Comparison of the Intestinal Microbiome of Italian Patients with Multiple Sclerosis and Their Household Relatives
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Collection, Genomic DNA Extraction, PCR Amplification and Sequencing
2.3. Raw Data Processing
3. Results
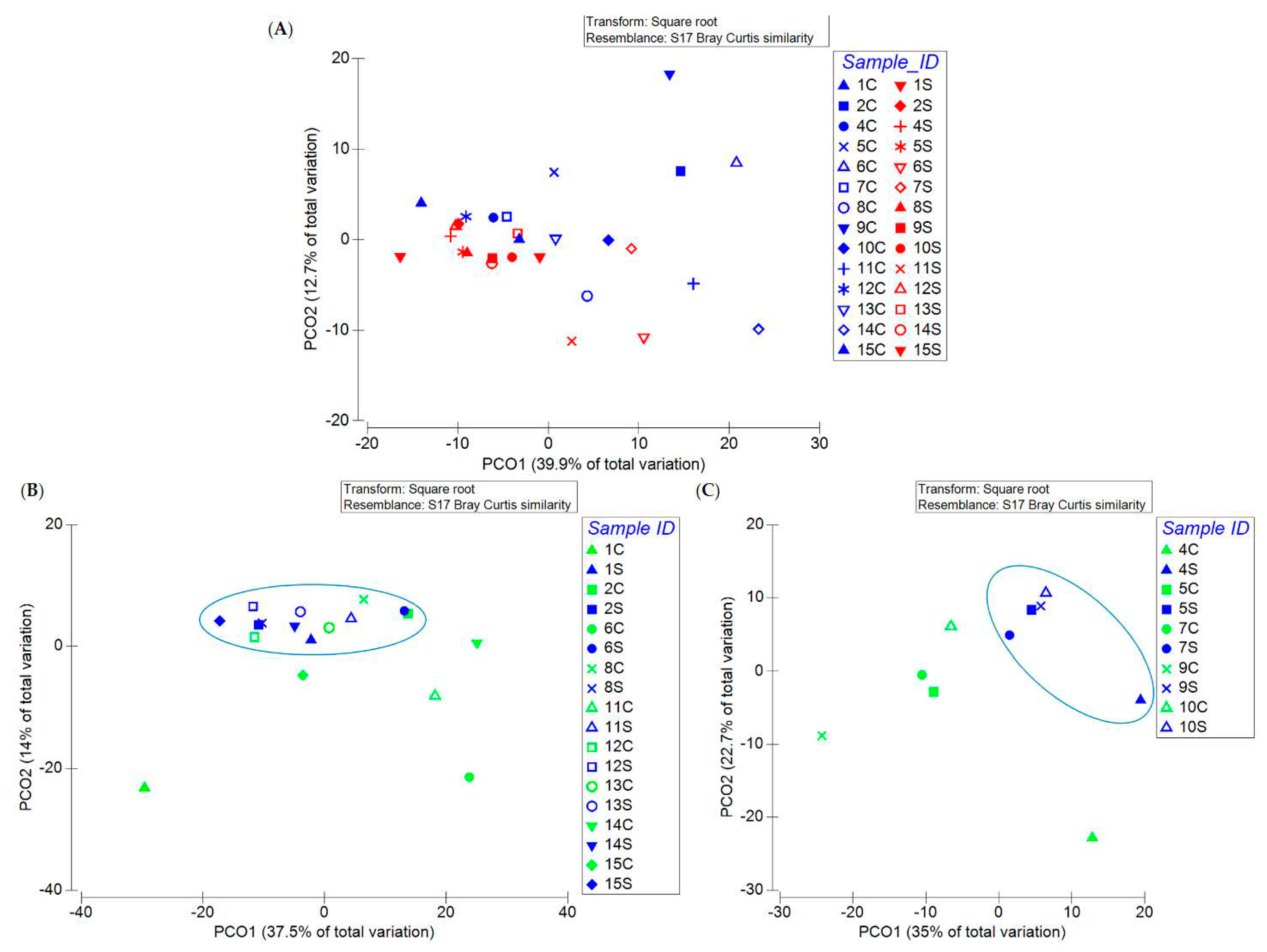
3.1. Bacterial Communities’ Diversity
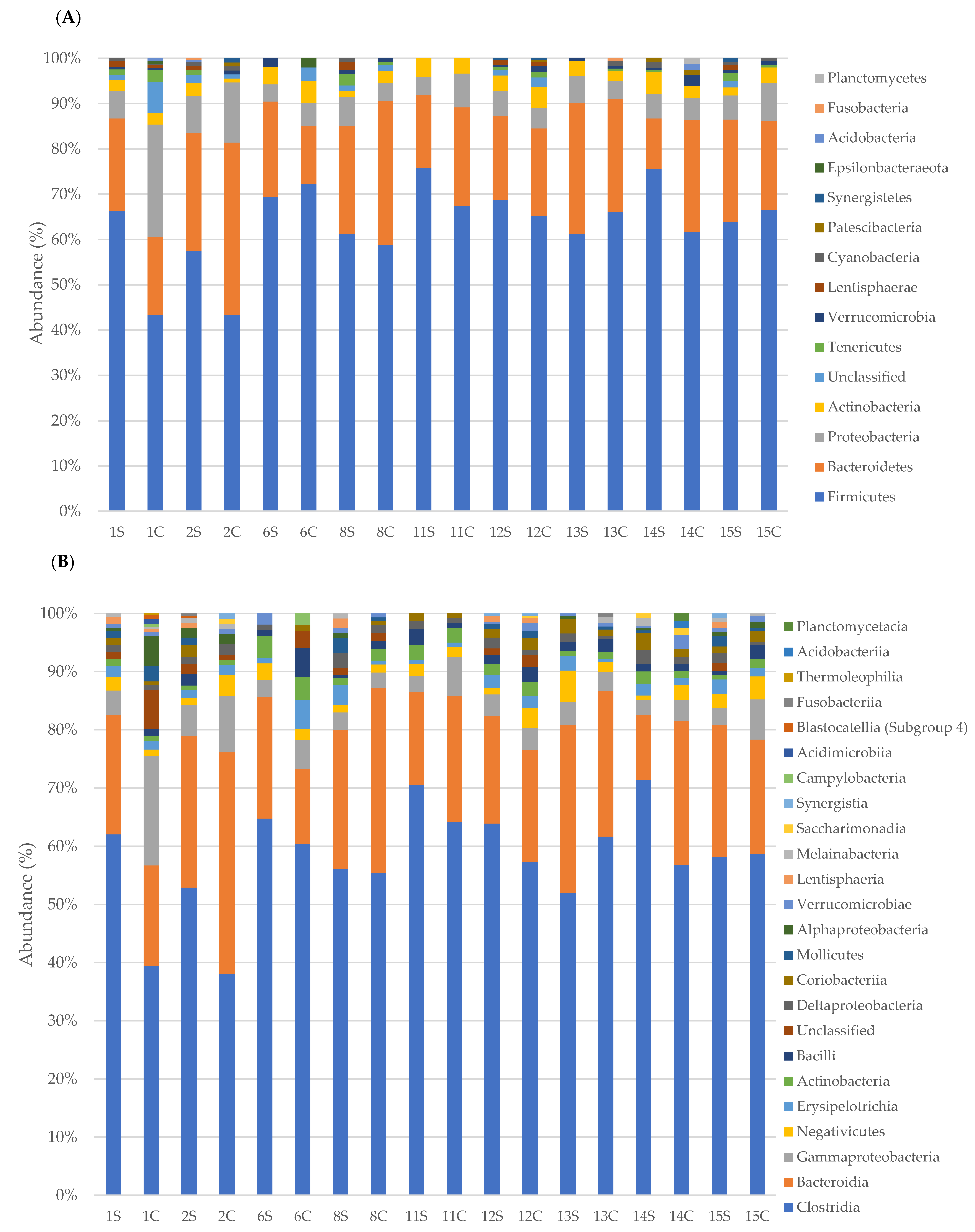
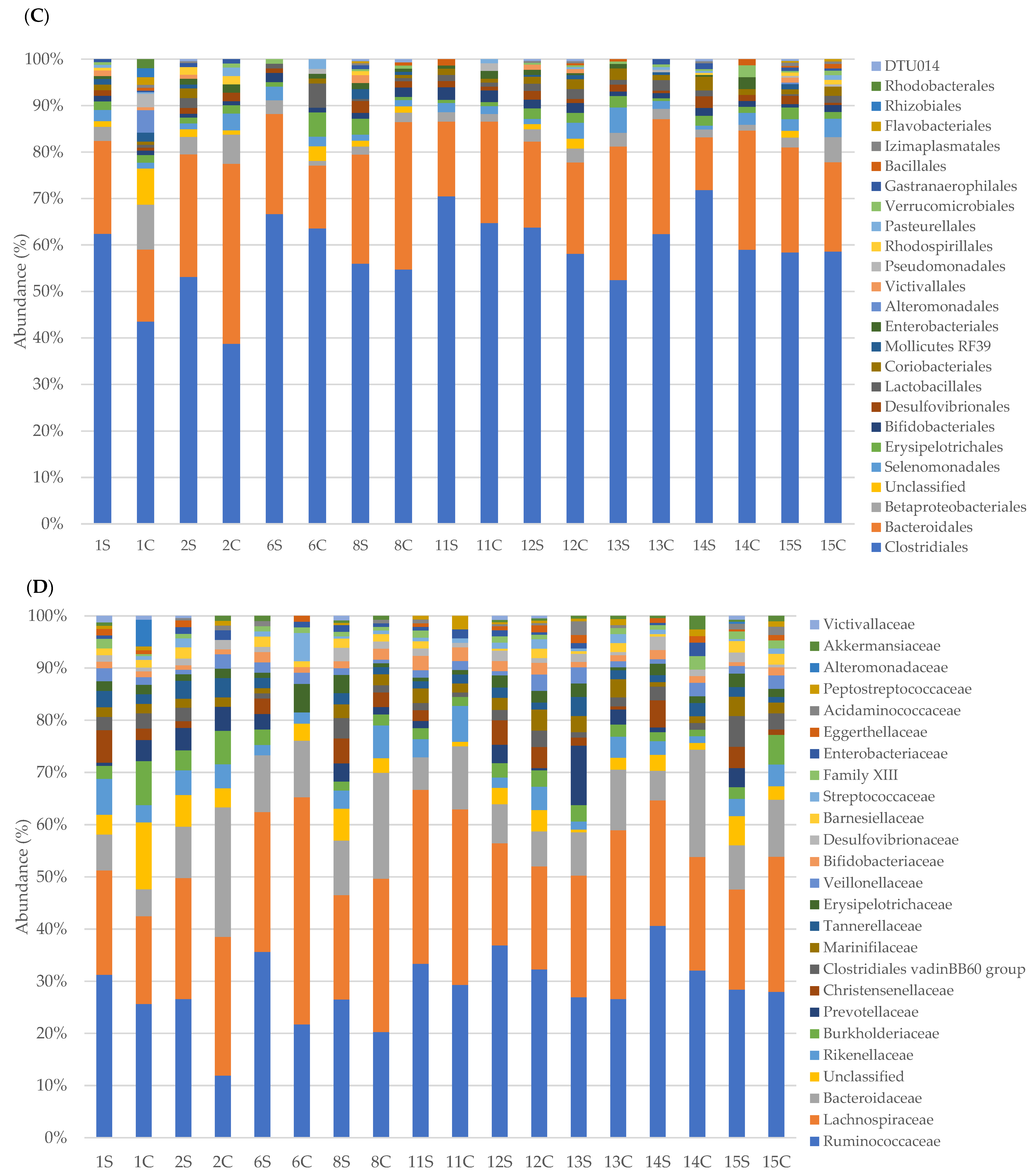
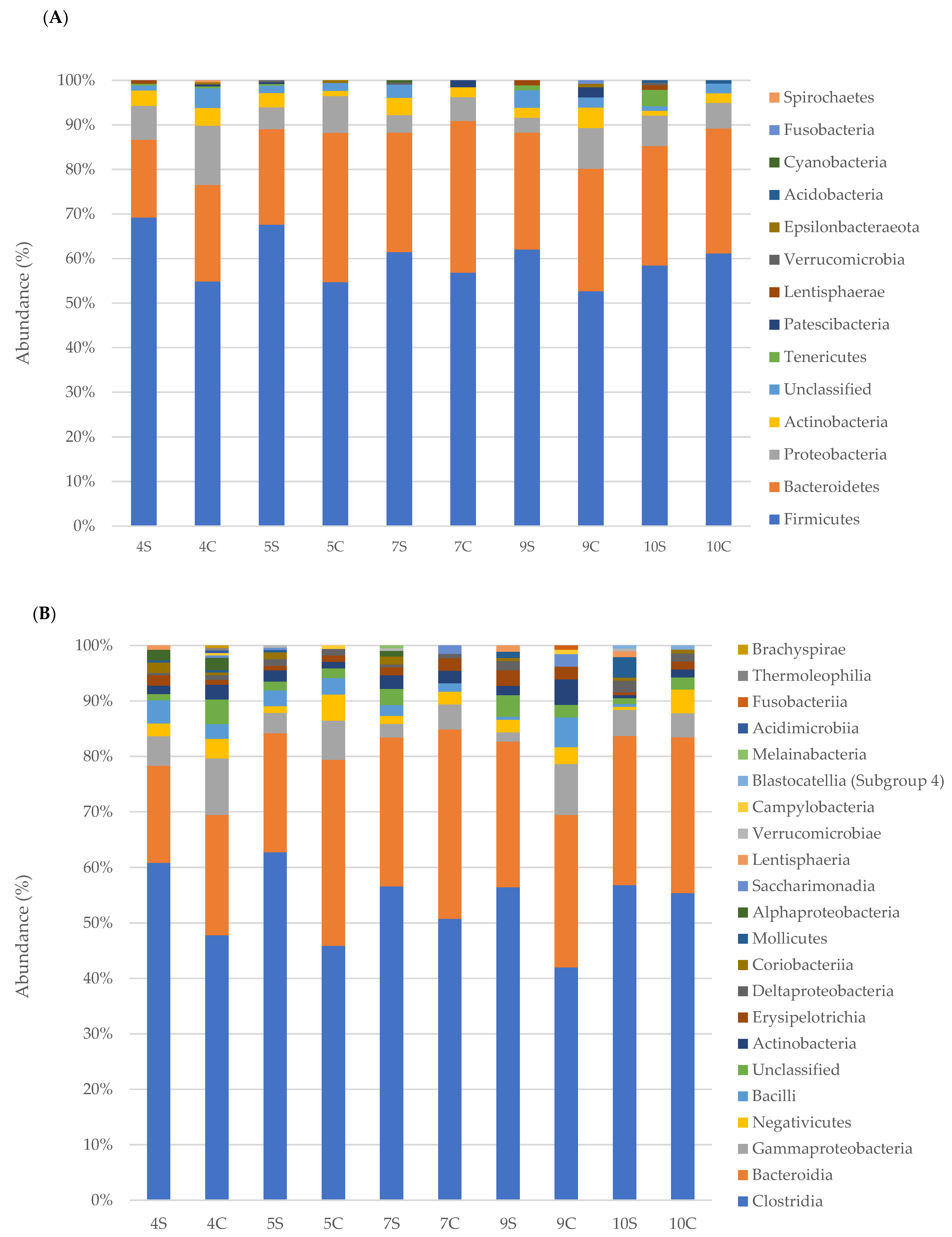
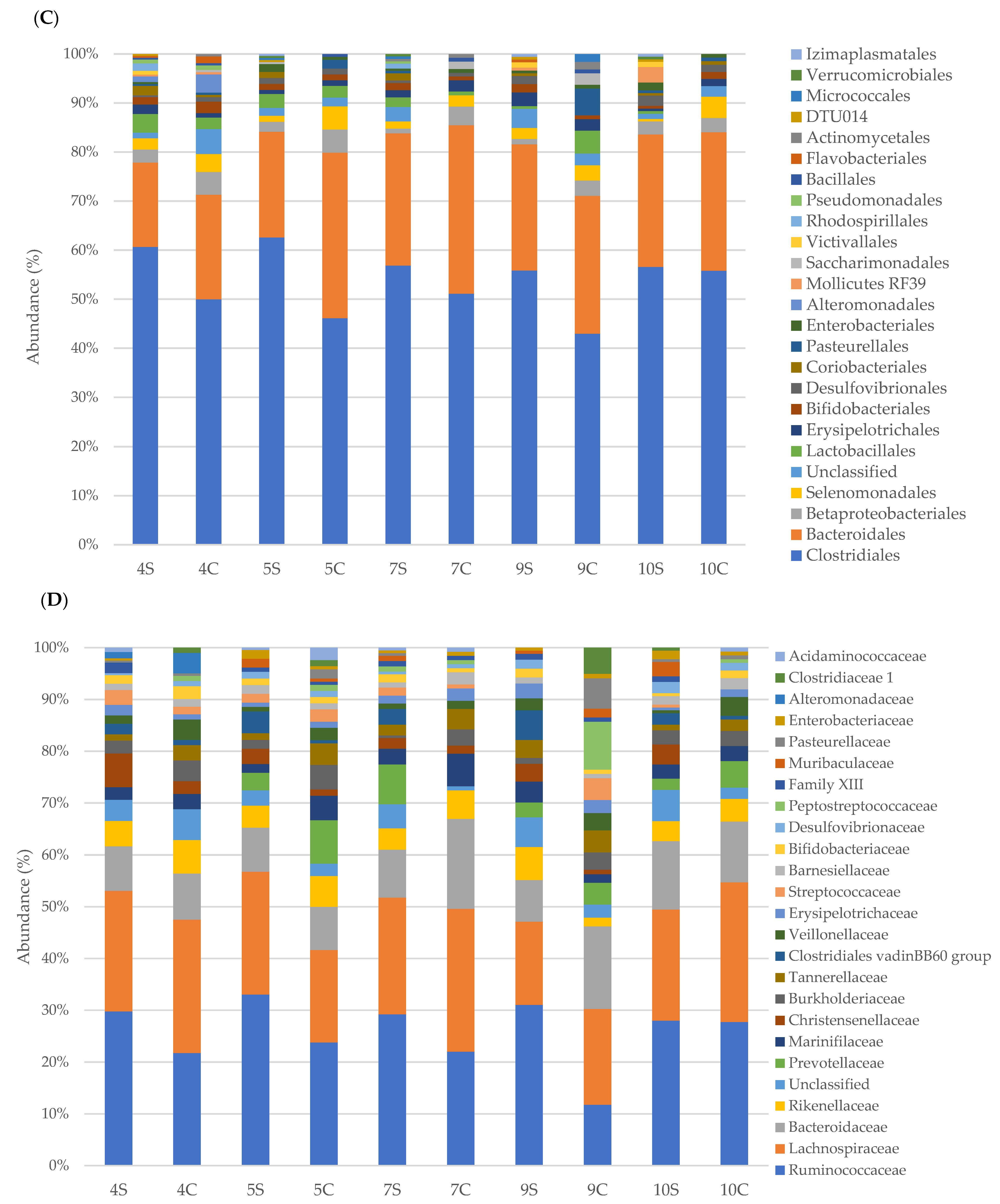
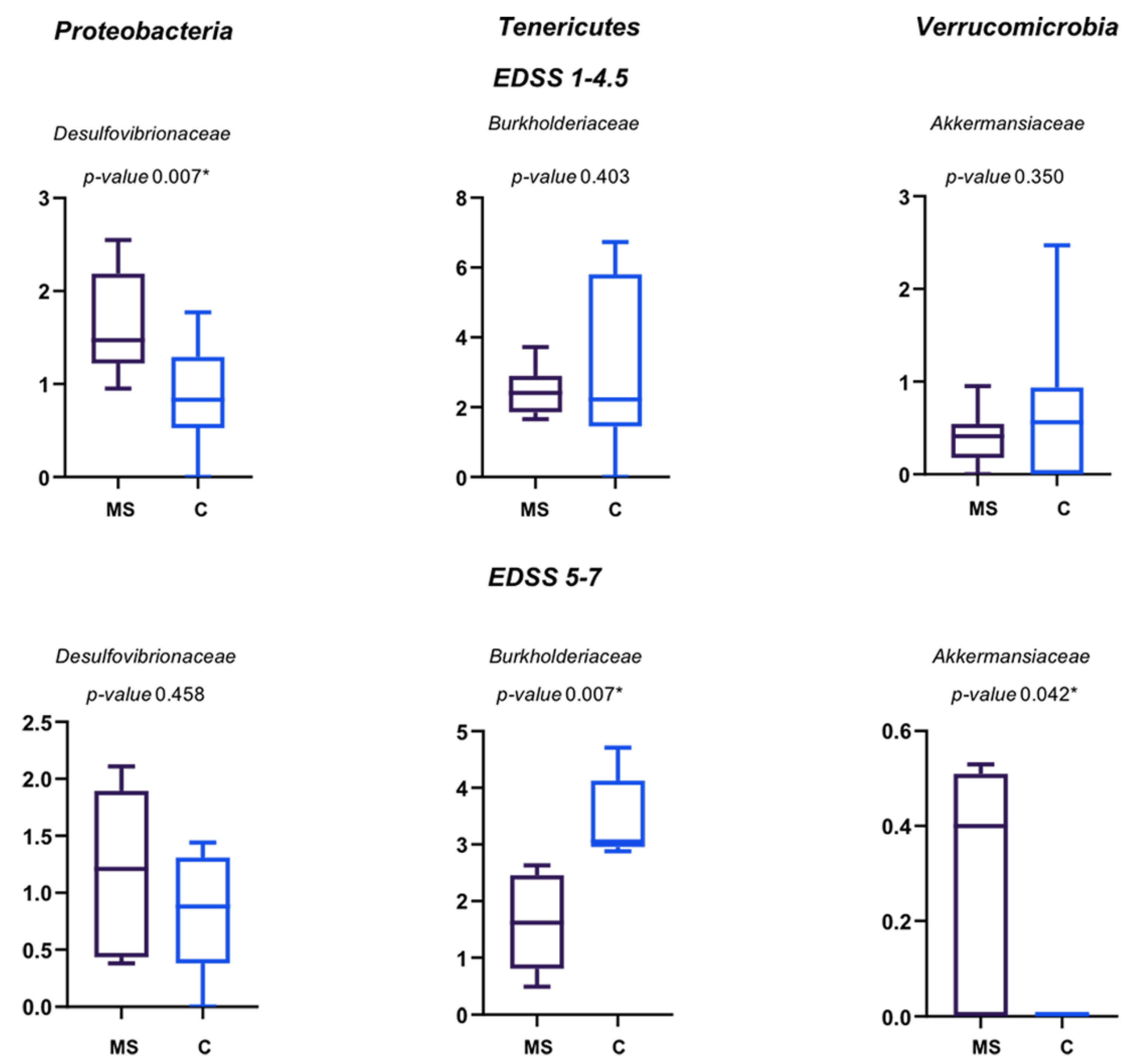
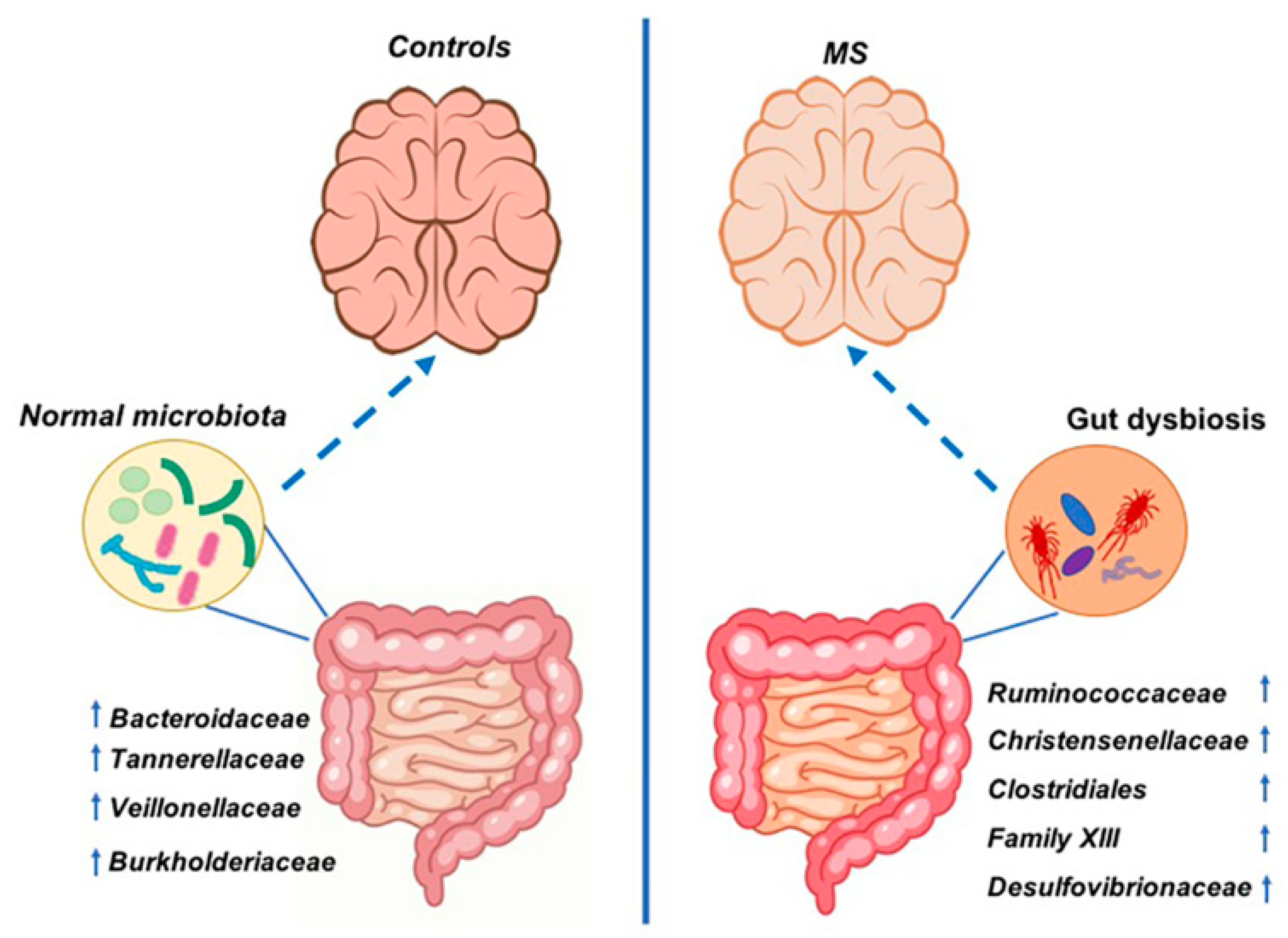
3.2. Faecal Bacterial Communities’ Taxonomic Composition
3.3. Phenotypic and Metabolic Inference
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ransohoff, R.M.; Hafler, D.A.; Lucchinetti, C.F. Multiple sclerosis-a quiet revolution. Nat. Rev. Neurol. 2015, 11, 134–142. [Google Scholar] [CrossRef] [Green Version]
- Baecher-Allan, C.; Kaskow, B.J.; Weiner, H.L. Multiple Sclerosis: Mechanisms and Immunotherapy. Neuron 2018, 97, 742–768. [Google Scholar] [CrossRef] [Green Version]
- Fresegna, D.; Bullitta, S.; Musella, A.; Rizzo, F.R.; De Vito, F.; Guadalupi, L.; Caioli, S.; Balletta, S.; Sanna, K.; Dolcetti, E.; et al. Re-Examining the Role of TNF in MS Pathogenesis and Therapy. Cells 2020, 9, 2290. [Google Scholar] [CrossRef]
- Noto, D.; Miyake, S. Gut dysbiosis and multiple sclerosis. Clin. Immunol. 2020, 108380, in press. [Google Scholar] [CrossRef]
- Lublin, F.D.; Reingold, S.C.; Cohen, J.A.; Cutter, G.R.; Sorensen, P.S.; Thompson, A.J.; Wolinsky, J.S.; Balcer, L.J.; Banwell, B.; Barkhof, F.; et al. Defining the clinical course of multiple sclerosis: The 2013 revisions. Neurology 2014, 83, 278–286. [Google Scholar] [CrossRef] [Green Version]
