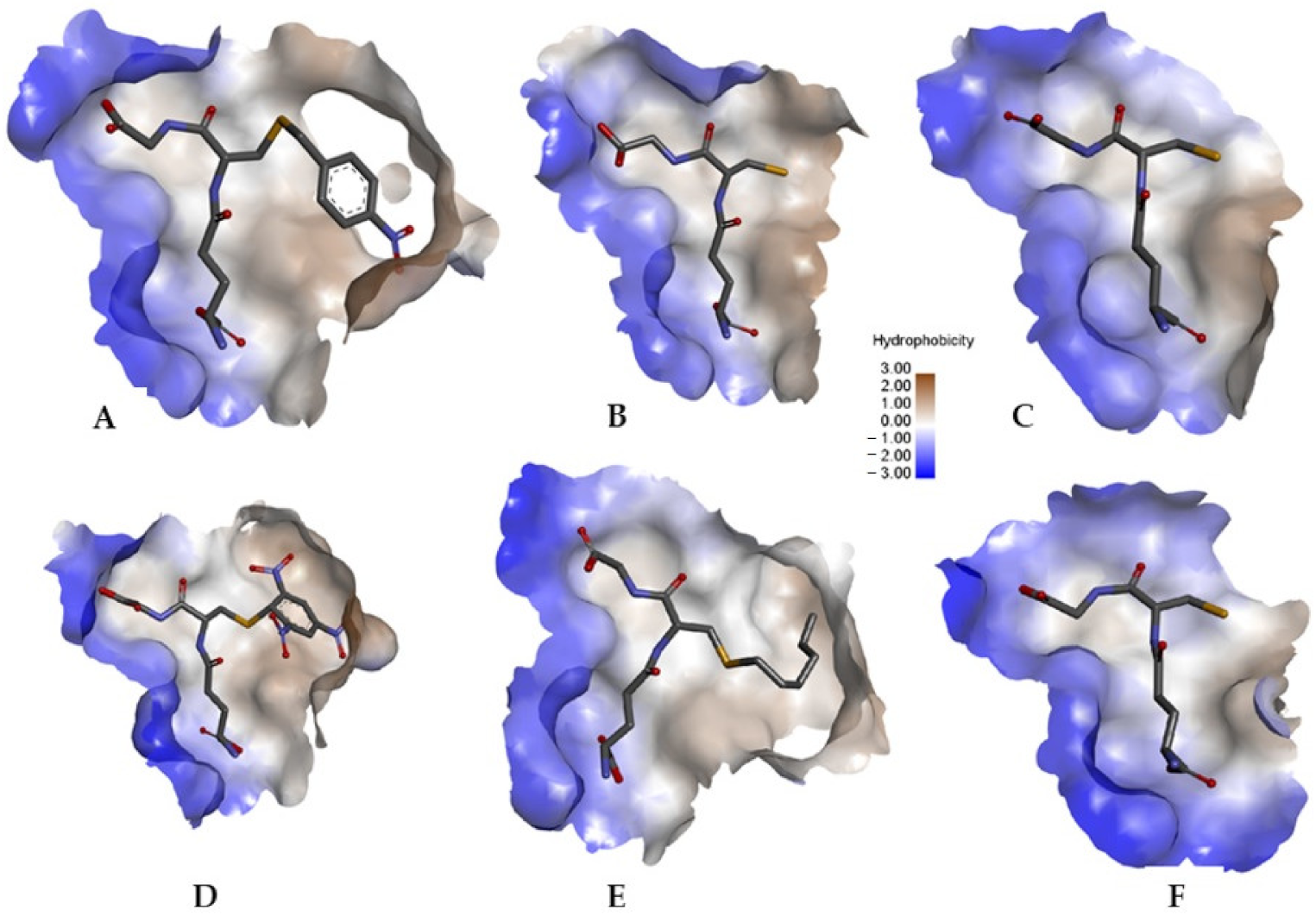
3.3. Structural Analysis of CdGSTM1-1 in Complex with GSH or S-hexyl-GSH
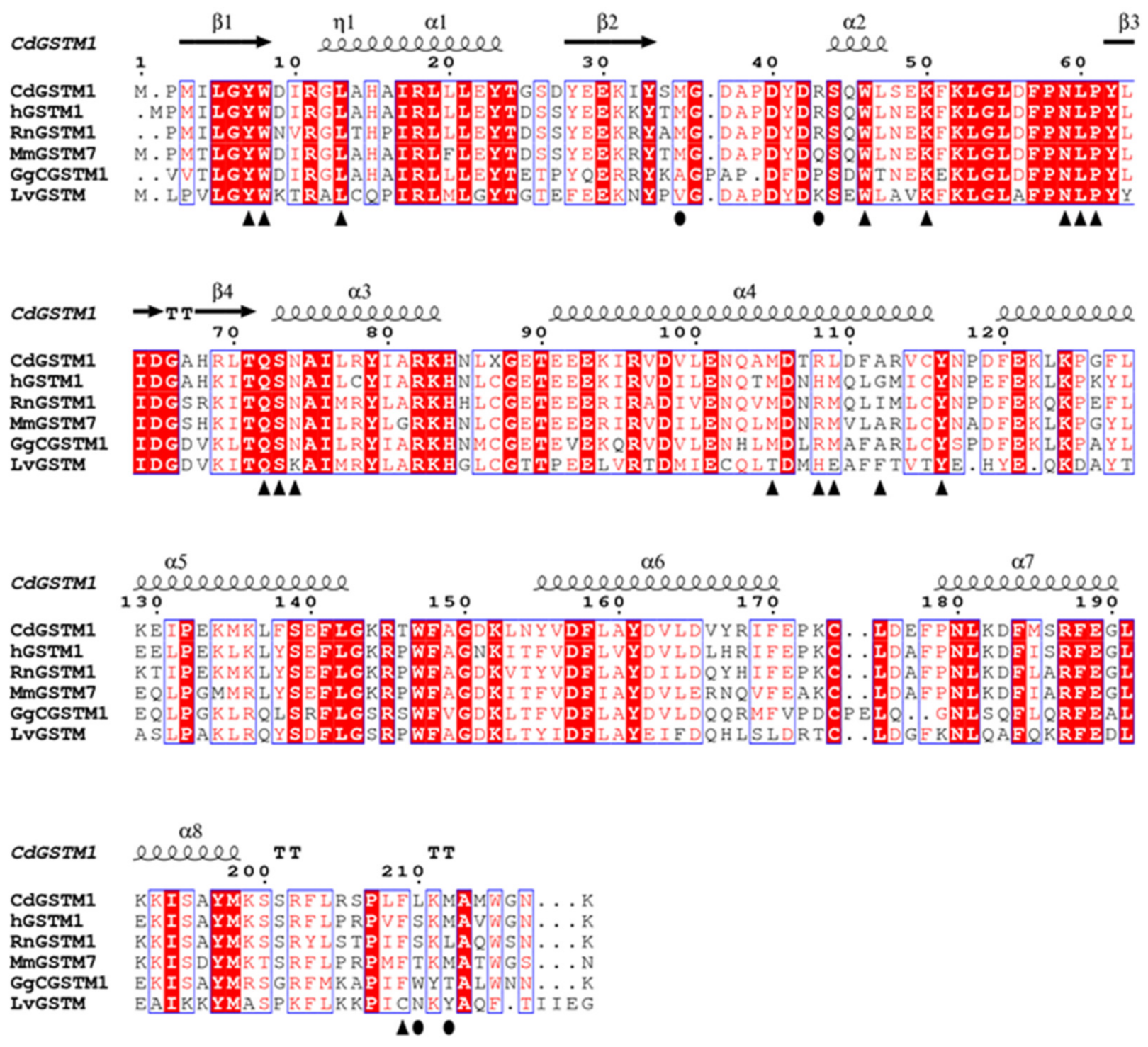
3.4. Structure Comparison with Human GSTM1-1 and Other GSTMs
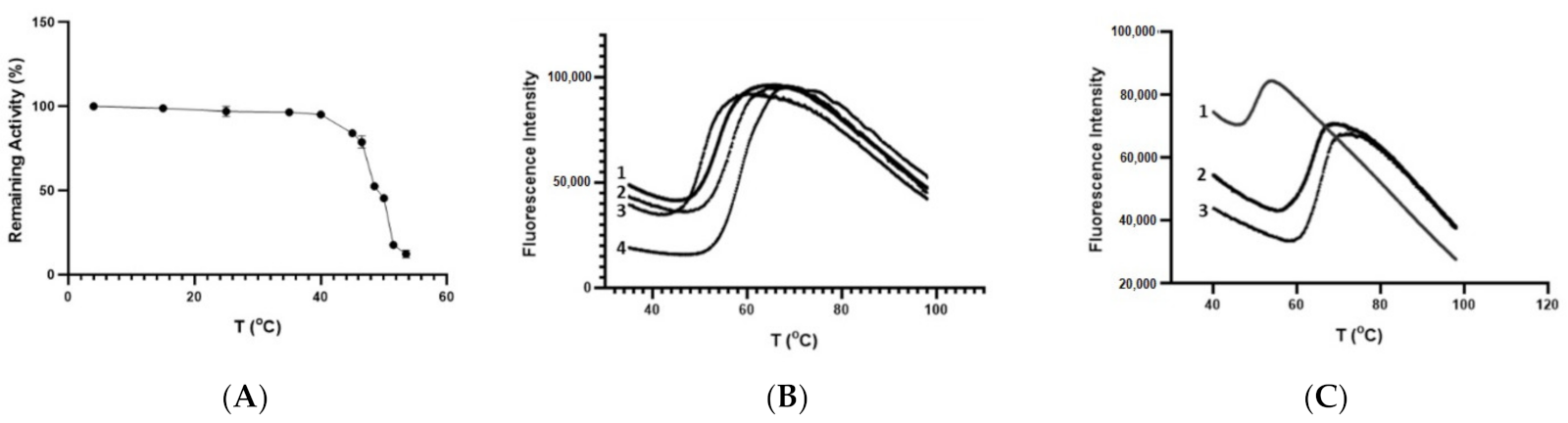
3.5. Thermal Stability of CdGSTM1-1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perperopoulou, F.; Ataya, F.S.; Fouad, D.; Malik, A.; Saeed, H.M.; Labrou, N.E. Biochemical characterization of the detoxifying enzyme glutathione transferase P1-1 from the camel Camelus dromedarius. Cell Biochem. Biophys. 2016, 74, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Sylvestre-Gonon, E.; Law, S.R.; Schwartz, M.; Robe, K.; Keech, O.; Didierjean, C.; Dubos, C.; Rouhier, N.; Hecker, A. Functional, Structural and Biochemical Features of Plant Serinyl-Glutathione Transferases. Front Plant Sci. 2019, 10, 608. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, S.; Antunes, S.C.; Correia, A.T.; Nunes, B. Oxytetracycline effects in specific biochemical pathways of detoxification, neurotransmission and energy production in Oncorhynchus mykiss. Ecotoxicol. Environ. Saf. 2018, 164, 100–108. [Google Scholar] [CrossRef]
- Mihaljević, I.; Bašica, B.; Maraković, N.; Kovačević, R.; Smital, T. Interaction of organotin compounds with three major glutathione S-transferases in zebrafish. Toxicology 2020, 62, 104713. [Google Scholar] [CrossRef] [PubMed]
- Labrou, N.E.; Papageorgiou, A.C.; Pavli, O.; Flemetakis, E. Plant GSTome: Structure and functional role in xenome network and plant stress response. Curr. Opin. Biotech. 2015, 32, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tian, J.; Hou, N.; Yu, N.; Zhang, Y.; Liu, Z. Identification, genomic organization and expression pattern of glutathione transferase in Pardosa pseudoannulata. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 32, 100626. [Google Scholar] [CrossRef]
