High SPIN4 Expression Is Linked to Advanced Nodal Status and Inferior Prognosis in Nasopharyngeal Carcinoma Patients
Abstract
1. Introduction
2. Patients and Methods
2.1. Evaluation of the Gene Expression Profiles in Nasopharyngeal Carcinoma
2.2. Patient Enrollment and Follow-Up
2.3. Histopathological and Immunohistochemical Appraisals
2.4. Statistical Analysis
3. Results
3.1. SPIN4 Is Identified as the Most Significantly Differentially Expressed Gene in Relation to Advanced Nodal Status
3.2. Clinicopathological Characteristics of Nasopharyngeal Carcinoma Patients in Our Cohort
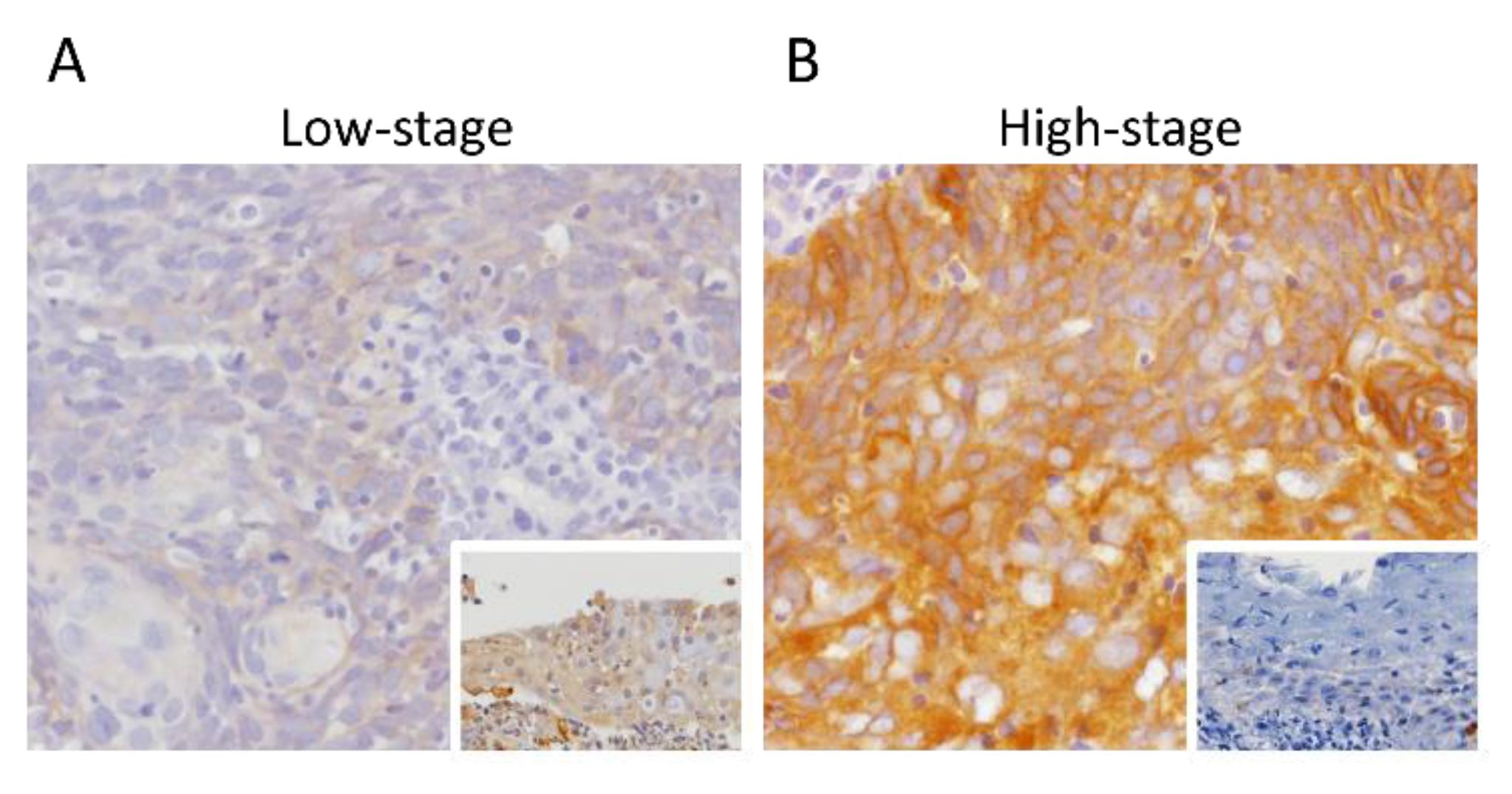
3.3. SPIN4 Immunoexpression and Its Associations with Clinicopathological Variables
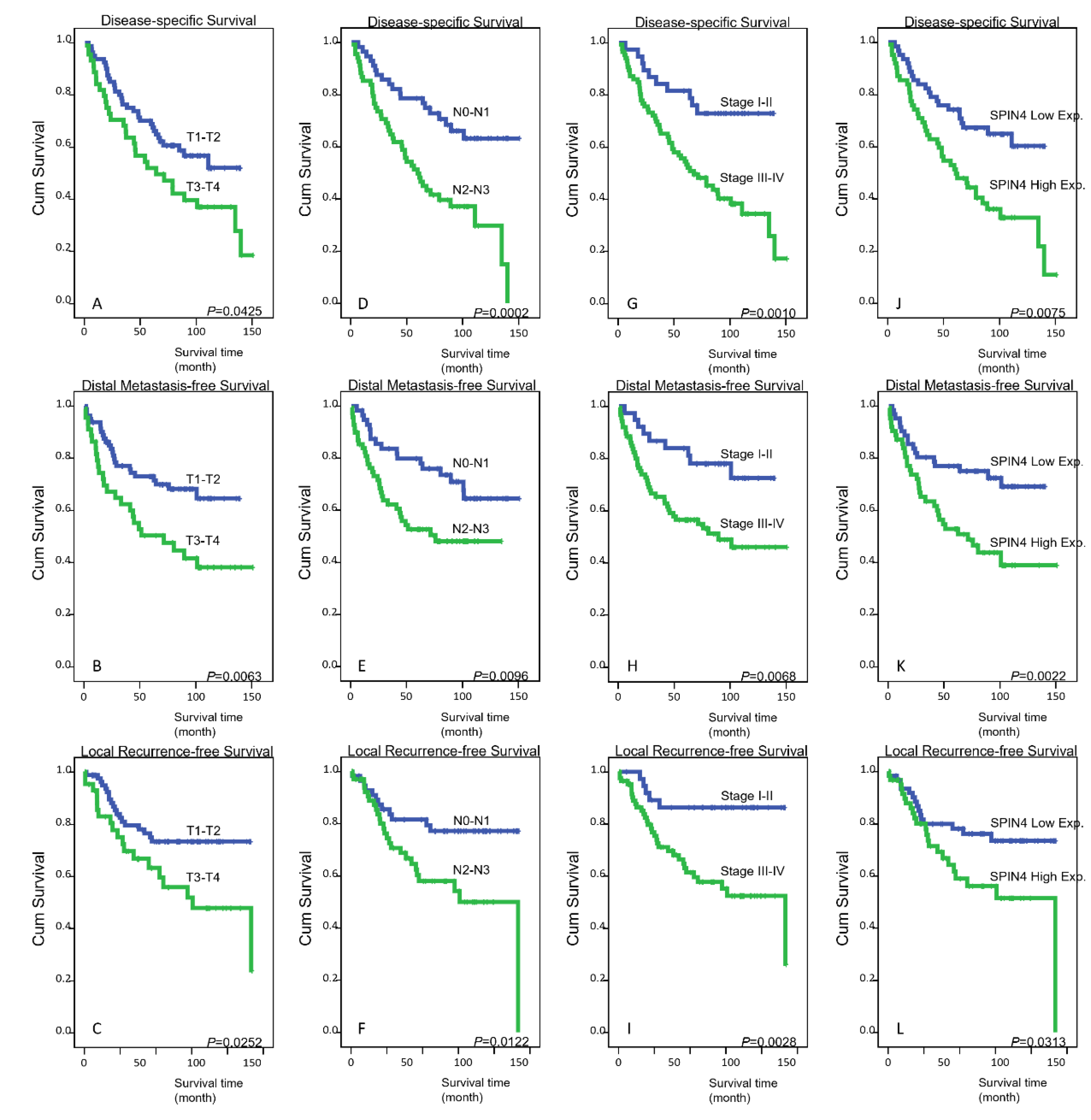
3.4. Survival and Prognostic Impact of SPIN4 Expression in Nasopharyngeal Carcinoma
3.5. High SPIN4 Level May Link Tight Junctions to Cancer Cell Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, L.; Li, C.; Pan, L. Nasopharyngeal carcinoma: A review of current updates. Exp. Ther. Med. 2018, 15, 3687–3692. [Google Scholar] [CrossRef]
- Hwang, J.S.G.; Chang, Y.-L.; To, K.-F.; Mai, H.-Q.; Feng, Y.-F.; Chang, E.T.; Wang, C.-P.; Kam, M.K.M.; Cheah, S.-L.; Lee, M.; et al. A new prognostic histopathologic classification of nasopharyngeal carcinoma. Chin. J. Cancer 2016, 35, 41. [Google Scholar] [CrossRef]
- Tsao, S.W.; Tsang, C.M.; Lo, K.W. Epstein–Barr virus infection and nasopharyngeal carcinoma. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160270. [Google Scholar] [CrossRef]
- Tsang, C.M.; Lui, V.W.Y.; Bruce, J.P.; Pugh, T.J.; Lo, K.W. Translational genomics of nasopharyngeal cancer. Semin. Cancer Biol. 2020, 61, 84–100. [Google Scholar] [CrossRef] [PubMed]
- Perri, F.; Scarpati, G.D.V.; Caponigro, F.; Ionna, F.; Longo, F.; Buonopane, S.; Muto, P.; Di Marzo, M.; Pisconti, S.; Solla, R. Management of recurrent nasopharyngeal carcinoma: Current perspectives. OncoTargets Ther. 2019, 12, 1583–1591. [Google Scholar] [CrossRef]
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity 2010, 105, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef]
