CTSE Overexpression Is an Adverse Prognostic Factor for Survival among Rectal Cancer Patients Receiving CCRT
Abstract
:1. Introduction
2. Patients and Methods
2.1. Transcriptomic Profiling of a Public Rectal Cancer Dataset
2.2. Patient Eligibility and Enrollment
2.3. Histopathological and Immunohistochemical Assessments
2.4. Statistical Analysis
3. Results
3.1. CTSE Is Identified as the Most Significant Differentially Expressed Gene Related to Digestion
3.2. Clinicopathological Characteristics of Rectal Carcinoma Patients in Our Cohort
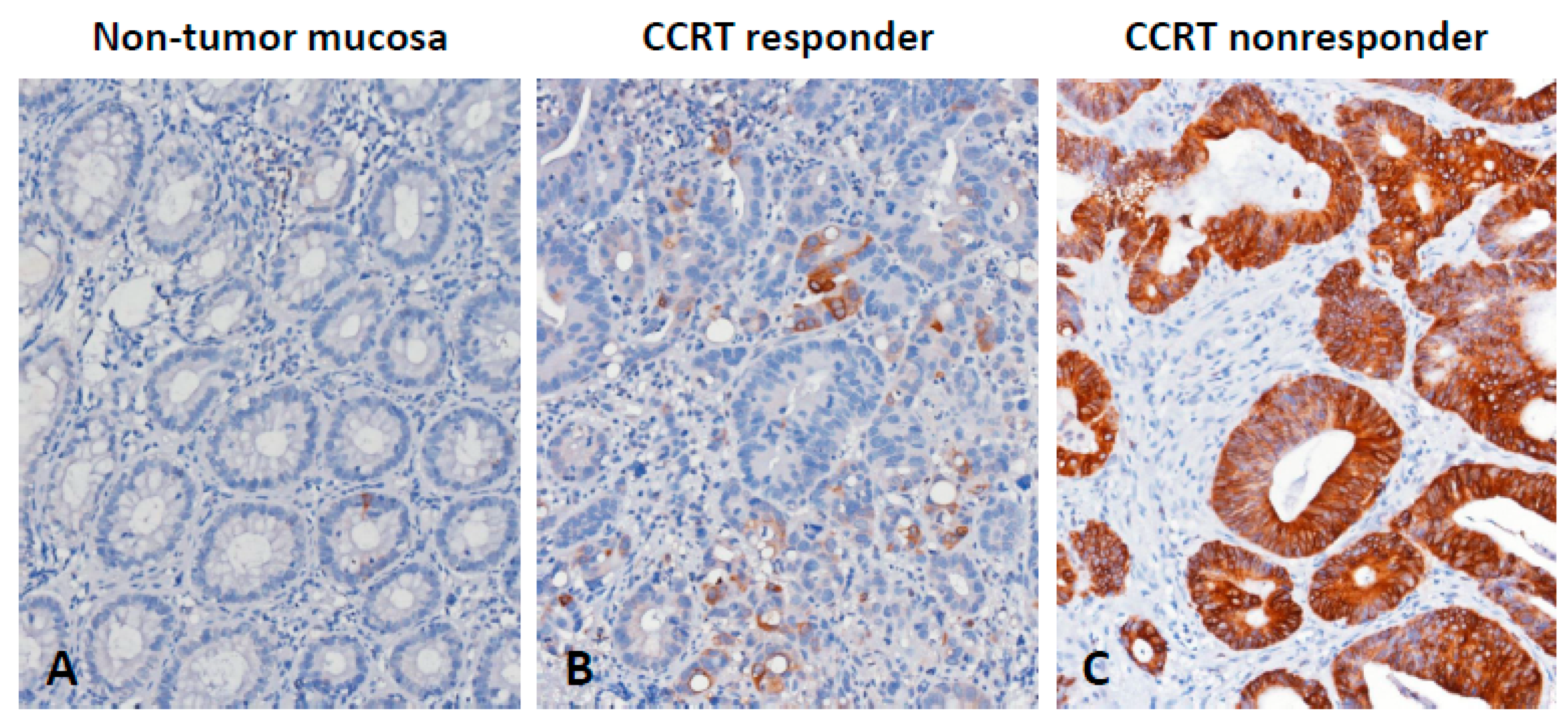
3.3. CTSE Immunoexpression and Its Correlations with Clinicopathological Factors
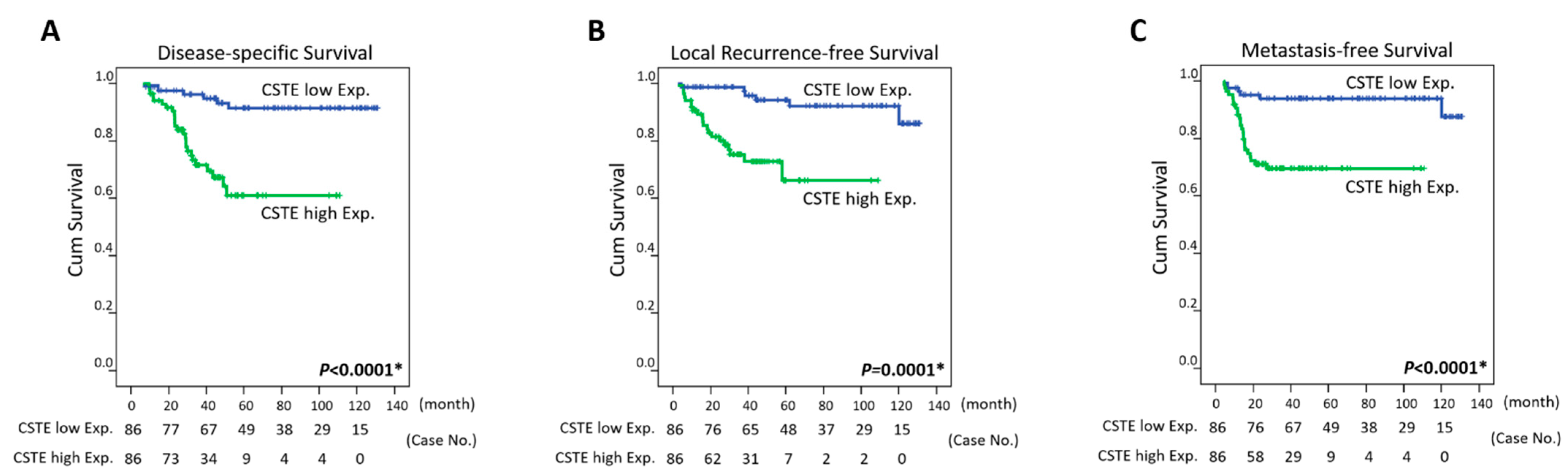
3.4. Survival and Prognostic Implications of CTSE Expression in Rectal Carcinoma
3.5. CTSE Overexpression may form a Defensive Mucous Barrier against CCRT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wong, M.C.; Ding, H.; Wang, J.; Chan, P.S.; Huang, J. Prevalence and risk factors of colorectal cancer in Asia. Intest. Res. 2019, 17, 317–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ermund, A.; Schütte, A.; Johansson, M.E.; Gustafsson, J.K.; Hansson, G.C. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G341–G347. [Google Scholar] [CrossRef]
- Johansson, M.E.; Hansson, G.C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 2016, 16, 639–649. [Google Scholar] [CrossRef]
- Norkina, O.; Burnett, T.G.; De Lisle, R.C. Bacterial Overgrowth in the Cystic Fibrosis Transmembrane Conductance Regulator Null Mouse Small Intestine. Infect. Immun. 2004, 72, 6040–6049. [Google Scholar] [CrossRef] [Green Version]
- Rao, C.V.; Janakiram, N.B.; Mohammed, A. Molecular Pathways: Mucins and Drug Delivery in Cancer. Clin. Cancer Res. 2017, 23, 1373–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azuma, T.; Liu, W.G.; Laan, D.J.V.; Bowcock, A.M.; Taggart, R.T. Human gastric cathepsin E gene. Multiple transcripts result from alternative polyadenylation of the primary transcripts of a single gene locus at 1q31-q32. J. Biol. Chem. 1992, 267, 1609–1614. [Google Scholar] [CrossRef]
- Tsukuba, T.; Okamoto, K.; Okamoto, Y.; Yanagawa, M.; Kohmura, K.; Yasuda, Y.; Uchi, H.; Nakahara, T.; Furue, M.; Nakayama, K.; et al. Association of cathepsin E deficiency with development of atopic dermatitis. J. Biochem. 2003, 134, 893–902. [Google Scholar] [CrossRef]
- Matsuo, K.; Kobayashi, I.; Tsukuba, T.; Kiyoshima, T.; Ishibashi, Y.; Miyoshi, A.; Yamamoto, K.; Sakai, H. Immunohistochemical localization of cathepsins D and E in human gastric cancer: A possible correlation with local invasive and metastatic activities of carcinoma cells. Hum. Pathol. 1996, 27, 184–190. [Google Scholar] [CrossRef]
