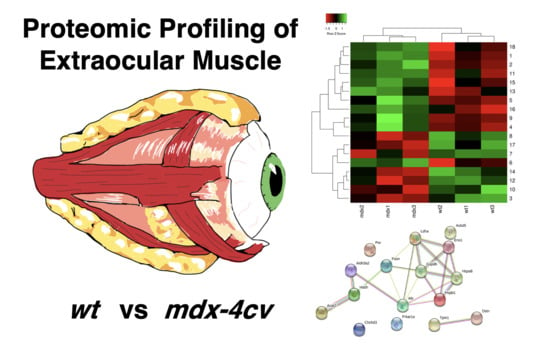
Mass Spectrometric Profiling of Extraocular Muscle and Proteomic Adaptations in the mdx-4cv Model of Duchenne Muscular Dystrophy
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Extraocular Muscle Specimens
2.3. Label-Free Liquid Chromatography Mass Spectrometry and Proteomic Data Analysis
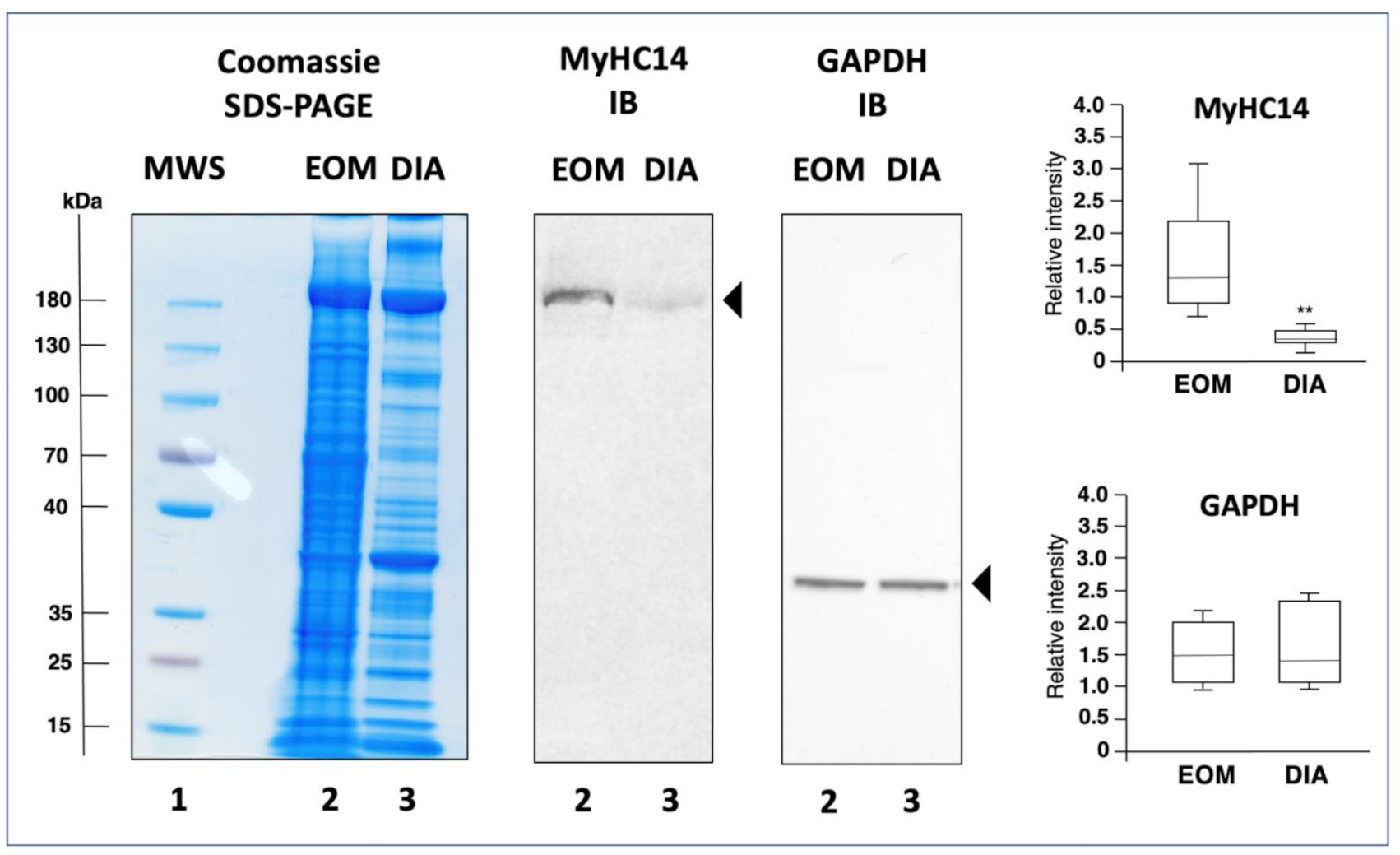
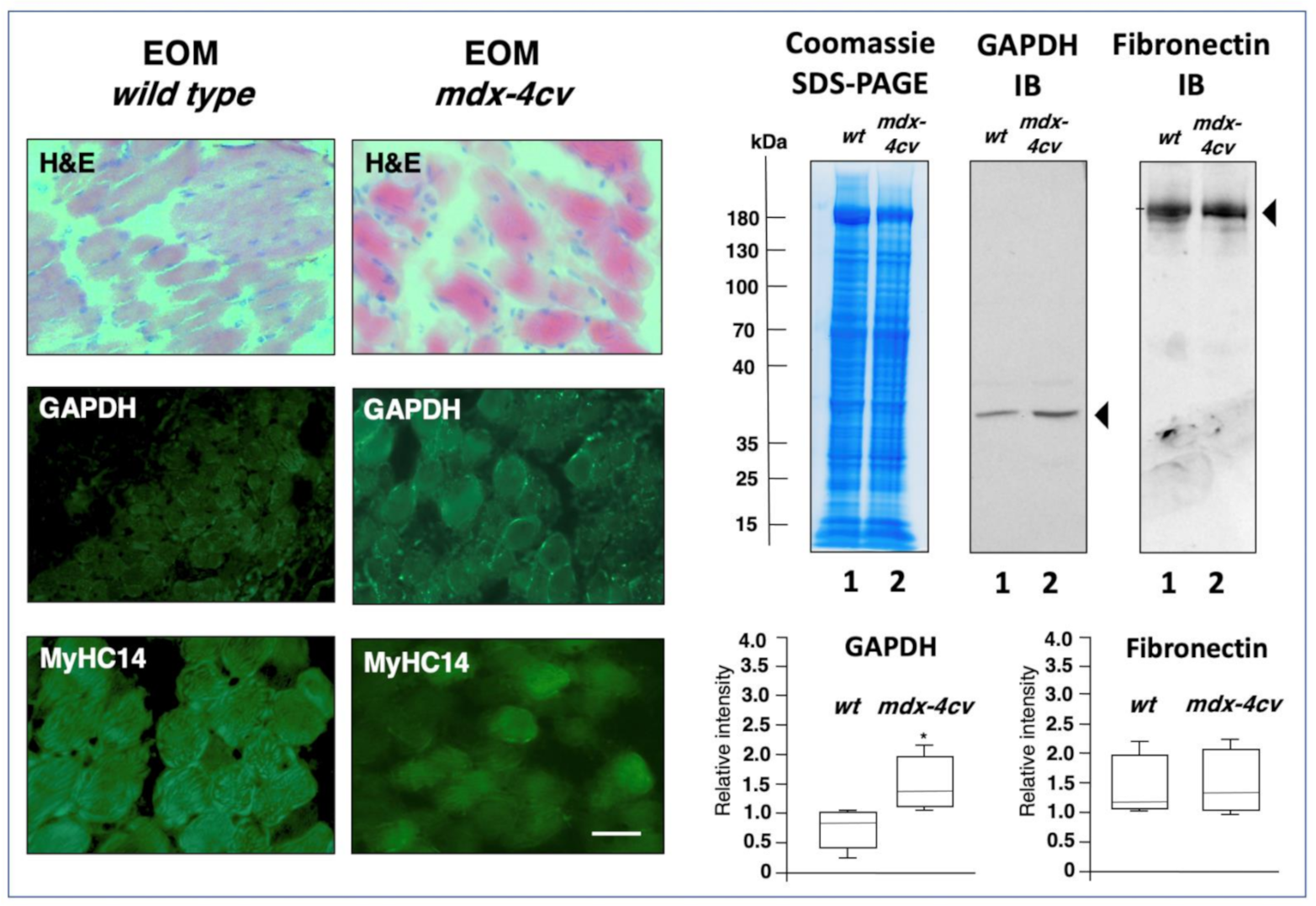
2.4. Comparative Immunoblot Analysis
2.5. Immunofluorescence Microscopy
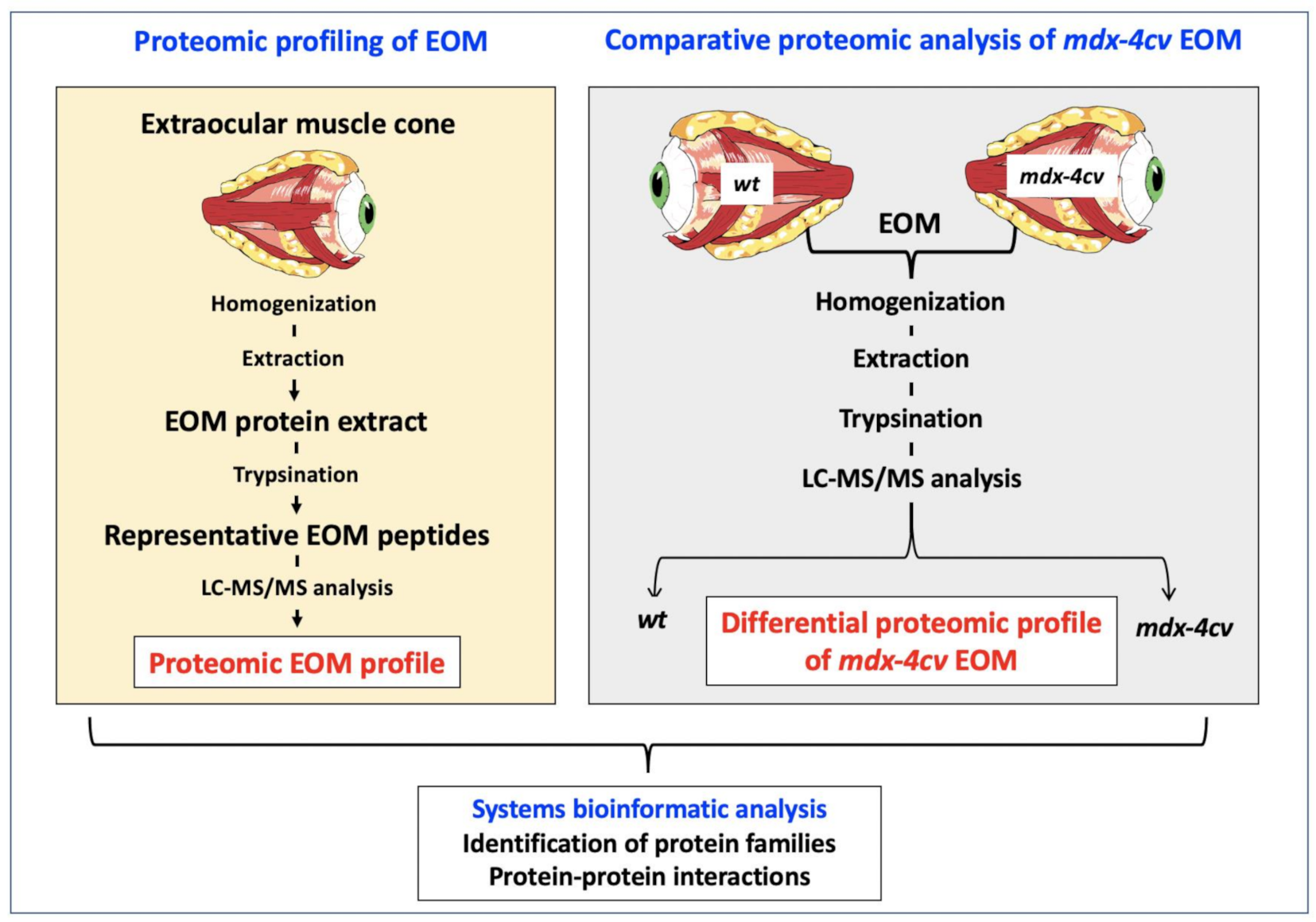
3. Results and Discussion
3.1. The Proteomic Profile of Extraocular Muscle
3.2. Proteomic Profile of the Sarcomere from Extraocular Muscle

3.3. Mass Spectrometric Identification of Proteins Specifically Expressed in Extraocular Muscle
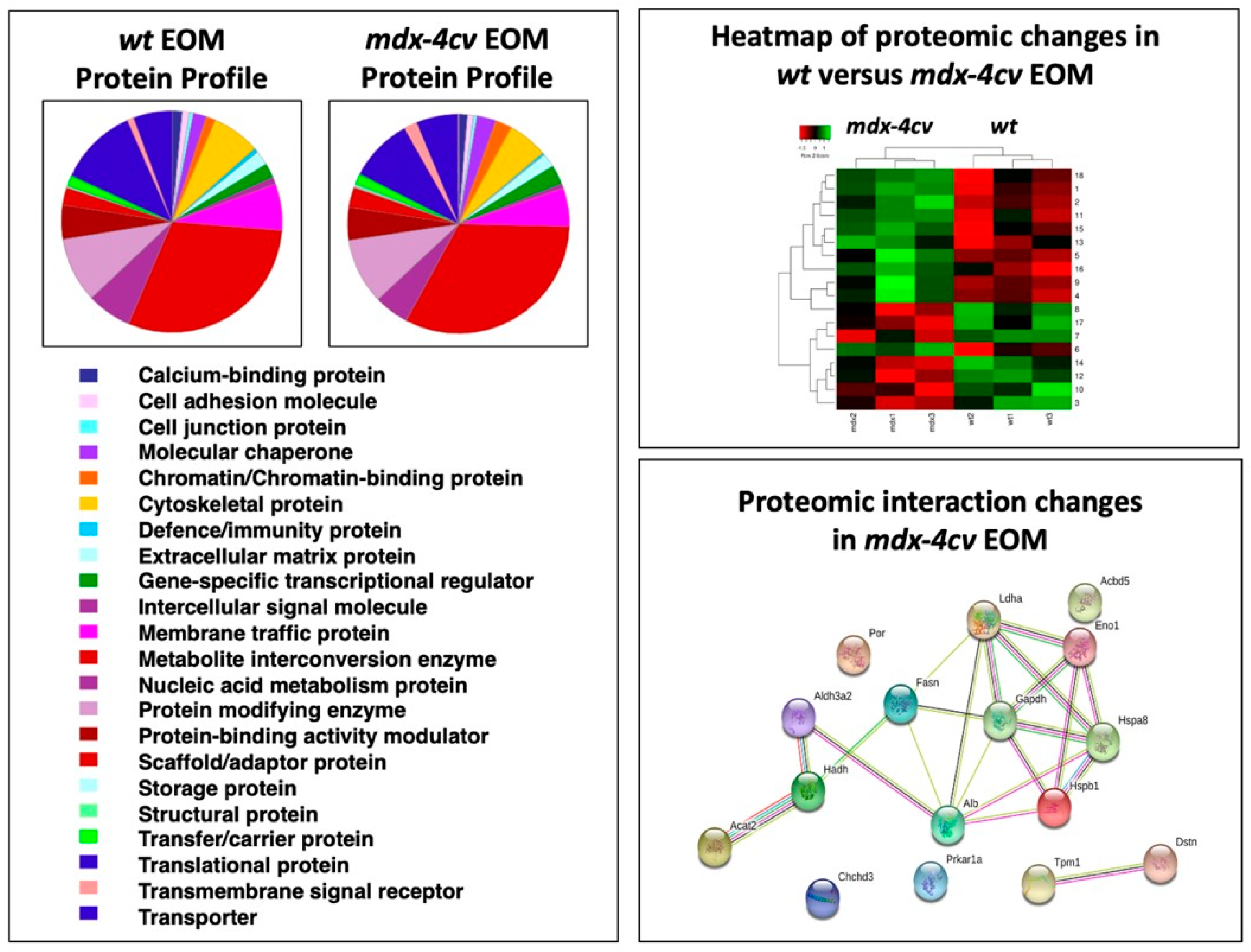
3.4. Comparative Proteomic Profiling of Extraocular Muscle from the Dystrophic mdx-4cv Model of Duchenne Muscular Dystrophy
| Accession | Protein | Gene | Peptides | Annova (p) | Fold Change |
|---|---|---|---|---|---|
| P16858 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 14 | 0.01427 | +18.45 |
| P14602 | Heat shock protein 1, beta | HSPB1 | 25 | 0.04444 | +15.62 |
| P63017 | Heat shock cognate 71 kDa protein | HSPA8 | 24 | 0.00894 | +10.22 |
| G3XA25 | Acetyl-CoA acetyltransferase 2 | ACAT2 | 24 | 0.04001 | +7.56 |
| P06151 | Lactate dehydrogenase 1, A chain | LDHA | 14 | 0.02856 | +5.93 |
| P19536 | Hydroxyacyl-coenzyme A dehydrogenase | HADH | 22 | 0.04369 | +3.38 |
| P17182 | Alpha-enolase | ENO1 | 19 | 0.04825 | +3.13 |
| P47740 | Aldehyde dehydrogenase family 3 member A2 | ALDH3A2 | 15 | 0.04301 | +2.94 |
| E9QNH7 | Acyl-CoA-binding domain-containing protein 5 | ACBD5 | 6 | 0.03134 | +2.43 |
| P07724 | Albumin | ALB | 45 | 0.03520 | +1.76 |
| P19096 | Fatty acid synthase | FASN | 119 | 0.02425 | +1.69 |
| G5E8R1 | Tropomyosin alpha-1 chain | TPM1 | 11 | 0.03985 | −1.98 |
| Q9DB20 | ATP synthase subunit O, mitochondrial | ATP5PO | 7 | 0.03449 | −2.07 |
| P37040 | NADPH-cytochrome P450 reductase | POR | 12 | 0.04813 | −2.45 |
| Q9R0P5 | Destrin | DSTN | 9 | 0.03484 | −2.66 |
| Q9D3D9 | ATP synthase subunit delta, mitochondrial | ATP5F1D | 4 | 0.04228 | −2.71 |
| Q9CRB9 | MICOS complex subunit Mic19 | CHCHD3 | 11 | 0.04353 | −2.90 |
| Q9DBC7 | cAMP-dependent protein kinase type I-alpha | PRKAR1A | 2 | 0.02217 | −2.91 |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blaauw, B.; Schiaffino, S.; Reggiani, C. Mechanisms modulating skeletal muscle phenotype. Compr. Physiol. 2013, 3, 1645–1687. [Google Scholar] [CrossRef]
- Verma, M.; Fitzpatrick, K.; McLoon, L.K. Extraocular Muscle Repair and Regeneration. Curr. Ophthalmol. Rep. 2017, 5, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Haladaj, R.; Polguj, M.; Tubbs, R.S. Comparison of the Superior and Inferior Rectus Muscles in Humans: An Anatomical Study with Notes on Morphology, Anatomical Variations, and Intramuscular Innervation Patterns. BioMed Res. Int. 2020, 2020, 9037693. [Google Scholar] [CrossRef] [PubMed]
- Spencer, R.F.; Porter, J.D. Biological organization of the extraocular muscles. Prog. Brain Res. 2006, 151, 43–80. [Google Scholar] [CrossRef]