- Klineova, S.; Lublin, F.D. Clinical Course of Multiple Sclerosis. Cold Spring Harb. Perspect. Med. 2018, 8, a028928. [Google Scholar] [CrossRef]
- Fletcher, J.M.; Lalor, S.J.; Sweeney, C.M.; Tubridy, N.; Mills, K.H. T cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Clin. Exp. Immunol. 2010, 162, 1–11. [Google Scholar] [CrossRef]
- Legroux, L.; Arbour, N. Multiple Sclerosis and T Lymphocytes: An Entangled Story. J. Neuroimmune Pharmacol. Off. J. Soc. NeuroImmune Pharmacol. 2015, 10, 528–546. [Google Scholar] [CrossRef] [Green Version]
- Cosorich, I.; Dalla-Costa, G.; Sorini, C.; Ferrarese, R.; Messina, M.J.; Dolpady, J.; Radice, E.; Mariani, A.; Testoni, P.A.; Canducci, F.; et al. High frequency of intestinal TH17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci. Adv. 2017, 3, e1700492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balasa, R.; Barcutean, L.; Balasa, A.; Motataianu, A.; Roman-Filip, C.; Manu, D. The action of TH17 cells on blood brain barrier in multiple sclerosis and experimental autoimmune encephalomyelitis. Hum. Immunol. 2020, 81, 237–243. [Google Scholar] [CrossRef]
- Handel, A.E.; Giovannoni, G.; Ebers, G.C.; Ramagopalan, S.V. Environmental factors and their timing in adult-onset multiple sclerosis. Nat. Rev. Neurol. 2010, 6, 156–166. [Google Scholar] [CrossRef]
- Fung, T.C.; Olson, C.A.; Hsiao, E.Y. Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 2017, 20, 145–155. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Kho, Z.Y.; Lal, S.K. The Human Gut Microbiome—A Potential Controller of Wellness and Disease. Front. Microbiol. 