- Uno, K.; Azuma, T.; Nakajima, M.; Yasuda, K.; Hayakumo, T.; Mukai, H.; Sakai, T.; Kawai, K. Clinical significance of cathepsin E in pancreatic juice in the diagnosis of pancreatic ductal adenocarcinoma. J. Gastroenterol. Hepatol. 2000, 15, 1333–1338. [Google Scholar] [PubMed]
- Tenti, P.; Romagnoli, S.; Silini, E.M.; Zappatore, R.; Giunta, P.; Stella, G.; Carnevali, L. Cervical Adenocarcinomas Express Markers Common to Gastric, Intestinal, and Pancreatobiliary Epithelial Cells. Pathol. Res. Pr. 1994, 190, 342–349. [Google Scholar] [CrossRef]
- Ullmann, R.; Morbini, P.; Halbwedl, I.; Bongiovanni, M.; Gogg-Kammerer, M.; Papotti, M.; Gabor, S.; Renner, H.; Popper, H.H. Protein expression profiles in adenocarcinomas and squamous cell carcinomas of the lung generated using tissue microarrays. J. Pathol. 2004, 203, 798–807. [Google Scholar] [CrossRef]
- Dworak, O.; Keilholz, L.; Hoffmann, A. Pathological features of rectal cancer after preoperative radiochemotherapy. Int. J. Colorectal. Dis. 1997, 12, 19–23. [Google Scholar] [CrossRef]
- Chan, T.-C.; Wu, W.-J.; Li, W.-M.; Shiao, M.-S.; Shiue, Y.-L.; Li, C.-F. SLC14A1 prevents oncometabolite accumulation and recruits HDAC1 to transrepress oncometabolite genes in urothelial carcinoma. Theranostics 2020, 10, 11775–11793. [Google Scholar] [CrossRef]
- Lee, Y.-Y.; Wei, Y.-C.; Tian, Y.-F.; Sun, D.-P.; Sheu, M.-J.; Yang, C.-C.; Lin, L.-C.; Lin, C.-Y.; Hsing, C.-H.; Li, W.-S.; et al. Overexpression of Transcobalamin 1 is an Independent Negative Prognosticator in Rectal Cancers Receiving Concurrent Chemoradiotherapy. J. Cancer 2017, 8, 1330–1337. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.T.; Heneghan, H.; Winter, D.C. Systematic review and meta-analysis of outcomes following pathological complete response to neoadjuvant chemoradiotherapy for rectal cancer. BJS 2012, 99, 918–928. [Google Scholar] [CrossRef]
- Guillem, J.G.; Chessin, D.B.; Cohen, A.M.; Shia, J.; Mazumdar, M.; Enker, W.; Paty, P.B.; Weiser, M.R.; Klimstra, D.; Saltz, L.; et al. Long-term Oncologic Outcome Following Preoperative Combined Modality Therapy and Total Mesorectal Excision of Locally Advanced Rectal Cancer. Ann. Surg. 2005, 241, 829–838. [Google Scholar] [CrossRef]
- Mastrogamvraki, N.; Zaravinos, A. Signatures of co-deregulated genes and their transcriptional regulators in colorectal cancer. NPJ Syst. Biol. Appl. 2020, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Busquets, L.; Guillen, H.; DeFord, M.E.; Suckow, M.A.; Navari, R.M.; Castellino, F.J.; Prorok, M. Cathepsin E is a specific marker of dysplasia in APC mouse intestine. Tumour. Biol. 2006, 27, 36–42. [Google Scholar] [CrossRef]