- van der Pol, C.B.; Chakraborty, S.; Gao, J.; Nguyen, T.; Torres, C.; Glikstein, R. Imaging anatomy and pathology of extraocular muscles in adults. Can. Assoc. Radiol. J. 2014, 65, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Büttner-Ennever, J.A. Anatomy of the oculomotor system. Dev. Ophthalmol. 2007, 40, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sambasivan, R.; Gayraud-Morel, B.; Dumas, G.; Cimper, C.; Paisant, S.; Kelly, R.G.; Tajbakhsh, S. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev. Cell. 2009, 16, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, A.L.; Lewandoski, M.; Rudnicki, M.A.; Gage, P.J. Pitx2 is an upstream activator of extraocular myogenesis and survival. Dev. Biol. 2011, 349, 395–405. [Google Scholar] [CrossRef]
- Jacoby, J.; Chiarandini, D.J.; Stefani, E. Electrical properties and innervation of fibers in the orbital layer of rat extraocular muscles. J. Neurophysiol. 1989, 61, 116–125. [Google Scholar] [CrossRef]
- Hernández, R.G.; Calvo, P.M.; Blumer, R.; de la Cruz, R.R.; Pastor, A.M. Functional diversity of motoneurons in the oculomotor system. Proc. Natl. Acad. Sci. USA 2019, 116, 3837–3846. [Google Scholar] [CrossRef]
- Liu, J.X.; Domellöf, F.P. A Novel Type of Multiterminal Motor Endplate in Human Extraocular Muscles. Investig. Ophthalmol. Vis. Sci. 2018, 59, 539–548. [Google Scholar] [CrossRef]
- Bruenech, J.R.; Kjellevold Haugen, I.B. How does the structure of extraocular muscles and their nerves affect their function? Eye (Lond.) 2015, 29, 177–183. [Google Scholar] [CrossRef]
- McLoon, L.K.; Rowe, J.; Wirtschafter, J.; McCormick, K.M. Continuous myofiber remodeling in uninjured extraocular myofibers: Myonuclear turnover and evidence for apoptosis. Muscle Nerve 2004, 29, 707–715. [Google Scholar] [CrossRef]
- Hebert, S.L.; Daniel, M.L.; McLoon, L.K. The role of Pitx2 in maintaining the phenotype of myogenic precursor cells in the extraocular muscles. PLoS ONE 2013, 8, e58405. [Google Scholar] [CrossRef]
- Sekulic-Jablanovic, M.; Ullrich, N.D.; Goldblum, D.; Palmowski-Wolfe, A.; Zorzato, F.; Treves, S. Functional characterization of orbicularis oculi and extraocular muscles. J. Gen. Physiol. 2016, 147, 395–406. [Google Scholar] [CrossRef]
- Asmussen, G.; Punkt, K.; Bartsch, B.; Soukup, T. Specific metabolic properties of rat oculorotatory extraocular muscles can be linked to their low force requirements. Investig. Ophthalmol. Vis. Sci. 2008, 49, 4865–4871. [Google Scholar] [CrossRef]
- Hoh, J.F.Y. Myosin heavy chains in extraocular muscle fibres: Distribution, regulation and function. Acta Physiol. (Oxf.) 2021, 231, e13535. [Google Scholar] [CrossRef]
- Khurana, T.S.; Prendergast, R.A.; Alameddine, H.S.; Tome, F.M.; Fardeau, M.; Arahata, K.; Sugita, H.; Kunkel, L.M. Absence of extraocular muscle pathology in Duchenne’s muscular dystrophy: Role for calcium homeostasis in extraocular muscle sparing. J. Exp. Med. 1995, 182, 467–475. [Google Scholar] [CrossRef]
- Andrade, F.H.; Porter, J.D.; Kaminski, H.J. Eye muscle sparing by the muscular dystrophies: Lessons to be learned? Microsc. Res. Tech. 2000, 48, 192–203. [Google Scholar] [CrossRef]
- Kallestad, K.M.; Hebert, S.L.; McDonald, A.A.; Daniel, M.L.; Cu, S.R.; McLoon, L.K. Sparing of extraocular muscle in aging and muscular dystrophies: A myogenic precursor cell hypothesis. Exp. Cell Res. 2011, 317, 873–885. [Google Scholar] [CrossRef]
- Thompson, R.; Spendiff, S.; Roos, A.; Bourque, P.R.; Warman Chardon, J.; Kirschner, J.; Horvath, R.; Lochmüller, H. Advances in the diagnosis of inherited neuromuscular diseases and implications for therapy development. Lancet Neurol. 2020, 19, 522–532. [Google Scholar] [CrossRef]