- Park, J.C.; Kim, D.H.; Lee, M.C.; Han, J.; Kim, H.J.; Hagiwara, A.; Hwang, U.K.; Park, H.G.; Lee, J.S. Genome-wide identification of the entire 90 glutathione S-transferase (GST) subfamily genes in four rotifer Brachionus species and transcriptional modulation in response to endocrine disrupting chemicals. Comp. Biochem. Physiol. Part D Genom. Proteom. 2018, 28, 183–195. [Google Scholar] [CrossRef]
- Tan, H.M.; Low, W.Y. Rapid birth-death evolution and positive selection in detoxification-type glutathione S-transferases in mammals. PLoS ONE 2018, 13, e0209336. [Google Scholar] [CrossRef]
- Allocati, N.; Masulli, M.; Di Ilio, C.; Federici, L. Glutathione transferases: Substrates, inhibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis 2018, 7, 8. [Google Scholar] [CrossRef]
- Axarli, I.; Muleta, A.W.; Vlachakis, D.; Kossida, S.; Kotzia, G.; Maltezos, A.; Dhavala, P.; Papageorgiou, A.C.; Labrou, N.E. Directed evolution of Tau class glutathione transferases reveals a site that regulates catalytic efficiency and masks co-operativity. Biochem. J. 2016, 473, 559–570. [Google Scholar] [CrossRef]
- Lopez-Gonzalez, V.; La-Rocca, S.; Arbildi, P.; Fernandez, V. Characterization of catalytic and non-catalytic activities of EgGST2-3, a heterodimeric glutathione transferase from Echinococcus granulosus. Acta Trop. 2018, 180, 69–75. [Google Scholar] [CrossRef]
- Dubey, J.P.; Schuster, R.K.; Kinne, J. Gametogony of Eimeria cameli in the small intestine of one-humped camel (Camelus dromedarius). Parasitol. Res. 2018, 117, 3633–3638. [Google Scholar] [CrossRef]
- Zidan, M.; Pabst, R. Histological characterization of the lingual tonsils of the one-humped camel (Camelus dromedarius). Cell Tissue Res. 2020, 380, 107–113. [Google Scholar] [CrossRef]
- Malik, A.; Khan, J.M.; Alamery, S.F.; Fouad, D.; Labrou, N.E.; Daoud, M.S.; Abdelkader, M.O.; Ataya, F.S. Monomeric Camelus dromedarius GSTM1 at low pH is structurally more thermostable than its native dimeric form. PLoS ONE 2018, 13, e0205274. [Google Scholar] [CrossRef]