- Yuan, J.; Jiang, Y.-Y.; Mayakonda, A.; Huang, M.; Ding, L.-W.; Lin, H.; Yu, F.; Lu, Y.; Loh, T.K.S.; Chow-Castro, M.; et al. Super-Enhancers Promote Transcriptional Dysregulation in Nasopharyngeal Carcinoma. Cancer Res. 2017, 77, 6614–6626. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Yang, X.; Cheng, B.; Chen, X.; Zhang, T.; He, Q.; Li, B.; Li, Y.; Tang, X.; Wen, X.; et al. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat. Commun. 2017, 8, 14053. [Google Scholar] [CrossRef]
- Yin, L.; Chen, J.; Ma, C.; Pei, S.; Du, M.; Zhang, Y.; Feng, Y.; Yin, R.; Bian, X.; He, X.; et al. Hsa_circ_0046263 functions as a ceRNA to promote nasopharyngeal carcinoma progression by upregulating IGFBP3. Cell Death Dis. 2020, 11, 562. [Google Scholar] [CrossRef]
- Bae, N.; Gao, M.; Li, X.; Premkumar, T.; Sbardella, G.; Chen, J.; Bedford, M.T. A transcriptional coregulator, SPIN·DOC, attenuates the coactivator activity of Spindlin1. J. Biol. Chem. 2017, 292, 20808–20817. [Google Scholar] [CrossRef]
- Zhang, X.; Thielert, M.; Li, H.; Cravatt, B.F. SPIN4 Is a Principal Endogenous Substrate of the E3 Ubiquitin Ligase DCAF16. Biochemistry 2021, 60, 637–642. [Google Scholar] [CrossRef]
- Dai, W.; Cheung, A.K.L.; Ko, J.; Cheng, Y.; Zheng, H.; Ngan, R.K.C.; Ng, W.T.; Lee, A.W.M.; Yau, C.C.; Lee, V.; et al. Comparative methylome analysis in solid tumors reveals aberrant methylation at chromosome 6p in nasopharyngeal carcinoma. Cancer Med. 2015, 4, 1079–1090. [Google Scholar] [CrossRef]
- Ferreira, W.; Amorim, C.; Burbano, R.; Villacis, R.; Marchi, F.; Medina, T.; de Lima, M.; de Oliveira, E. Genomic and transcriptomic characterization of the human glioblastoma cell line AHOL1. Braz. J. Med. Biol. Res. 2021, 54, e9571. [Google Scholar] [CrossRef]
- Kang, Y.; He, W.; Ren, C.; Qiao, J.; Guo, Q.; Hu, J.; Xu, H.; Jiang, X.; Wang, L. Advances in targeted therapy mainly based on signal pathways for nasopharyngeal carcinoma. Signal Transduct. Target. Ther. 2020, 5, 245. [Google Scholar] [CrossRef] [PubMed]