- Murphy, S.; Ohlendieck, K. The biochemical and mass spectrometric profiling of the dystrophin complexome from skeletal muscle. Comput. Struct. Biotechnol. J. 2015, 14, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Dowling, P.; Gargan, S.; Murphy, S.; Zweyer, M.; Sabir, H.; Swandulla, D.; Ohlendieck, K. The Dystrophin Node as Integrator of Cytoskeletal Organization, Lateral Force Transmission, Fiber Stability and Cellular Signaling in Skeletal Muscle. Proteomes 2021, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.G.; Whitehead, N.P.; Froehner, S.C. Absence of Dystrophin Disrupts Skeletal Muscle Signaling: Roles of Ca2+, Reactive Oxygen Species, and Nitric Oxide in the Development of Muscular Dystrophy. Physiol. Rev. 2016, 96, 253–305. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Tajrishi, M.M.; Ogura, Y.; Kumar, A. Wasting mechanisms in muscular dystrophy. Int. J. Biochem. Cell Biol. 2013, 45, 2266–2279. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, E.; Bönnemann, C.G.; Muntoni, F. Muscular dystrophies. Lancet 2019, 394, 2025–2038. [Google Scholar] [CrossRef]
- Starosta, A.; Konieczny, P. Therapeutic aspects of cell signaling and communication in Duchenne muscular dystrophy. Cell. Mol. Life Sci. 2021, in press. [Google Scholar] [CrossRef]
- Ohlendieck, K.; Swandulla, D. Molecular pathogenesis of Duchenne muscular dystrophy-related fibrosis. Pathologe 2017, 38, 21–29. [Google Scholar] [CrossRef]
- Birnkrant, D.J.; Bushby, K.; Bann, C.M.; Apkon, S.D.; Blackwell, A.; Brumbaugh, D.; Case, L.E.; Clemens, P.R.; Hadjiyannakis, S.; Pandya, S.; et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: Diagnosis, and neuromuscular, rehabilitation, endocrine, and gastrointestinal and nutritional management. Lancet Neurol. 2018, 17, 251–267. [Google Scholar] [CrossRef]
- Meyers, T.A.; Townsend, D. Cardiac Pathophysiology and the Future of Cardiac Therapies in Duchenne Muscular Dystrophy. Int. J. Mol. Sci. 2019, 20, 4098. [Google Scholar] [CrossRef]
- Carr, S.J.; Zahedi, R.P.; Lochmüller, H.; Roos, A. Mass spectrometry-based protein analysis to unravel the tissue pathophysiology in Duchenne muscular dystrophy. Proteom. Clin. Appl. 2018, 12. [Google Scholar] [CrossRef]
- Dowling, P.; Murphy, S.; Zweyer, M.; Raucamp, M.; Swandulla, D.; Ohlendieck, K. Emerging proteomic biomarkers of X-linked muscular dystrophy. Expert Rev. Mol. Diagn. 2019, 19, 739–755. [Google Scholar] [CrossRef]
- Holland, A.; Murphy, S.; Dowling, P.; Ohlendieck, K. Pathoproteomic profiling of the skeletal muscle matrisome in dystrophinopathy associated myofibrosis. Proteomics 2016, 16, 345–366. [Google Scholar] [CrossRef]
- Fraterman, S.; Zeiger, U.; Khurana, T.S.; Wilm, M.; Rubinstein, N.A. Quantitative proteomics profiling of sarcomere associated proteins in limb and extraocular muscle allotypes. Mol. Cell. Proteom. 2007, 6, 728–737. [Google Scholar] [CrossRef]
- Fraterman, S.; Zeiger, U.; Khurana, T.S.; Rubinstein, N.A.; Wilm, M. Combination of peptide OFFGEL fractionation and label-free quantitation facilitated proteomics profiling of extraocular muscle. Proteomics 2007, 7, 3404–3416. [Google Scholar] [CrossRef]
- Zhang, S.Z.; Xie, H.Q.; Xu, Y.; Li, X.Q.; Wei, R.Q.; Zhi, W.; Deng, L.; Qiu, L.; Yang, Z.M. Regulation of cell proliferation by fast Myosin light chain 1 in myoblasts derived from extraocular muscle, diaphragm and gastrocnemius. Exp. Biol. Med. (Maywood) 2008, 233, 1374–1384. [Google Scholar] [CrossRef]