- Albarakati, N.; Khayyat, D.; Dallol, A.; Al-Maghrabi, J.; Nedjadi, T. The prognostic impact of GSTM1/GSTP1 genetic variants in bladder cancer. BMC Cancer 2019, 19, 991. [Google Scholar] [CrossRef]
- Georgakis, N.D.; Karagiannopoulos, D.A.; Thireou, T.N.; Eliopoulos, E.E.; Labrou, N.E.; Tsoungas, P.G.; Koutsilieris, M.N.; Clonis, Y.D. Concluding the trilogy: The interaction of 2, 2′-dihydroxy-benzophenones and their carbonyl N-analogues with human glutathione transferase M1-1 face to face with the P1-1 and A1-1 isoenzymes involved in MDR. Chem. Biol. Drug Des. 2017, 90, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Karageorgi, S.; Prescott, J.; Wong, J.Y.; Lee, I.M.; Buring, J.E.; De Vivo, I. GSTM1 and GSTT1 copy number variation in population-based studies of endometrial cancer risk. Cancer Epidemiol. Prev. Biomark. 2011, 20, 1447–1452. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gui, Y.; Zhang, L.; Lv, W.; Zhang, W.; Zhao, J.; Hu, X. NFE2L2 variations reduce antioxidant response in patients with Parkinson disease. Oncotarget 2016, 7, 10756. [Google Scholar] [CrossRef] [PubMed]
- Pouliou, F.; Perperopoulou, F.; Labrou, N.E. Comparative analysis of two stress-inducible tau class glutathione transferases from Glycine max revealed significant catalytic and structural diversification. Protein Pept. Lett. 2017, 24, 922–935. [Google Scholar] [CrossRef] [PubMed]
- Axarli, I.; Dhavala, P.; Papageorgiou, A.C.; Labrou, N.E. Crystallographic and functional characterization of the fluorodifen-inducible glutathione transferase from Glycine max reveals an active site topography suited for diphenylether herbicides and a novel L-site. J. Mol. Biol. 2009, 385, 984–1002. [Google Scholar] [CrossRef]
- Chronopoulou, E.G.; Papageorgiou, A.C.; Ataya, F.; Nianiou-Obeidat, I.; Madesis, P.; Labrou, N.E. Expanding the plant GSTome through directed evolution: DNA shuffling for the generation of new synthetic enzymes with engineered catalytic and binding properties. Front. Plant Sci. 2018, 9, 1737. [Google Scholar] [CrossRef]