- Wolden, S.L.; Zelefsky, M.J.; Kraus, D.H.; Rosenzweig, K.E.; Chong, L.M.; Shaha, A.R.; Zhang, H.; Harrison, L.B.; Shah, J.P.; Pfister, D.G. Accelerated Concomitant Boost Radiotherapy and Chemotherapy for Advanced Nasopharyngeal Carcinoma. J. Clin. Oncol. 2001, 19, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-C.; Jan, J.-S.; Hsu, C.-Y.; Liang, W.-M.; Jiang, R.-S.; Wang, W.-Y. Phase III Study of Concurrent Chemoradiotherapy Versus Radiotherapy Alone for Advanced Nasopharyngeal Carcinoma: Positive Effect on Overall and Progression-Free Survival. J. Clin. Oncol. 2003, 21, 631–637. [Google Scholar] [CrossRef]
- Lee, Y.-E.; He, H.-L.; Chen, T.-J.; Lee, S.-W.; Chang, I.-W.; Hsing, C.-H.; Li, C.-F. The prognostic impact of RAP2A expression in patients with early and locoregionally advanced nasopharyngeal carcinoma in an endemic area. Am. J. Transl. Res. 2015, 7, 912–921. [Google Scholar]
- Chan, T.-C.; Wu, W.-J.; Li, W.-M.; Shiao, M.-S.; Shiue, Y.-L.; Li, C.-F. SLC14A1 prevents oncometabolite accumulation and recruits HDAC1 to transrepress oncometabolite genes in urothelial carcinoma. Theranostics 2020, 10, 11775–11793. [Google Scholar] [CrossRef] [PubMed]
- Budwit-Novotny, D.A.; Cox, E.B.; Soper, J.T.; Mutch, D.G.; Creasman, W.T.; Flowers, J.L.; Mccarty, K.S. Immunohistochemical analyses of estrogen receptor in endometrial adenocarcinoma using a monoclonal antibody. Cancer Res. 1986, 46, 5419–5425. [Google Scholar]
- Mao, Y.-P.; Liang, S.-B.; Liu, L.-Z.; Chen, Y.; Sun, Y.; Tang, L.-L.; Tian, L.; Lin, A.-H.; Liu, M.-Z.; Li, L.; et al. The N Staging System in Nasopharyngeal Carcinoma with Radiation Therapy Oncology Group Guidelines for Lymph Node Levels Based on Magnetic Resonance Imaging. Clin. Cancer Res. 2008, 14, 7497–7503. [Google Scholar] [CrossRef] [PubMed]
- Zihni, C.; Mills, C.; Matter, K.; Balda, M. Tight junctions: From simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 2016, 17, 564–580. [Google Scholar] [CrossRef]
- Ke, L.; Zhou, H.; Wang, C.; Xiong, G.; Xiang, Y.; Ling, Y.; Khabir, A.; Tsao, G.S.; Zeng, Y.; Zeng, M.; et al. Nasopharyngeal carcinoma super-enhancer–driven ETV6 correlates with prognosis. Proc. Natl. Acad. Sci. USA 2017, 114, 9683–9688. [Google Scholar] [CrossRef]