- Lewis, C.; Ohlendieck, K. Proteomic profiling of naturally protected extraocular muscles from the dystrophin-deficient mdx mouse. Biochem. Biophys. Res. Commun. 2010, 396, 1024–1029. [Google Scholar] [CrossRef]
- Matsumura, C.Y.; Menezes de Oliveira, B.; Durbeej, M.; Marques, M.J. Isobaric Tagging-Based Quantification for Proteomic Analysis: A Comparative Study of Spared and Affected Muscles from mdx Mice at the Early Phase of Dystrophy. PLoS ONE 2013, 8, e65831. [Google Scholar] [CrossRef]
- Murphy, S.; Zweyer, M.; Raucamp, M.; Henry, M.; Meleady, P.; Swandulla, D.; Ohlendieck, K. Proteomic profiling of the mouse diaphragm and refined mass spectrometric analysis of the dystrophic phenotype. J. Muscle Res. Cell. Motil. 2019, 40, 9–28. [Google Scholar] [CrossRef]
- Dowling, P.; Gargan, S.; Zweyer, M.; Henry, M.; Meleady, P.; Swandulla, D.; Ohlendieck, K. Proteome-wide Changes in the mdx-4cv Spleen due to Pathophysiological Cross Talk with Dystrophin-Deficient Skeletal Muscle. iScience 2020, 23, 101500. [Google Scholar] [CrossRef]
- Dowling, P.; Gargan, S.; Zweyer, M.; Henry, M.; Meleady, P.; Swandulla, D.; Ohlendieck, K. Protocol for the Bottom-Up Proteomic Analysis of Mouse Spleen. STAR Protoc. 2020, 1, 100196. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.; Zweyer, M.; Henry, M.; Meleady, P.; Mundegar, R.R.; Swandulla, D.; Ohlendieck, K. Proteomic analysis of the sarcolemma-enriched fraction from dystrophic mdx-4cv skeletal muscle. J. Proteom. 2019, 191, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Dowling, P.; Gargan, S.; Zweyer, M.; Sabir, H.; Henry, M.; Meleady, P.; Swandulla, D.; Ohlendieck, K. Proteomic profiling of the interface between the stomach wall and the pancreas in dystrophinopathy. Eur. J. Transl. Myol. 2021, 31, 9627. [Google Scholar] [CrossRef]
- Antharavally, B.S.; Mallia, K.A.; Rangaraj, P.; Haney, P.; Bell, P.A. Quantitation of proteins using a dye-metal-based colorimetric protein assay. Anal. Biochem. 2009, 385, 342–345. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Dowling, P.; Zweyer, M.; Raucamp, M.; Henry, M.; Meleady, P.; Swandulla, D.; Ohlendieck, K. Proteomic and cell biological profiling of the renal phenotype of the mdx-4cv mouse model of Duchenne muscular dystrophy. Eur. J. Cell Biol. 2020, 99, 151059. [Google Scholar] [CrossRef]
- Murphy, S.; Zweyer, M.; Mundegar, R.R.; Henry, M.; Meleady, P.; Swandulla, D.; Ohlendieck, K. Concurrent Label-Free Mass Spectrometric Analysis of Dystrophin Isoform Dp427 and the Myofibrosis Marker Collagen in Crude Extracts from mdx-4cv Skeletal Muscles. Proteomes 2015, 3, 298–327. [Google Scholar] [CrossRef]
- Stuelsatz, P.; Yablonka-Reuveni, Z. Isolation of Mouse Periocular Tissue for Histological and Immunostaining Analyses of the Extraocular Muscles and Their Satellite Cells. Methods Mol. Biol. 2016, 1460, 101–127. [Google Scholar] [CrossRef]
- Holland, A.; Ohlendieck, K. Proteomic profiling of the contractile apparatus from skeletal muscle. Expert Rev. Proteom. 2013, 10, 239–257. [Google Scholar] [CrossRef]
- Thul, P.J.; Lindskog, C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018, 27, 233–244. [Google Scholar] [CrossRef]
- Deshmukh, A.S.; Murgia, M.; Nagaraj, N.; Treebak, J.T.; Cox, J.; Mann, M. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol. Cell. Proteom. 2015, 14, 841–853. [Google Scholar] [CrossRef]