2018, 9, 1835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stojanov, S.; Berlec, A.; Strukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Rajilic-Stojanovic, M.; Figueiredo, C.; Smet, A.; Hansen, R.; Kupcinskas, J.; Rokkas, T.; Andersen, L.; Machado, J.C.; Ianiro, G.; Gasbarrini, A.; et al. Systematic review: Gastric microbiota in health and disease. Aliment. Pharmacol. Ther. 2020, 51, 582–602. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Fassarella, M.; Blaak, E.E.; Penders, J.; Nauta, A.; Smidt, H.; Zoetendal, E.G. Gut microbiome stability and resilience: Elucidating the response to perturbations in order to modulate gut health. Gut 2021, 70, 595–605. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Waubant, E.; Chehoud, C.; Kuczynski, J.; DeSantis, T.Z.; Warrington, J.; Venkatesan, A.; Fraser, C.M.; Mowry, E.M. Gut microbiota in multiple sclerosis: Possible influence of immunomodulators. J. Investig. Med. 2015, 63, 729–734. [Google Scholar] [CrossRef]
- Miyake, S.; Kim, S.; Suda, W.; Oshima, K.; Nakamura, M.; Matsuoka, T.; Chihara, N.; Tomita, A.; Sato, W.; Kim, S.W.; et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS ONE 2015, 10, e0137429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jangi, S.; Gandhi, R.; Cox, L.M.; Li, N.; von Glehn, F.; Yan, R.; Patel, B.; Mazzola, M.A.; Liu, S.; Glanz, B.L.; et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016, 7, 12015. [Google Scholar] [CrossRef]
- Tremlett, H.; Fadrosh, D.W.; Faruqi, A.A.; Zhu, F.; Hart, J.; Roalstad, S.; Graves, J.; Lynch, S.; Waubant, E.; Centers, U.S.N.o.P.M. Gut microbiota in early pediatric multiple sclerosis: A case-control study. Eur. J. Neurol. 2016, 23, 1308–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berer, K.; Gerdes, L.A.; Cekanaviciute, E.; Jia, X.; Xiao, L.; Xia, Z.; Liu, C.; Klotz, L.; Stauffer, U.; Baranzini, S.E.; et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. USA 2017, 114, 10719–10724. [Google Scholar] [CrossRef] [Green Version]