- Cai, M.-Y.; Tong, Z.-T.; Zhu, W.; Wen, Z.-Z.; Rao, H.-L.; Kong, L.-L.; Guan, X.-Y.; Kung, H.-F.; Zeng, Y.-X.; Xie, D. H3K27me3 Protein Is a Promising Predictive Biomarker of Patients’ Survival and Chemoradioresistance in Human Nasopharyngeal Carcinoma. Mol. Med. 2011, 17, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Leong, M.M.L.; Cheung, A.K.L.; Dai, W.; Tsao, S.W.; Tsang, C.M.; Dawson, C.W.; Mun Yee Ko, J.; Lung, M.L. EBV infection is associated with histone bivalent switch modifications in squamous epithelial cells. Proc. Natl. Acad. Sci. USA 2019, 116, 14144–14153. [Google Scholar] [CrossRef]
- Zhao, X.; Zeng, H.; Lei, L.; Tong, X.; Yang, L.; Yang, Y.; Li, S.; Zhou, Y.; Luo, L.; Huang, J.; et al. Tight junctions and their regulation by non-coding RNAs. Int. J. Biol. Sci. 2021, 17, 712–727. [Google Scholar] [CrossRef]
- Lee, T.I.; Young, R.A. Transcriptional Regulation and Its Misregulation in Disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef] [PubMed]
- Bushweller, J.H. Targeting transcription factors in cancer—From undruggable to reality. Nat. Rev. Cancer 2019, 19, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Heyn, H.; Esteller, M. DNA methylation profiling in the clinic: Applications and challenges. Nat. Rev. Genet. 2012, 13, 679–692. [Google Scholar] [CrossRef]
- Deville, S.S.; Cordes, N. The Extracellular, Cellular, and Nuclear Stiffness, a Trinity in the Cancer Resistome—A Review. Front. Oncol. 2019, 9, 1376. [Google Scholar] [CrossRef]
- Liu, C.; Guo, P.; Zhou, L.; Wang, Y.; Tian, S.; Ding, Y.; Wu, J.; Zhu, J.; Wang, Y. A practical method to screen and identify functioning biomarkers in nasopharyngeal carcinoma. Sci. Rep. 2021, 11, 7294. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Semba, S.; Khan, R.I.; Bochimoto, H.; Watanabe, T.; Fujiya, M.; Kohgo, Y.; Liu, Y.; Taniguchi, T. Focal adhesion kinase regulates intestinal epithelial barrier function via redistribution of tight junction. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2013, 1832, 151–159. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, M.; Luo, J.; Zhou, H. Radiotherapy targeting cancer stem cells “awakens” them to induce tumour relapse and metastasis in oral cancer. Int. J. Oral Sci. 2020, 12, 19. [Google Scholar] [CrossRef]