- Gregorieff, A.; Stange, D.; Kujala, P.; Begthel, H.; Born, M.V.D.; Korving, J.; Peters, P.J.; Clevers, H. The Ets-Domain Transcription Factor Spdef Promotes Maturation of Goblet and Paneth Cells in the Intestinal Epithelium. Gastroenterology 2009, 137, 1333–1345.e3. [Google Scholar] [CrossRef]
- Park, S.-W.; Zhen, G.; Verhaeghe, C.; Nakagami, Y.; Nguyenvu, L.T.; Barczak, A.J.; Killeen, N.; Erle, D.J. The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc. Natl. Acad. Sci. USA 2009, 106, 6950–6955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuru, A.; Fujimoto, N.; Takahashi, S.; Saito, M.; Nakamura, D.; Iwano, M.; Iwawaki, T.; Kadokura, H.; Ron, D.; Kohno, K. Negative feedback by IRE1β optimizes mucin production in goblet cells. Proc. Natl. Acad. Sci. USA 2013, 110, 2864–2869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gum, J.R.; Hicks, J.W.; Toribara, N.W.; Siddiki, B.; Kim, Y.S. Molecular cloning of human intestinal mucin (MUC2) cDNA. Identification of the amino terminus and overall sequence similarity to prepro-von Willebrand factor. J. Biol. Chem. 1994, 269, 2440–2446. [Google Scholar] [CrossRef]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Birchenough, G.M.; Johansson, M.E.V.; Gustafsson, J.K.; Bergström, J.H.; Hansson, G.C. New developments in goblet cell mucus secretion and function. Mucosal Immunol. 2015, 8, 712–719. [Google Scholar] [CrossRef] [Green Version]
- Saegusa, C.; Tanaka, T.; Tani, S.; Itohara, S.; Mikoshiba, K.; Fukuda, M. Decreased basal mucus secretion by Slp2-a-deficient gastric surface mucous cells. Genes Cells 2006, 11, 623–631. [Google Scholar] [CrossRef] [PubMed]
- McGuckin, M.A.; Hasnain, S.Z. Goblet cells as mucosal sentinels for immunity. Mucosal Immunol. 2017, 10, 1118–1121. [Google Scholar] [CrossRef] [PubMed]
- Ambort, D.; Johansson, M.E.; Gustafsson, J.K.; Nilsson, H.E.; Ermund, A.; Johansson, B.R.; Koeck, P.J.B.; Hebert, H.; Hansson, G.C. Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proc. Natl. Acad. Sci. USA 2012, 109, 5645–5650. [Google Scholar] [CrossRef] [Green Version]
- Gustafsson, J.K.; Ermund, A.; Ambort, D.; Johansson, M.E.V.; Nilsson, H.; Thorell, K.; Hebert, H.; Sjövall, H.; Hansson, G.C. Bicarbonate and functional CFTR channel are required for proper mucin secretion and link cystic fibrosis with its mucus phenotype. J. Exp. Med. 2012, 209, 1263–1272. [Google Scholar] [CrossRef] [Green Version]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef]
- Miller, S.A.; Ghobashi, A.H.; O’Hagan, H.M. Consensus molecular subtyping of colorectal cancers is influenced by goblet cell content. Cancer Genet. 2021, 254–255, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, C. The relationship between intestinal goblet cells and the immune response. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, D.H.; Gustafsson, J.K.; Knoop, K.A.; McDonald, K.G.; Bidani, S.S.; Davis, J.E.; Floyd, A.N.; Hogan, S.P.; Hsieh, C.-S.; Newberry, R.D. Goblet cell associated antigen passages support the induction and maintenance of oral tolerance. Mucosal Immunol. 2020, 13, 271–282. [Google Scholar] [CrossRef] [PubMed]