- Ling, Z.; Cheng, Y.; Yan, X.; Shao, L.; Liu, X.; Zhou, D.; Zhang, L.; Yu, K.; Zhao, L. Alterations of the Fecal Microbiota in Chinese Patients With Multiple Sclerosis. Front. Immunol. 2020, 11, 590783. [Google Scholar] [CrossRef] [PubMed]
- Reynders, T.; Devolder, L.; Valles-Colomer, M.; Van Remoortel, A.; Joossens, M.; De Keyser, J.; Nagels, G.; D’Hooghe, M.; Raes, J. Gut microbiome variation is associated to Multiple Sclerosis phenotypic subtypes. Ann. Clin. Transl. Neurol. 2020, 7, 406–419. [Google Scholar] [CrossRef]
- Takewaki, D.; Suda, W.; Sato, W.; Takayasu, L.; Kumar, N.; Kimura, K.; Kaga, N.; Mizuno, T.; Miyake, S.; Hattori, M.; et al. Alterations of the gut ecological and functional microenvironment in different stages of multiple sclerosis. Proc. Natl. Acad. Sci. USA 2020, 117, 22402–22412. [Google Scholar] [CrossRef]
- Chen, J.; Chia, N.; Kalari, K.R.; Yao, J.Z.; Novotna, M.; Paz Soldan, M.M.; Luckey, D.H.; Marietta, E.V.; Jeraldo, P.R.; Chen, X.; et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 2016, 6, 28484. [Google Scholar] [CrossRef] [Green Version]
- Lax, S.; Smith, D.P.; Hampton-Marcell, J.; Owens, S.M.; Handley, K.M.; Scott, N.M.; Gibbons, S.M.; Larsen, P.; Shogan, B.D.; Weiss, S.; et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 2014, 345, 1048–1052. [Google Scholar] [CrossRef] [Green Version]