- Skopelitou, K.; Muleta, A.W.; Papageorgiou, A.C.; Chronopoulou, E.G.; Pavli, O.; Flemetakis, E.; Skaracis, G.N.; Labrou, N.E. Characterization and functional analysis of a recombinant tau class glutathione transferase GmGSTU2-2 from Glycine max. Int. J. Biol. Macromol. 2017, 94, 802–812. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Chronopoulou, E.G.; Vlachakis, D.; Papageorgiou, A.C.; Ataya, F.S.; Labrou, N.E. Structure-based design and application of an engineered glutathione transferase for the development of an optical biosensor for pesticides determination. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2019, 1863, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Keegan, R.M.; Winn, M.D. MrBUMP: An automated pipeline for molecular replacement. Acta Cryst. D Biol. Cryst. 2008, 64, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Mannervik, B.; Danielson, U.H. Glutathione transferases--structure and catalytic activity. CRC Crit. Rev. Biochem. 1988, 23, 283–337. [Google Scholar] [CrossRef] [PubMed]
- Hurst, R.; Bao, Y.; Jemth, P.; Mannervik, B.; Williamson, G. Phospholipid hydroperoxide glutathione peroxidase activity of human glutathione transferases. Biochem. J. 1998, 332, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kolm, R.H.; Mannervik, B.; Talalay, P. Reversible conjugation of isothiocyanates with glutathione catalyzed by human glutathione transferases. Biochem. Biophys. Res. Commun. 1995, 206, 748–755. [Google Scholar] [CrossRef] [PubMed]
- Miękus, N.; Marszałek, K.; Podlacha, M.; Iqbal, A.; Puchalski, C.; Świergiel, A.H. Health benefits of plant-derived sulfur compounds, glucosinolates, and organosulfur compounds. Molecules 2020, 25, 3804. [Google Scholar] [CrossRef]