- He, Y.-F.; Zhang, M.-Y.; Wu, X.; Sun, X.-J.; Xu, T.; He, Q.-Z.; Di, W. High MUC2 Expression in Ovarian Cancer Is Inversely Associated with the M1/M2 Ratio of Tumor-Associated Macrophages and Patient Survival Time. PLoS ONE 2013, 8, e79769. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Probe | Comparison log Ratio | Comparison p-Value | Gene Symbol | Gene Name | Biological Process | Molecular Function |
|---|---|---|---|---|---|---|
| 1555854_at | 0.1462 | 0.0069 | AKR1C1 | aldo-keto reductase family 1; member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase), aldo-keto reductase family 1; member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase; type III) | bile acid and bile salt transport, bile acid metabolic process, cholesterol absorption, cholesterol homeostasis, digestion, electron transport, lipid metabolic process, prostaglandin metabolic process, protein homooligomerization, steroid metabolic process, xenobiotic metabolic process | 20-alpha-hydroxysteroid dehydrogenase activity, 3-alpha-hydroxysteroid dehydrogenase (A-specific) activity, 3-alpha-hydroxysteroid dehydrogenase (B-specific) activity, aldo-keto reductase activity, bile acid binding, carboxylic acid binding, oxidoreductase activity, trans-1;2-dihydrobenzene-1;2-diol dehydrogenase activity |
| 204151_x_at | 0.7096 | 0.003 | AKR1C1 | aldo-keto reductase family 1; member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase), aldo-keto reductase family 1; member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase; type III) | bile acid and bile salt transport, bile acid metabolic process, cholesterol absorption, cholesterol homeostasis, digestion, electron transport, lipid metabolic process, prostaglandin metabolic process, protein homooligomerization, steroid metabolic process, xenobiotic metabolic process | 20-alpha-hydroxysteroid dehydrogenase activity, 3-alpha-hydroxysteroid dehydrogenase (A-specific) activity, 3-alpha-hydroxysteroid dehydrogenase (B-specific) activity, aldo-keto reductase activity, bile acid binding, carboxylic acid binding, oxidoreductase activity, trans-1;2-dihydrobenzene-1;2-diol dehydrogenase activity |
| 205009_at | 0.9983 | 0.0022 | TFF1 | trefoil factor 1 | carbohydrate metabolic process, cell differentiation, defense response, digestion, negative regulation of cell proliferation | 20-alpha-hydroxysteroid dehydrogenase activity, 3-alpha-hydroxysteroid dehydrogenase (A-specific) activity, 3-alpha-hydroxysteroid dehydrogenase (B-specific) activity, aldo-keto reductase activity, bile acid binding, carboxylic acid binding, oxidoreductase activity, trans-1;2-dihydrobenzene-1;2-diol dehydrogenase activity |