- Cignarella, F.; Cantoni, C.; Ghezzi, L.; Salter, A.; Dorsett, Y.; Chen, L.; Phillips, D.; Weinstock, G.M.; Fontana, L.; Cross, A.H.; et al. Intermittent Fasting Confers Protection in CNS Autoimmunity by Altering the Gut Microbiota. Cell Metab. 2018, 27, 1222–1235. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Nishida, A.; Fujimoto, T.; Fujii, M.; Shioya, M.; Imaeda, H.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Andoh, A. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn’s Disease. Digestion 2016, 93, 59–65. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glockner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Clarke, K.R.; Gorley, R.N. PRIMER v6: User Manual/Tutorial (Plymouth Routines in Multivariate Ecological Research); Primer-E Ltd.: Plymouth, UK, 2006; Available online: https://www.scienceopen.com/document?vid=2cd68314-640b-4288-8316-532e8932d7a1 (accessed on 20 May 2021).
- Arndt, D.; Xia, J.; Liu, Y.; Zhou, Y.; Guo, A.C.; Cruz, J.A.; Sinelnikov, I.; Budwill, K.; Nesbo, C.L.; Wishart, D.S. METAGENassist: A comprehensive web server for comparative metagenomics. Nucleic Acids Res. 2012, 40, W88–W95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009, 1, 6ra14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.J.; Lauber, C.; Costello, E.K.; Lozupone, C.A.; Humphrey, G.; Berg-Lyons, D.; Caporaso, J.G.; Knights, D.; Clemente, J.C.; Nakielny, S.; et al. Cohabiting family members share microbiota with one another and with their dogs. eLife 2013, 2, e00458. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Di Rienzi, S.C.; Poole, A.C.; Koren, O.; Walters, W.A.; Caporaso, J.G.; Knight, R.; Ley, R.E. Conducting a microbiome study. Cell 2014, 158, 250–262. [Google Scholar] [CrossRef] [Green Version]
- Abeles, S.R.; Jones, M.B.; Santiago-Rodriguez, T.M.; Ly, M.; Klitgord, N.; Yooseph, S.; Nelson, K.E.; Pride, D.T. Microbial diversity in individuals and their household contacts following typical antibiotic courses. Microbiome 2016, 4, 39. [Google Scholar] [CrossRef] [Green Version]