- Deshmukh, A.S.; Steenberg, D.E.; Hostrup, M.; Birk, J.B.; Larsen, J.K.; Santos, A.; Kjøbsted, R.; Hingst, J.R.; Schéele, C.C.; Murgia, M.; et al. Deep muscle-proteomic analysis of freeze-dried human muscle biopsies reveals fiber type-specific adaptations to exercise training. Nat. Commun. 2021, 12, 304. [Google Scholar] [CrossRef]
- Murphy, S.; Ohlendieck, K. Proteomic profiling of large myofibrillar proteins from dried and long-term stored polyacrylamide gels. Anal. Biochem. 2018, 543, 8–11. [Google Scholar] [CrossRef]
- Dowling, P.; Zweyer, M.; Swandulla, D.; Ohlendieck, K. Characterization of Contractile Proteins from Skeletal Muscle Using Gel-Based Top-Down Proteomics. Proteomes 2019, 7, 25. [Google Scholar] [CrossRef]
- Henderson, C.A.; Gomez, C.G.; Novak, S.M.; Mi-Mi, L.; Gregorio, C.C. Overview of the Muscle Cytoskeleton. Compr. Physiol. 2017, 7, 891–944. [Google Scholar] [CrossRef]
- Rossi, A.C.; Mammucari, C.; Argentini, C.; Reggiani, C.; Schiaffino, S. Two novel/ancient myosins in mammalian skeletal muscles: MYH14/7b and MYH15 are expressed in extraocular muscles and muscle spindles. J. Physiol. 2010, 588, 353–364. [Google Scholar] [CrossRef]
- Lee, L.A.; Karabina, A.; Broadwell, L.J.; Leinwand, L.A. The ancient sarcomeric myosins found in specialized muscles. Skelet. Muscle 2019, 9, 7. [Google Scholar] [CrossRef]
- Sitbon, Y.H.; Yadav, S.; Kazmierczak, K.; Szczesna-Cordary, D. Insights into myosin regulatory and essential light chains: A focus on their roles in cardiac and skeletal muscle function, development and disease. J. Muscle Res. Cell. Motil. 2020, 41, 313–327. [Google Scholar] [CrossRef]
- Leber, Y.; Ruparelia, A.A.; Kirfel, G.; van der Ven, P.F.; Hoffmann, B.; Merkel, R.; Bryson-Richardson, R.J.; Fürst, D.O. Filamin C is a highly dynamic protein associated with fast repair of myofibrillar microdamage. Hum. Mol. Genet. 2016, 25, 2776–2788. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.Y.; Ackermann, M.A.; Kontrogianni-Konstantopoulos, A. The sarcomeric M-region: A molecular command center for diverse cellular processes. Biomed. Res. Int. 2015, 2015, 714197. [Google Scholar] [CrossRef] [PubMed]
- Briggs, M.M.; Schachat, F. The superfast extraocular myosin (MYH13) is localized to the innervation zone in both the global and orbital layers of rabbit extraocular muscle. J. Exp. Biol. 2002, 205, 3133–3142. [Google Scholar] [CrossRef] [PubMed]
- Squire, J. Special Issue: The Actin-Myosin Interaction in Muscle: Background and Overview. Int. J. Mol. Sci. 2019, 20, 5715. [Google Scholar] [CrossRef]
- Banks, G.B.; Combs, A.C.; Chamberlain, J.S. Sequencing protocols to genotype mdx, mdx(4cv), and mdx(5cv) mice. Muscle Nerve 2010, 42, 268–270. [Google Scholar] [CrossRef]
- Bengtsson, N.E.; Hall, J.K.; Odom, G.L.; Phelps, M.P.; Andrus, C.R.; Hawkins, R.D.; Hauschka, S.D.; Chamberlain, J.R.; Chamberlain, J.S. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun. 2017, 8, 14454. [Google Scholar] [CrossRef]
- Tichy, E.D.; Mourkioti, F. A new method of genotyping MDX4CV mice by PCR-RFLP analysis. Muscle Nerve 2017, 56, 522–524. [Google Scholar] [CrossRef]
- Holland, A.; Dowling, P.; Meleady, P.; Henry, M.; Zweyer, M.; Mundegar, R.R.; Swandulla, D.; Ohlendieck, K. Label-free mass spectrometric analysis of the mdx-4cv diaphragm identifies the matricellular protein periostin as a potential factor involved in dystrophinopathy-related fibrosis. Proteomics 2015, 15, 2318–2331. [Google Scholar] [CrossRef]