| 205869_at | 0.1705 | 0.0076 | PRSS1 | protease; serine; 1 (trypsin 1) | digestion, proteolysis | growth factor activity, protein binding |
| 205927_s_at | 1.7848 | <0.0001 | CTSE | cathepsin E | antigen processing and presentation of exogenous peptide antigen via MHC class II, digestion, proteolysis | calcium ion binding, hydrolase activity, metal ion binding, peptidase activity, serine-type endopeptidase activity, trypsin activity |
| 206023_at | 0.8363 | 0.0076 | NMU | neuromedin U | G-protein coupled receptor protein signaling pathway, digestion, neuropeptide signaling pathway, regulation of smooth muscle contraction, signal transduction | aspartic-type endopeptidase activity, cathepsin E activity, hydrolase activity, pepsin A activity, peptidase activity |
| 207080_s_at | 0.7389 | 0.0092 | PYY | peptide YY | G-protein coupled receptor protein signaling pathway, cell motility, cell proliferation, cell-cell signaling, cytoskeleton organization and biogenesis, digestion, feeding behavior | receptor binding |
| 208596_s_at | 0.6909 | 0.0091 | UGT1A1 | UDP glucuronosyltransferase 1 family; polypeptide A1, UDP glucuronosyltransferase 1 family; polypeptide A10, UDP glucuronosyltransferase 1 family; polypeptide A3, UDP glucuronosyltransferase 1 family; polypeptide A4, UDP glucuronosyltransferase 1 family; polypeptide A5, UDP glucuronosyltransferase 1 family; polypeptide A6, UDP glucuronosyltransferase 1 family; polypeptide A7, UDP glucuronosyltransferase 1 family; polypeptide A8, UDP glucuronosyltransferase 1 family; polypeptide A9 | bilirubin conjugation, digestion, estrogen metabolic process, metabolic process, xenobiotic metabolic process | hormone activity |
| 213921_at | 0.8083 | 0.002 | SST | somatostatin | G-protein coupled receptor protein signaling pathway, cell surface receptor linked signal transduction, cell-cell signaling, digestion, induction of apoptosis by hormones, negative regulation of cell proliferation, regulation of cell migration, response to nutrient, synaptic transmission | UDP-glycosyltransferase activity, glucuronosyltransferase activity, transferase activity, transferase activity; transferring glycosyl groups, transferase activity; transferring hexosyl groups |
| 214476_at | 0.4628 | 0.0024 | TFF2 | trefoil factor 2 (spasmolytic protein 1) | defense response, digestion | hormone activity |