- iMSMS Consortium. Household paired design reduces variance and increases power in multi-city gut microbiome study in multiple sclerosis. Mult. Scler. 2020, 27, 366–379. [Google Scholar] [CrossRef]
- Hill-Burns, E.M.; Debelius, J.W.; Morton, J.T.; Wissemann, W.T.; Lewis, M.R.; Wallen, Z.D.; Peddada, S.D.; Factor, S.A.; Molho, E.; Zabetian, C.P.; et al. Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Mov. Disord. 2017, 32, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Tan, C.; Zhu, J.; Zeng, X.; Gao, X.; Wu, Q.; Chen, Q.; Wang, H.; Zhou, H.; He, Y.; et al. Dysbiosis of the intestinal microbiota in neurocritically ill patients and the risk for death. Crit. Care 2019, 23, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghaly, S.; Kaakoush, N.O.; Lloyd, F.; Gordon, L.; Forest, C.; Lawrance, I.C.; Hart, P.H. Ultraviolet Irradiation of Skin Alters the Faecal Microbiome Independently of Vitamin D in Mice. Nutrients 2018, 10, 1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riccio, P.; Rossano, R. Diet, Gut Microbiota, and Vitamins D + A in Multiple Sclerosis. Neurotherapeutics 2018, 15, 75–91. [Google Scholar] [CrossRef] [Green Version]
- Rumah, K.R.; Linden, J.; Fischetti, V.A.; Vartanian, T. Isolation of Clostridium perfringens type B in an individual at first clinical presentation of multiple sclerosis provides clues for environmental triggers of the disease. PLoS ONE 2013, 8, e76359. [Google Scholar] [CrossRef]
- Loubinoux, J.; Bronowicki, J.P.; Pereira, I.A.; Mougenel, J.L.; Faou, A.E. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol. Ecol. 2002, 40, 107–112. [Google Scholar] [CrossRef] [PubMed]



| Cohort | n. Subjects | ID Family | Sex (M/F) | Age Range | EDSS |