- Koeplinger, K.A.; Zhao, Z.; Peterson, T.; Leone, J.W.; Schwende, F.S.; Heinrikson, R.L.; Tomasselli, A.G. Activated sulfonamides are cleaved by glutathione-S-transferases. Drug Metab. Dispos. 1999, 27, 986–991. [Google Scholar] [PubMed]
- Lederer, B.; Böger, P. A ligand function of glutathione S-transferase. Z Naturforsch. C J. Biosci. 2005, 60, 166–171. [Google Scholar] [CrossRef]
- Dixon, D.P.; Edwards, R. Protein-Ligand Fishing in planta for Biologically Active Natural Products Using Glutathione Transferases. Front. Plant Sci. 2018, 9, 1659. [Google Scholar] [CrossRef]
- Patskovsky, Y.V.; Patskovska, L.N.; Listowsky, I. Functions of His107 in the catalytic mechanism of human glutathione S-transferase hGSTM1a-1a. Biochemistry 1999, 38, 1193–1202. [Google Scholar] [CrossRef]
- Patskovsky, Y.; Patskovska, L.; Almo, S.C.; Listowsky, I. Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a. Biochemistry 2006, 45, 3852–3862. [Google Scholar] [CrossRef]
- Ji, X.; Armstrong, R.N.; Gilliland, G.L. Snapshots along the reaction coordinate of an SNAr reaction catalyzed by glutathione transferase. Biochemistry 1993, 32, 12949–12954. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.J.; Kuan, I.C.; Tam, M.F.; Hsiao, C.D. The three-dimensional structure of an avian class-mu glutathione S-transferase, cGSTM1-1 at 1.94 A resolution. J. Mol. Biol. 1998, 278, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Martínez, A.B.; Sotelo-Mundo, R.R.; Rudiño-Piñera, E. Crystal structure of a class-mu glutathione S-transferase from whiteleg shrimp Litopenaeus vannamei: Structural changes in the xenobiotic binding H-site may alter the spectra of molecules bound. J. Biochem. Mol. Toxicol. 2017, 31, 2. [Google Scholar] [CrossRef]
- Axarli, I.; Georgiadou, C.; Dhavala, P.; Papageorgiou, A.C.; Labrou, N.E. Investigation of the role of conserved residues Ser13, Asn48 and Pro49 in the catalytic mechanism of the tau class glutathione transferase from Glycine max. Biochim. Biophys. Acta 2010, 1804, 662–667. [Google Scholar] [CrossRef] [PubMed]