- Watters, A.; Rom, S.; Hill, J.; Dematatis, M.K.; Zhou, Y.; Merkel, S.F.; Andrews, A.; Cenna, J.M.; Potula, R.; Skuba, A.; et al. Identification and Dynamic Regulation of Tight Junction Protein Expression in Human Neural Stem Cells. Stem Cells Dev. 2015, 24, 1377–1389. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef]
- Le, Q.T.; Colevas, A.D.; O’Sullivan, B.; Lee, A.W.M.; Lee, N.; Ma, B.; Siu, L.L.; Waldron, J.; Lim, C.-M.; Riaz, N.; et al. Current Treatment Landscape of Nasopharyngeal Carcinoma and Potential Trials Evaluating the Value of Immunotherapy. J. Natl. Cancer Inst. 2019, 111, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Kwong, D.L.-W.; Dai, W.; Wu, P.; Li, S.; Yan, Q.; Zhang, Y.; Zhang, B.; Fang, X.; Liu, L.; et al. Comprehensive single-cell sequencing reveals the stromal dynamics and tumor-specific characteristics in the microenvironment of nasopharyngeal carcinoma. Nat. Commun. 2021, 12, 1540. [Google Scholar] [CrossRef]
- Salvador, E.; Burek, M.; Förster, C.Y. Tight Junctions and the Tumor Microenvironment. Curr. Pathobiol. Rep. 2016, 4, 135–145. [Google Scholar] [CrossRef]
- Tang, T.; Huang, X.; Zhang, G.; Hong, Z.; Bai, X.; Liang, T. Advantages of targeting the tumor immune microenvironment over blocking immune checkpoint in cancer immunotherapy. Signal Transduct. Target. Ther. 2021, 6, 72. [Google Scholar] [CrossRef]
- Qu, X.; Jeldres, C.; Glaskova, L.; Friedman, C.; Schroeder, S.; Nelson, P.S.; Porter, C.; Fang, M. Identification of Combinatorial Genomic Abnormalities Associated with Prostate Cancer Early Recurrence. J. Mol. Diagn. 2016, 18, 215–224. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Di Oto, E.; Biserni, G.B.; Varga, Z.; Morandi, L.; Cucchi, M.C.; Masetti, R.; Foschini, M.P. X chromosome gain is related to increased androgen receptor expression in male breast cancer. Virchows Arch. 2018, 473, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, N.; Shiotsu, T.; Hes, O.; Michal, M.; Shuin, T.; Lee, G.