- Murphy, S.; Dowling, P.; Zweyer, M.; Mundegar, R.R.; Henry, M.; Meleady, P.; Swandulla, D.; Ohlendieck, K. Proteomic analysis of dystrophin deficiency and associated changes in the aged mdx-4cv heart model of dystrophinopathy-related cardiomyopathy. J. Proteom. 2016, 145, 24–36. [Google Scholar] [CrossRef]
- Murphy, S.; Zweyer, M.; Henry, M.; Meleady, P.; Mundegar, R.R.; Swandulla, D.; Ohlendieck, K. Label-free mass spectrometric analysis reveals complex changes in the brain proteome from the mdx-4cv mouse model of Duchenne muscular dystrophy. Clin. Proteom. 2015, 12, 27. [Google Scholar] [CrossRef]
- Murphy, S.; Zweyer, M.; Henry, M.; Meleady, P.; Mundegar, R.R.; Swandulla, D.; Ohlendieck, K. Proteomic profiling of liver tissue from the mdx-4cv mouse model of Duchenne muscular dystrophy. Clin. Proteom. 2018, 15, 34. [Google Scholar] [CrossRef]
- Murphy, S.; Dowling, P.; Zweyer, M.; Henry, M.; Meleady, P.; Mundegar, R.R.; Swandulla, D.; Ohlendieck, K. Proteomic profiling of mdx-4cv serum reveals highly elevated levels of the inflammation-induced plasma marker haptoglobin in muscular dystrophy. Int. J. Mol. Med. 2017, 39, 1357–1370. [Google Scholar] [CrossRef]
- Murphy, S.; Zweyer, M.; Mundegar, R.R.; Swandulla, D.; Ohlendieck, K. Proteomic identification of elevated saliva kallikrein levels in the mdx-4cv mouse model of Duchenne muscular dystrophy. Biochem. Biophys. Rep. 2018, 18, 100541. [Google Scholar] [CrossRef]
- Gargan, S.; Dowling, P.; Zweyer, M.; Swandulla, D.; Ohlendieck, K. Identification of marker proteins of muscular dystrophy in the urine proteome from the mdx-4cv model of dystrophinopathy. Mol. Omics 2000, 16, 268–278. [Google Scholar] [CrossRef]
- Formicola, L.; Marazzi, G.; Sassoon, D.A. The extraocular muscle stem cell niche is resistant to ageing and disease. Front. Aging Neurosci. 2014, 6, 328. [Google Scholar] [CrossRef]
- Zeiger, U.; Mitchell, C.H.; Khurana, T.S. Superior calcium homeostasis of extraocular muscles. Exp. Eye Res. 2010, 91, 613–622. [Google Scholar] [CrossRef]
- Dowling, P.; Culligan, K.; Ohlendieck, K. Distal mdx muscle groups exhibiting up-regulation of utrophin and rescue of dystrophin-associated glycoproteins exemplify a protected phenotype in muscular dystrophy. Naturwissenschaften 2002, 89, 75–78. [Google Scholar] [CrossRef][Green Version]
- Dowling, P.; Lohan, J.; Ohlendieck, K. Comparative analysis of Dp427-deficient mdx tissues shows that the milder dystrophic phenotype of extraocular and toe muscle fibres is associated with a persistent expression of beta-dystroglycan. Eur. J. Cell Biol. 2003, 82, 222–230. [Google Scholar] [CrossRef][Green Version]
- Pertille, A.; de Carvalho, C.L.; Matsumura, C.Y.; Neto, H.S.; Marques, M.J. Calcium-binding proteins in skeletal muscles of the mdx mice: Potential role in the pathogenesis of Duchenne muscular dystrophy. Int. J. Exp. Pathol. 2010, 91, 63–71. [Google Scholar] [CrossRef]
- McDonald, A.A.; Kunz, M.D.; McLoon, L.K. Dystrophic changes in extraocular muscles after gamma irradiation in mdx:utrophin(+/−) mice. PLoS ONE 2014, 9, e86424. [Google Scholar] [CrossRef]
- McDonald, A.A.; Hebert, S.L.; McLoon, L.K. Sparing of the extraocular muscles in mdx mice with absent or reduced utrophin expression: A life span analysis. Neuromuscul. Disord. 2015, 25, 873–887. [Google Scholar] [CrossRef]