| Electrophile Substrates | U/mg |
|---|---|
| 1-Chloro-2,4-dinitrobenzene | 12.6 |
| 1-Bromo-2,4-dinitrobenzene | 14.3 |
| 1-Fluoro-2,4-dinitrobenzene | 8.0 |
| 1-Iodo-2,4-dinitrobenzene | 1.1 |
| p-Nitrobenzyl chloride | 10.1 |
| 4-Chloro-7-nitrobenzofurazan | - |
| Fluorodifen | 1.1 |
| 2,3-Dichloro-4-[2-methylene-butyryl]phenoxy)acetic acid (Ethacrynic acid) | - |
| trans-4-Phenyl-3-buten-2-one | - |
| trans-2-Nonenal | 1.7 |
| Cumene hydroperoxide | - |
| tert-Butyl hydroperoxide | - |
| Allyl isothiocyanate | 5.4 |
| Phenethyl isothiocyanate | 5.6 |
| 2-Hydroxyethyl disulphide (2,2-dithiodiethanol) | - |
| Sulphanilamide | 4.3 |
| Epoxy-3-(p-nitrophenoxy)-propane | - |
| Bromosulfalein | - |
| GSH Analogues a | |
| Glutathione reduced ethyl ester | 6.6 |
| γ-Glu-Cys | - |
| Cys-Gly | - |
| Cysteine | - |
| N-Acetyl-L-cysteine | - |
| Kinetic Parameters | CDNB | GSH |
|---|---|---|
| Km (mM) | 0.84 ± 0.0994 | 0.0867 ± 0.0117 |
| kcat (min−1) | 1608 ± 82 | |
| kcat /KmCDNB (mM−1min−1) | 1914.3 ± 370 | |
| Data Collection | CdGSTM1-GTB | CdGSTM1-GSH |
|---|---|---|
| Wavelength (Å) | 1.0322 | 1.0322 |
| Resolution range (Å) | 66.68–2.05 (2.11–2.05) | 95.9–2.55 (2.64–2.55) |
| Space group | P21 | P21 |
| Unit cell | 50.4 177.4 59.4 90 115.0 90 | 50.8 150.2 191.8 90.0 90.0 90.0 |
| Total observations | 192,763 (11609) | 323,629 (30,311) |
| Unique reflections | 55,058 (4146) | 49,030 (4404) |
| Multiplicity | 3.5 (2.8) | 6.6 (6.9) |
| Completeness (%) | 93.5 (90.5) | 100 (100) |
| Mean I/sigma(I) | 5.0 (1.4) | 10.3 (0.9) |
| Wilson B-factor (Å2) | 33.18 | 49.03 |
| Rmeas | 0.175 (1.593) | 0.188 (2.369) |
| Rpim | 0.106 (0.969) | 0.099 (1.238) |
| CC1/2 | 0.984 (0.305) | 0.997 (0.348) |
| Refinement | ||
| Reflections used in refinement | 54,874 (5265) | 48,915 (4780) |
| Reflections used for R-free | 2627 (242) | 2393 (213) |
| Rwork/Rfree | 0.195 (0.290/0.252 (0.359) | 0.206 (0.287)/0.273 (0.318) |