-H. Acquired cystic disease-associated renal cell carcinoma with gain of chromosomes 3, 7, and 16, gain of chromosome X, and loss of chromosome Y. Med. Mol. Morphol. 2010, 43, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Madrigal, I.; Rodríguez-Revenga, L.; Armengol, L.; Gonzalez, E.; Rodriguez, B.; Badenas, C.; Sánchez, A.; Martínez, F.; Guitart, M.; Fernández, I.; et al. X-chromosome tiling path array detection of copy number variants in patients with chromosome X-linked mental retardation. BMC Genom. 2007, 8, 443. [Google Scholar] [CrossRef] [PubMed]
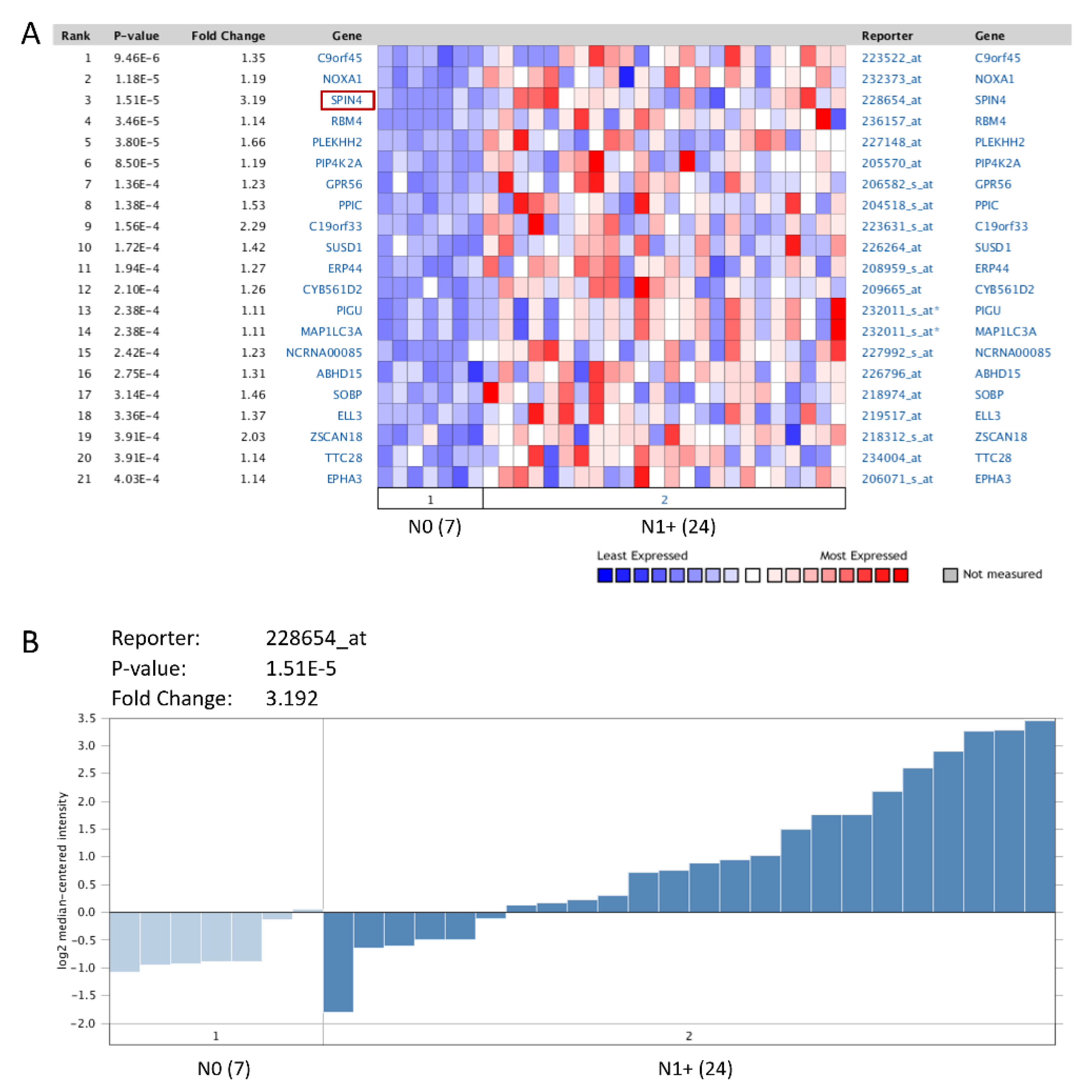
| Gene Rank | Gene Name | Gene Symbol | Reporter ID | p-Value | q-Value | Fold Change |
|---|---|---|---|---|---|---|
| 1 | chromosome 9 open reading frame 45 | C9orf45 | 223522_at | 9.46E-06 | 0.516966 | 1.350599 |
| 2 | NADPH oxidase activator 1 | NOXA1 | 232373_at | 1.18E-05 | 0.321552 | 1.190006 |
| 3 | spindlin family, member 4 | SPIN4 | 228654_at | 1.51E-05 | 0.274995 | 3.191617 |
| 4 | RNA binding motif protein 4 | RBM4 | 236157_at | 3.46E-05 | 0.473519 | 1.144074 |
| 5 | pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 | PLEKHH2 | 227148_at | 3.80E-05 | 0.415277 | 1.659764 |
| 6 | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha | PIP4K2A | 205570_at | 8.50E-05 | 0.774191 | 1.18786 |
| 7 | G protein-coupled receptor 56 | GPR56 | 206582_s_at | 1.36E-04 | 0.744635 | 1.227697 |
| 8 | peptidylprolyl isomerase C (cyclophilin C) | PPIC | 204518_s_at | 1.38E-04 | 0.685162 | 1.528623 |
| 9 | chromosome 19 open reading frame 33 | C19orf33 | 223631_s_at | 1.56E-04 | 0.710361 | 2.287249 |
| 10 | sushi domain containing 1 | SUSD1 | 226264_at | 1.72E-04 | 0.721422 | 1.420516 |
| 11 | endoplasmic reticulum protein 44 | ERP44 | 208959_s_at | 1.94E-04 | 0.663589 | 1.266438 |
| 12 | cytochrome b-561 domain containing 2 | CYB561D2 | 209665_at | 2.10E-04 | 0.674904 | 1.264451 |
| 13 | phosphatidylinositol glycan anchor biosynthesis, class U | PIGU | 232011_s_at | 2.38E-04 | 0.723024 | 1.110637 |