- Ohlendieck, K. Proteomics of skeletal muscle glycolysis. Biochim. Biophys. Acta 2010, 1804, 2089–2101. [Google Scholar] [CrossRef]
- Brinkmeier, H.; Ohlendieck, K. Chaperoning heat shock proteins: Proteomic analysis and relevance for normal and dystrophin-deficient muscle. Proteom. Clin. Appl. 2014, 8, 875–895. [Google Scholar] [CrossRef]
- Doran, P.; Martin, G.; Dowling, P.; Jockusch, H.; Ohlendieck, K. Proteome analysis of the dystrophin-deficient MDX diaphragm reveals a drastic increase in the heat shock protein cvHSP. Proteomics 2006, 6, 4610–4621. [Google Scholar] [CrossRef]
- van Westering, T.L.E.; Johansson, H.J.; Hanson, B.; Coenen-Stass, A.M.L.; Lomonosova, Y.; Tanihata, J.; Motohashi, N.; Yokota, T.; Takeda, S.; Lehtiö, J.; et al. Mutation-independent Proteomic Signatures of Pathological Progression in Murine Models of Duchenne Muscular Dystrophy. Mol. Cell. Proteom. 2020, 19, 2047–2068. [Google Scholar] [CrossRef]
- Van Pelt, D.W.; Kharaz, Y.A.; Sarver, D.C.; Eckhardt, L.R.; Dzierzawski, J.T.; Disser, N.P.; Piacentini, A.N.; Comerford, E.; McDonagh, B.; Mendias, C.L. Multiomics analysis of the mdx/mTR mouse model of Duchenne muscular dystrophy. Connect. Tissue Res. 2021, 62, 24–39. [Google Scholar] [CrossRef]
- Capitanio, D.; Moriggi, M.; Torretta, E.; Barbacini, P.; De Palma, S.; Viganò, A.; Lochmüller, H.; Muntoni, F.; Ferlini, A.; Mora, M.; et al. Comparative proteomic analyses of Duchenne muscular dystrophy and Becker muscular dystrophy muscles: Changes contributing to preserve muscle function in Becker muscular dystrophy patients. J. Cachexia Sarcopenia Muscle 2020, 11, 547–563. [Google Scholar] [CrossRef]
- Porter, J.D.; Merriam, A.P.; Khanna, S.; Andrade, F.H.; Richmonds, C.R.; Leahy, P.; Cheng, G.; Karathanasis, P.; Zhou, X.; Kusner, L.L.; et al. Constitutive properties, not molecular adaptations, mediate extraocular muscle sparing in dystrophic mdx mice. FASEB J. 2003, 17, 893–895. [Google Scholar] [CrossRef]
| Accession | Protein | Gene | Score | Coverage % | Peptides | Molecular Mass (kDa) |
|---|---|---|---|---|---|---|
| E9Q8K5 | Titin, muscle- specific | TTN | 171.5 | 2.14 | 52 | 3713.7 |
| Q5SX40 | Myosin-1 heavy chain, MyHC-IId, fast muscle | MYH1 | 459.6 | 31.10 | 63 | 223.2 |
| G3UW82 | Myosin-2 heavy chain, MyHC-IIa, fast muscle | MYH2 | 382.7 | 31.15 | 60 | 223.1 |
| P13541 | Myosin-3 heavy chain, MyHC-embryonic, muscle | MYH3 | 125.1 | 11.70 | 24 | 223.7 |
| Q5SX39 | Myosin-4, MyHC-IIb, fast muscle | MYH4 | 472.9 | 32.75 | 68 | 222.7 |
| Q91Z83 | Myosin-7 heavy chain, MyHC-I, slow muscle | MYH7 | 96.04 | 10.70 | 20 | 222.7 |
| P13542 | Myosin-8 heavy chain, MyHC-perinatal, muscle | MYH8 | 277.9 | 24.68 | 47 | 222.6 |
| Q8VDD5 | Myosin-9 heavy chain, MyHC-cellular, type A | MYH9 | 87.81 | 10.61 | 20 | 226.2 |
| Q61879 | Myosin-10 heavy chain, MyHC-cellular, type B | MYH10 | 20.79 | 2.68 | 5 | 228.8 |
| A0A2R8VHF9 | Myosin-11 heavy chain, smooth muscle | MYH11 | 255.1 | 20.07 | 44 | 223.2 |
| B1AR69 | Myosin-13 heavy chain, extraocular muscle | MYH13 | 217.41 | 17.03 | 31 | 223.4 |
| K3W4R2 | Myosin-14 heavy chain, MyHC-eom, developmental | MYH14 | 25.40 | 3.45 | 6 | 228.4 |
| E9Q264 | Myosin-15 heavy chain, extraocular muscle | MYH15 | 10.20 | 1.56 | 3 | 221.7 |
| P05977 | Myosin light chain MLC-1/3, muscle | MYL1 | 74.36 | 45.21 | 8 | 20.6 |
| A0A0U1RP93 | Myosin light chain MLC-2, muscle | MYLPF | 28.02 | 10.07 | 1 | 16.9 |