| 215125_s_at | 0.8769 | 0.0065 | UGT1A1 | UDP glucuronosyltransferase 1 family; polypeptide A1, UDP glucuronosyltransferase 1 family; polypeptide A10, UDP glucuronosyltransferase 1 family; polypeptide A3, UDP glucuronosyltransferase 1 family; polypeptide A4, UDP glucuronosyltransferase 1 family; polypeptide A5, UDP glucuronosyltransferase 1 family; polypeptide A6, UDP glucuronosyltransferase 1 family; polypeptide A7, UDP glucuronosyltransferase 1 family; polypeptide A8, UDP glucuronosyltransferase 1 family; polypeptide A9 | bilirubin conjugation, digestion, estrogen metabolic process, metabolic process, xenobiotic metabolic process | |
| 221305_s_at | 0.4507 | 0.0039 | UGT1A8 | UDP glucuronosyltransferase 1 family; polypeptide A8, UDP glucuronosyltransferase 1 family; polypeptide A9 | bilirubin conjugation, digestion, estrogen metabolic process, metabolic process, xenobiotic metabolic process | UDP-glycosyltransferase activity, glucuronosyltransferase activity, transferase activity, transferase activity; transferring glycosyl groups, transferase activity; transferring hexosyl groups |
| 206293_at | −0.3299 | 0.0003 | SULT2A1 | Sulfotransferase family; cytosolic; 2A; dehydroepiandrosterone (DHEA)-preferring; member 1 | bile acid catabolic process, digestion, lipid metabolic process, steroid metabolic process | bile-salt sulfotransferase activity, sulfotransferase activity, transferase activity |
| Parameter | No. | CTSE Expression | p-Value | ||
|---|---|---|---|---|---|
| Low Exp. | High Exp. | ||||
| Gender | Male | 108 | 57 | 51 | 0.334 |
| Female | 64 | 29 | 35 | ||
| Age | <70 | 106 | 56 | 50 | 0.347 |
| ≥70 | 66 | 30 | 36 | ||
| Pre-Tx tumor status (Pre-T) | T1-T2 | 81 | 54 | 27 | <0.001 * |
| T3-T4 | 91 | 32 | 59 | ||
| Pre-Tx nodal status (Pre-N) | N0 | 125 | 76 | 49 | <0.001 * |
| N1-N2 | 47 | 10 | 37 | ||
| Post-Tx tumor status (Post-T) | T1-T2 | 86 | 53 | 33 | 0.002 * |
| T3-T4 | 86 | 33 | 53 | ||
| Post-Tx nodal status (Post-N) | N0 | 123 | 72 | 51 | <0.001 * |
| N1-N2 | 49 | 14 | 35 | ||
| Vascular invasion | Absent | 157 | 85 | 72 | <0.001 * |
| Present | 15 | 1 | 14 | ||
| Perineural invasion | Absent | 167 | 86 | 81 | 0.023 * |
| Present | 5 | 0 | 5 | ||
| Tumor regression grade | Grade 0-1 | 37 | 10 | 27 | 0.003 * |
| Grade 2~3 | 118 | 64 | 54 | ||
| Grade 4 | 17 | 12 | 5 | ||
| Parameter | No. of Case | DSS | LRFS | MeFS | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of Event | p-Value | No. of Event | p-Value | No. of Event | p-Value | |||||
| Gender | Male | 108 | 20 | 0.9026 | 7 | 0.2250 | 17 | 0.3520 | ||
| Female | 64 | 11 | 20 | 14 | ||||||
| Age | <70 | 106 | 19 | 0.8540 | 18 | 0.6615 | 20 | 0.7427 | ||
| ≥70 | 66 | 12 | 9 | 11 | ||||||
| Pre-Tx tumor status (Pre-T) | T1-T2 | 81 | 10 | 0.0776 | 10 | 0.2261 | 11 | 0.1745 | ||
| T3-T4 | 91 | 21 | 17 | 20 | ||||||
| Pre-Tx nodal status (Pre-N) | N0 | 125 | 19 | 0.0711 | 15 | 0.0070 * | 19 | 0.0973 | ||
| N1-N2 | 47 | 21 | 12 | 12 | ||||||
| Post-Tx tumor status (Post-T) | T1-T2 | 86 | 7 | 0.0006 * | 7 | 0.0040 * | 8 | 0.0033 * | ||