| 14 | microtubule-associated protein 1 light chain 3 alpha | MAP1LC3A | 232011_s_at | 2.38E-04 | 0.723024 | 1.110637 |
| 15 | non-protein coding RNA 85 | NCRNA00085 | 227992_s_at | 2.42E-04 | 0.695161 | 1.233108 |
| 16 | abhydrolase domain containing 15 | ABHD15 | 226796_at | 2.75E-04 | 0.750663 | 1.305885 |
| 17 | sine oculis binding protein homolog (Drosophila) | SOBP | 218974_at | 3.14E-04 | 0.781121 | 1.46335 |
| 18 | elongation factor RNA polymerase II-like 3 | ELL3 | 219517_at | 3.36E-04 | 0.797915 | 1.369774 |
| 19 | zinc finger and SCAN domain containing 18 | ZSCAN18 | 218312_s_at | 3.91E-04 | 0.854255 | 2.034354 |
| 20 | tetratricopeptide repeat domain 28 | TTC28 | 234004_at | 3.91E-04 | 0.82273 | 1.14447 |
| Parameters | Category | SPIN4 Exp. | p-Value | |
|---|---|---|---|---|
| Low | High | |||
| Gender | Male | 46 | 49 | 0.524 |
| Female | 16 | 13 | ||
| Age (years) | <60 years | 46 | 52 | 0.186 |
| ≥60 years | 16 | 14 | ||
| Primary tumor (T) | T1–T2 | 44 | 36 | 0.133 |
| T3–T4 | 18 | 26 | ||
| Nodal status (N) | N0–N1 | 45 | 11 | <0.001 * |
| N2–N3 | 17 | 51 | ||
| Stage | I–II | 32 | 6 | <0.001 * |
| III–IV | 30 | 56 | ||
| Histological grade | Keratinizing | 3 | 2 | 0.814 |
| Nonkeratinizing | 28 | 26 | ||
| Undifferentiated | 31 | 34 | ||
| EBER | Negative | 0 | 1 | 0.315 |
| Positive | 62 | 61 | ||
| Parameters | Category | No. of Case | DSS | DMeFS | LRFS | |||
|---|---|---|---|---|---|---|---|---|
| No. of Event | p-Value | No. of Event | p-Value | No. of Event | p-Value | |||
| Gender | Male | 95 | 48 | 0.8104 | 39 | 0.5524 | 31 | 0.3313 |
| Female | 29 | 15 | 11 | 7 | ||||
| Age (years) | <60 years | 98 | 52 | 0.5857 | 42 | 0.4146 | 30 | 0.8804 |
| ≥60 years | 26 | 11 | 8 | 8 | ||||
| Primary tumor (T) | T1–T2 | 80 | 34 | 0.0425 * | 25 | 0.0063 * | 19 | 0.0252 * |
| T3–T4 | 44 | 29 | 25 | 19 | ||||
| Nodal status (N) | N0–N1 | 56 | 19 | 0.0002 * | 17 | 0.0096 * | 12 | 0.0122 * |
| N2–N3 | 68 | 44 | 33 | 26 | ||||
| Stage | I–II | 38 | 10 | 0.0010 * | 9 | 0.0068 * | 5 | 0.0028 * |
| III–IV | 86 | 53 | 41 | 33 | ||||