|---|---|---|---|---|---|
| Controls | 15 | - | 8/7 | 21–69 | - |
| RRMS | 9 | 1,2,6,8,11, 12,13,14,15 | 3/6 | 28–65 | 1–4.5 |
| RRMS | 5 | 4,5,7,9,10 | 1/4 | 54–66 | 5–7 |
| PPMS | 1 | 3 | 0/1 | 57 | 7.5 |
| Sample | S | Good’s Coverage | Chao1 | ACE | α | 1-D | H’ | e |
|---|---|---|---|---|---|---|---|---|
| 1S | 27 | 1.00 | 96.87 | 89.33 | 6.14 | 0.14 | 2.48 | 0.75 |
| 1C | 60 | 0.99 | 99.39 | 90.94 | 5.7 | 0.08 | 3.13 | 0.76 |
| 2S | 29 | 0.99 | 102.45 | 93.46 | 8.34 | 0.13 | 2.51 | 0.74 |
| 2C | 20 | 1.00 | 102.76 | 94.44 | 5.65 | 0.14 | 2.32 | 0.77 |
| 3S | 31 | 1.00 | 104.16 | 95.97 | 3.12 | 0.09 | 2.80 | 0.81 |
| 3C | 21 | 0.99 | 106.54 | 97.32 | 10.9 | 0.18 | 2.14 | 0.70 |
| 4S | 35 | 0.99 | 107.76 | 98.98 | 7.51 | 0.13 | 2.55 | 0.71 |
| 4C | 39 | 1.00 | 111.43 | 101.82 | 5.79 | 0.10 | 2.78 | 0.76 |
| 5S | 31 | 0.99 | 115.32 | 104.3 | 7.96 | 0.16 | 2.41 | 0.70 |
| 5C | 26 | 1.00 | 117 | 105.95 | 6.53 | 0.10 | 2.61 | 0.80 |
| 6S | 23 | 0.99 | 117.71 | 106.6 | 4.56 | 0.19 | 2.16 | 0.69 |
| 6C | 19 | 1.00 | 117.52 | 107.63 | 5.31 | 0.20 | 2.07 | 0.70 |
| 7S | 25 | 1.00 | 117.46 | 108.67 | 5.28 | 0.14 | 2.31 | 0.71 |
| 7C | 32 | 1.00 | 118.21 | 109.54 | 6.40 | 0.14 | 2.50 | 0.72 |
| 8S | 26 | 0.99 | 118.66 | 110.2 | 9.03 | 0.12 | 2.51 | 0.77 |
| 8C | 25 | 1.00 | 119.18 | 111.56 | 5.92 | 0.15 | 2.28 | 0.70 |
| 9S | 24 | 0.99 | 121.68 | 113.24 | 7.48 | 0.13 | 2.47 | 0.77 |
| 9C | 29 | 1.00 | 122.43 | 114.51 | 4.51 | 0.07 | 2.86 | 0.85 |
| 10S | 29 | 0.99 | 123.61 | 115.65 | 6.55 | 0.13 | 2.48 | 0.73 |
| 10C | 21 | 1.00 | 126.93 | 118.21 | 6.61 | 0.16 | 2.24 | 0.73 |
| 11S | 26 | 0.99 | 43.27 | 41.56 | 5.73 | 0.21 | 2.13 | 0.65 |
| 11C | 20 | 1.00 | 59.09 | 50.84 | 6 | 0.20 | 2.03 | 0.68 |