| T3-T4 | 86 | 24 | 20 | 23 | ||||||
| Post-Tx nodal status (Post-N) | N0 | 123 | 21 | 0.5998 | 16 | 0.1320 | 20 | 0.4634 | ||
| N1-N2 | 49 | 10 | 11 | 11 | ||||||
| Vascular invasion | Absent | 157 | 25 | 0.0184* | 21 | 0.0028 * | 27 | 0.4470 | ||
| Present | 15 | 6 | 6 | 4 | ||||||
| Perineural invasion | Absent | 167 | 29 | 0.2559 | 25 | 0.0940 | 30 | 0.9083 | ||
| Present | 5 | 2 | 2 | 1 | ||||||
| Tumor regression grade | Grade 0-1 | 37 | 13 | 0.0038 * | 10 | 0.0090* | 14 | 0.0006 * | ||
| Grade 2~3 | 118 | 17 | 17 | 16 | ||||||
| Grade 4 | 17 | 1 | 0 | 1 | ||||||
| Down stage after CCRT | Non-Sig. | 150 | 29 | 0.1651 | 24 | 0.5961 | 30 | 0.0853 | ||
| Sig. (≥2) | 22 | 2 | 3 | 1 | ||||||
| CTSE expression | Low Exp. | 86 | 6 | <0.0001 * | 6 | 0.0001* | 6 | <0.0001 * | ||
| High Exp. | 86 | 25 | 25 | 21 | ||||||
| Parameter | DSS | LRFS | MeFS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| H.R | 95% CI | p-Value | H.R | 95% CI | p-Value | H.R | 95% CI | p-Value | |
| Tumor regression grade | 1.754 | 0.867–3.546 | 0.118 | 2.132 | 0.994–9.132 | 0.052 | 2.049 | 1.022–4.115 | 0.043 * |
| CTSE expression | 3.901 | 1.514–10.046 | 0.005 * | 3.748 | 1.243–11.306 | 0.019 * | 4.123 | 1.538–11.059 | 0.005 * |
| Vascular invasion | 1.701 | 0.660–4.384 | 0.272 | 2.083 | 0.763–5.682 | 0.152 | - | - | - |
| Post-Tx tumor status (Post-T) | 2.335 | 0.955–5.706 | 0.063 | 1.806 | 0.727–4.489 | 0.203 | 1.827 | 0.781–4.271 | 0.164 |
| Pre-Tx nodal status (Pre-N) | - | - | - | 1.358 | 0.570–3.237 | 0.489 | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chou, C.-L.; Chen, T.-J.; Tian, Y.-F.; Chan, T.-C.; Yeh, C.-F.; Li, W.-S.; Tsai, H.-H.; Li, C.-F.; Lai, H.-Y. CTSE Overexpression Is an Adverse Prognostic Factor for Survival among Rectal Cancer Patients Receiving CCRT. Life 2021, 11, 646. https://doi.org/10.3390/life11070646
Chou C-L, Chen T-J, Tian Y-F, Chan T-C, Yeh C-F, Li W-S, Tsai H-H, Li C-F, Lai H-Y. CTSE Overexpression Is an Adverse Prognostic Factor for Survival among Rectal Cancer Patients Receiving CCRT. Life. 2021; 11(7):646. https://doi.org/10.3390/life11070646
Chicago/Turabian StyleChou, Chia-Lin, Tzu-Ju Chen, Yu-Feng Tian, Ti-Chun Chan, Cheng-Fa Yeh, Wan-Shan Li, Hsin-Hwa Tsai, Chien-Feng Li, and Hong-Yue Lai. 2021. "CTSE Overexpression Is an Adverse Prognostic Factor for Survival among Rectal Cancer Patients Receiving CCRT" Life 11, no. 7: 646. https://doi.org/10.3390/life11070646
APA StyleChou, C.-L., Chen, T.-J., Tian, Y.-F., Chan, T.-C., Yeh, C.-F., Li, W.-S., Tsai, H.-H., Li, C.-F., & Lai, H.-Y. (2021). CTSE Overexpression Is an Adverse Prognostic Factor for Survival among Rectal Cancer Patients Receiving CCRT. Life, 11(7), 646. https://doi.org/10.3390/life11070646