| Histological grade | Keratinizing/non-keratinizing | 47 | 24 | 0.4120 | 17 | 0.2155 | 16 | 0.9323 |
| Undifferentiated | 77 | 39 | 33 | 22 | ||||
| EBER | Negative | 1 | 1 | 0.0567 | 1 | 0.0923 | 0 | 0.7314 |
| Positive | 123 | 62 | 49 | 38 | ||||
| SPIN4 exp. | Low exp. (H-score < median) | 66 | 25 | 0.0075 * | 17 | 0.0022 * | 15 | 0.0313 * |
| High exp. (H-score ≥ median) | 58 | 34 | 33 | 23 | ||||
| Parameter | Category | DSS | DMeFS | LRFS | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| H.R | 95% CI | p-Value | H.R | 95% CI | p-Value | H.R | 95% CI | p-Value | ||
| Stage | I–II | 1 | - | 0.028 * | 1 | - | 0.104 | 1 | - | 0.019 * |
| III–IV | 2.275 | 1.029–4.740 | 1.922 | 0.874–4.228 | 3.288 | 1.215−8.901 | ||||
| SPIN4 exp. | Low Exp. | 1 | - | 0.056 | 1 | - | 0.049 * | 1 | - | 0.345 |
| High Exp. | 1.731 | 0.986–3.040 | 1.905 | 1.003–3.617 | 1.394 | 0.700–2.776 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, S.-L.; Chan, T.-C.; Chen, T.-J.; Yang, C.-C.; Tsai, H.-H.; Yeh, C.-F.; Lee, S.-W.; Lai, H.-Y. High SPIN4 Expression Is Linked to Advanced Nodal Status and Inferior Prognosis in Nasopharyngeal Carcinoma Patients. Life 2021, 11, 912. https://doi.org/10.3390/life11090912
Chang S-L, Chan T-C, Chen T-J, Yang C-C, Tsai H-H, Yeh C-F, Lee S-W, Lai H-Y. High SPIN4 Expression Is Linked to Advanced Nodal Status and Inferior Prognosis in Nasopharyngeal Carcinoma Patients. Life. 2021; 11(9):912. https://doi.org/10.3390/life11090912
Chicago/Turabian StyleChang, Shih-Lun, Ti-Chun Chan, Tzu-Ju Chen, Ching-Chieh Yang, Hsin-Hwa Tsai, Cheng-Fa Yeh, Sung-Wei Lee, and Hong-Yue Lai. 2021. "High SPIN4 Expression Is Linked to Advanced Nodal Status and Inferior Prognosis in Nasopharyngeal Carcinoma Patients" Life 11, no. 9: 912. https://doi.org/10.3390/life11090912
APA StyleChang, S.-L., Chan, T.-C., Chen, T.-J., Yang, C.-C., Tsai, H.-H., Yeh, C.-F., Lee, S.-W., & Lai, H.-Y. (2021). High SPIN4 Expression Is Linked to Advanced Nodal Status and Inferior Prognosis in Nasopharyngeal Carcinoma Patients. Life, 11(9), 912. https://doi.org/10.3390/life11090912