| 12S | 31 | 0.99 | 127.94 | 119.14 | 8.58 | 0.17 | 2.42 | 0.70 |
| 12C | 35 | 0.99 | 128.72 | 119.86 | 6.82 | 0.13 | 2.61 | 0.73 |
| 13S | 29 | 1.00 | 68.26 | 61.7 | 7.03 | 0.13 | 2.49 | 0.74 |
| 13C | 28 | 1.00 | 73.39 | 69.46 | 6.42 | 0.17 | 2.28 | 0.68 |
| 14S | 29 | 1.00 | 80.15 | 74.66 | 8.31 | 0.20 | 2.21 | 0.65 |
| 14C | 21 | 0.99 | 82.23 | 78.6 | 3.85 | 0.17 | 2.17 | 0.71 |
| 15S | 32 | 0.99 | 86.94 | 84.29 | 8.81 | 0.12 | 2.59 | 0.74 |
| 15C | 29 | 1.00 | 91.01 | 85.36 | 7 | 0.14 | 2.45 | 0.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galluzzo, P.; Capri, F.C.; Vecchioni, L.; Realmuto, S.; Scalisi, L.; Cottone, S.; Nuzzo, D.; Alduina, R. Comparison of the Intestinal Microbiome of Italian Patients with Multiple Sclerosis and Their Household Relatives. Life 2021, 11, 620. https://doi.org/10.3390/life11070620
Galluzzo P, Capri FC, Vecchioni L, Realmuto S, Scalisi L, Cottone S, Nuzzo D, Alduina R. Comparison of the Intestinal Microbiome of Italian Patients with Multiple Sclerosis and Their Household Relatives. Life. 2021; 11(7):620. https://doi.org/10.3390/life11070620
Chicago/Turabian StyleGalluzzo, Paola, Fanny Claire Capri, Luca Vecchioni, Sabrina Realmuto, Luca Scalisi, Salvatore Cottone, Domenico Nuzzo, and Rosa Alduina. 2021. "Comparison of the Intestinal Microbiome of Italian Patients with Multiple Sclerosis and Their Household Relatives" Life 11, no. 7: 620. https://doi.org/10.3390/life11070620
APA StyleGalluzzo, P., Capri, F. C., Vecchioni, L., Realmuto, S., Scalisi, L., Cottone, S., Nuzzo, D., & Alduina, R. (2021). Comparison of the Intestinal Microbiome of Italian Patients with Multiple Sclerosis and Their Household Relatives. Life, 11(7), 620. https://doi.org/10.3390/life11070620