| Number of non-hydrogen atoms | 7762 | 7564 |
| Macromolecules | 7232 | 7232 |
| Ligands | 127 | 81 |
| Solvent | 403 | 251 |
| Protein residues | 868 | 872 |
| RMSD in bonds (Å) | 0.008 | 0.008 |
| RMSD in angles (°) | 0.92 | 0.96 |
| Ramachandran favored/allowed/outliers (%) | 96.1/3.4/0.5 | 95.9/3.4/0.7 |
| Rotamer outliers (%) | 0.0 | 0.0 |
| Clashscore | 8.6 | 5.8 |
| Average B-factor (Å2) | 55.6 | 59.1 |
| Macromolecules | 56.2 | 59.1 |
| Ligands | 47.2 | 74.0 |
| Solvent | 47.3 | 54.2 |
| Number of TLS groups | 4 | 30 |
| PDB id | 7opy | 7opz |
| µ-GSTs | Organisms | PDB Entries | RSMD Values |
|---|---|---|---|
| hGSTM1 | Homo sapiens | 1xw6 | 0.44 |
| hGSTM2 | Homo sapiens | 2ab6 | 0.53 |
| hGSTM4 | Homo sapiens | 4gtu | 0.73 |
| MuGSTM7 | Mus musculus | 2dc5 | 0.79 |
| GgGSTM1-1 | Gallus gallus | 1gsu | 0.72 |
| LvGSTMu | Litopenaeus vannamei | 5an1 | 0.85 |
| RrGSTMu | Rattus rattus | 6gst | 0.50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perperopoulou, F.; Poudel, N.; Papageorgiou, A.C.; Ataya, F.S.; Labrou, N.E. Structural and Functional Characterization of Camelus dromedarius Glutathione Transferase M1-1. Life 2022, 12, 106. https://doi.org/10.3390/life12010106
Perperopoulou F, Poudel N, Papageorgiou AC, Ataya FS, Labrou NE. Structural and Functional Characterization of Camelus dromedarius Glutathione Transferase M1-1. Life. 2022; 12(1):106. https://doi.org/10.3390/life12010106
Chicago/Turabian StylePerperopoulou, Fereniki, Nirmal Poudel, Anastassios C. Papageorgiou, Farid S. Ataya, and Nikolaos E. Labrou. 2022. "Structural and Functional Characterization of Camelus dromedarius Glutathione Transferase M1-1" Life 12, no. 1: 106. https://doi.org/10.3390/life12010106
APA StylePerperopoulou, F., Poudel, N., Papageorgiou, A. C., Ataya, F. S., & Labrou, N. E. (2022). Structural and Functional Characterization of Camelus dromedarius Glutathione Transferase M1-1. Life, 12(1), 106. https://doi.org/10.3390/life12010106