| Q60605 | Myosin light chain MLC-3, muscle | MYL6 | 56.91 | 54.30 | 7 | 16.9 |
| D3YU50 | Myosin-binding protein C, slow | MYBPC1 | 20.96 | 5.24 | 4 | 126.5 |
| A0A571BF46 | Nebulin | NEB | 15.55 | 1.32 | 8 | 866.5 |
| P68033 | Actin, alpha, skeletal muscle | ACTC1 | 382.05 | 66.84 | 26 | 42.0 |
| P60710 | Actin, beta, cytoplasmic | ACTB | 384.11 | 52.53 | 25 | 41.7 |
| E9Q452 | Tropomyosin alpha-1 | TPM1 | 119.92 | 40.93 | 14 | 32.5 |
| A2AIM4 | Tropomyosin beta | TPM2 | 112.31 | 41.55 | 16 | 33.0 |
| D3Z6I8 | Tropomyosin alpha-3 | TPM3 | 47.89 | 43.72 | 12 | 28.7 |
| A0A571BEU1 | Tropomyosin alpha-4 | TPM4 | 4.33 | 14.56 | 3 | 18.4 |
| P20801 | Troponin C, muscle | TNNC2 | 3.99 | 11.88 | 2 | 18.1 |
| A0A1B0GRY8 | Troponin I, fast muscle | TNNI2 | 23.66 | 13.95 | 2 | 20.2 |
| A2A6I8 | Troponin T, fast muscle | TNNT3 | 8.66 | 20.08 | 4 | 28.3 |
| A1BN54 | Alpha-actinin-1 | ACTN1 | 36.58 | 8.34 | 6 | 102.7 |
| Q9JI91 | Alpha-actinin-2 | ACTN2 | 13.09 | 7.83 | 6 | 103.8 |
| O88990 | Alpha-actinin-3 | ACTN3 | 11.30 | 5.11 | 4 | 103.0 |
| P57780 | Alpha-actinin-4 | ACTN4 | 65.92 | 20.39 | 14 | 104.9 |
| P31001 | Desmin | DES | 27.89 | 15.99 | 8 | 53.5 |
| P20152 | Vimentin | VIM | 72.37 | 24.89 | 8 | 53.7 |
| E9Q3W4 | Plectin | PLEC | 22.89 | 3.17 | 12 | 498.8 |
| P23927 | Alpha-B-crystallin | CRYAB | 14.36 | 36.57 | 7 | 20.1 |
| E9PYJ9 | LIM domain-binding protein 3 | LDB3 | 10.34 | 3.98 | 2 | 72.3 |
| Q9JIF9 | Myotilin | MYOT | 10.23 | 6.45 | 3 | 55.3 |
| A0A1L1STC6 | Nesprin-1 | SYNE1 | 4.29 | 0.31 | 2 | 1009.3 |
| A0A571BDS0 | Xin actin-binding repeat-containing protein 1 | XIRP1 | 2.03 | 1.05 | 2 | 196.6 |
| Q8VHX6 | Filamin-C, muscle-specific | FNLC | 21.04 | 2.53 | 6 | 290.9 |
| Z4YJF5 | Myomesin-1 | MYOM1 | 2.09 | 0.83 | 1 | 175.4 |
| A2ABU4 | Myomesin-3, slow, extraocular muscle | MYOM3 | 5.42 | 2.08 | 2 | 161.7 |
| P07310 | Creatine kinase, muscle-type | CKM | 48.98 | 29.92 | 11 | 43.0 |
| Q9R0Y5 | Adenylate kinase, AK1 | AK1 | 17.30 | 16.49 | 2 | 21.5 |
| P97447 | Skeletal muscle LIM-protein 1 | FHL1 | 10.47 | 5.89 | 2 | 61.8 |
| P21550 | Beta-enolase, muscle-specific | ENO3 | 152.38 | 42.63 | 18 | 47.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gargan, S.; Dowling, P.; Zweyer, M.; Reimann, J.; Henry, M.; Meleady, P.; Swandulla, D.; Ohlendieck, K. Mass Spectrometric Profiling of Extraocular Muscle and Proteomic Adaptations in the mdx-4cv Model of Duchenne Muscular Dystrophy. Life 2021, 11, 595. https://doi.org/10.3390/life11070595
Gargan S, Dowling P, Zweyer M, Reimann J, Henry M, Meleady P, Swandulla D, Ohlendieck K. Mass Spectrometric Profiling of Extraocular Muscle and Proteomic Adaptations in the mdx-4cv Model of Duchenne Muscular Dystrophy. Life. 2021; 11(7):595. https://doi.org/10.3390/life11070595
Chicago/Turabian StyleGargan, Stephen, Paul Dowling, Margit Zweyer, Jens Reimann, Michael Henry, Paula Meleady, Dieter Swandulla, and Kay Ohlendieck. 2021. "Mass Spectrometric Profiling of Extraocular Muscle and Proteomic Adaptations in the mdx-4cv Model of Duchenne Muscular Dystrophy" Life 11, no. 7: 595. https://doi.org/10.3390/life11070595
APA StyleGargan, S., Dowling, P., Zweyer, M., Reimann, J., Henry, M., Meleady, P., Swandulla, D., & Ohlendieck, K. (2021). Mass Spectrometric Profiling of Extraocular Muscle and Proteomic Adaptations in the mdx-4cv Model of Duchenne Muscular Dystrophy. Life, 11(7), 595. https://doi.org/10.3390/life11070595