Abstract
Advanced melanoma is a relentless tumor with a high metastatic potential. The combat of melanoma by using the targeted therapy is impeded because several major driver mutations fuel its growth (predominantly BRAF and NRAS). Both these mutated oncogenes strongly activate the MAPK (MEK/ERK) pathway. Therefore, specific inhibitors of these oncoproteins or MAPK pathway components or their combination have been used for tumor eradication. After a good initial response, resistant cells develop almost universally and need the drug for further expansion. Multiple mechanisms, sometimes very distant from the MAPK pathway, are responsible for the development of resistance. Here, we review many of the mechanisms causing resistance and leading to the dismal final outcome of mutated BRAF and NRAS therapy. Very heterogeneous events lead to drug resistance. Due to this, each individual mechanism would be in fact needed to be determined for a personalized therapy to treat patients more efficiently and causally according to molecular findings. This procedure is practically impossible in the clinic. Other approaches are therefore needed, such as combined treatment with more drugs simultaneously from the beginning of the therapy. This could eradicate tumor cells more rapidly and greatly diminish the possibility of emerging mechanisms that allow the evolution of drug resistance.
1. Introduction
The incidence of melanoma has been slowly increasing worldwide in the last decades. Melanoma is derived from melanocytes, which originate from the neural crest precursor cells. Melanomas share many similar phenotypes with neural crest cell-derived melanocytes and express similar transcription factors [1]. A common problem in melanoma biology and therapy is tumor microheterogeneity. Different parts of a tumor possess distinct biological properties, which reflect the genetic and epigenetic diversity of the tumor cell subpopulations. These phenotypically distinct micropopulations also mirror the varying sensitivity to anticancer drugs. A hierarchy of tumoral cells exists in neoplasms. This is caused by both genetic and epigenetic mechanisms and results in a clonal evolution [2]. Driver mutations hit the genes critical for tumor progression and are considered the most important tumor promoters suitable for targeted therapy. Approximately 60% of melanomas harbor an activating BRAF (v-raf murine sarcoma viral oncogene homolog B1) mutation V600E (changing valine to glutamic acid and being highly predominant over other infrequent BRAF mutations). About 15% of melanomas contain NRAS (neuroblastoma rat sarcoma viral oncogene) mutations [3,4,5,6,7,8,9,10,11,12]. In mucosal melanomas, the BRAF and NRAS mutations are infrequent, which may reflect the lack of sunlight exposure [13]. Both mutational types are mutually exclusive, and each mutation substantially accelerates the activity of the MEK/ERK signaling pathway. It has long been known that BRAF mutation is already present in benign nevi, conferring the oncogene-induced senescence on nevus cells [14,15]. Given the importance of driver mutations, targeted drugs were developed to inhibit the aberrant activities of mutated BRAF and NRAS genes. Despite fundamental advances in the development of inhibitors focused on blocking the MAPK pathway in BRAF-mutated cancers, resistance remains a major hurdle to the enduring success of these therapies [11,16,17,18,19].
Large drug armamentarium exists to target mutated BRAF protein (dabrafenib, vemurafenib, encorafenib, sorafenib, GDC-0879, PLX8394) and the MEK/ERK pathway (MEK1/2 inhibitors: selumetinib, trametinib, binimetinib, cobimetinib; ERK1/2 inhibitors: ulixertinib, SCH772984, AZD0364, SCH772984). The number of the targeting NRAS-mutated drugs is limited, e.g., STK19(D89N) or pan-RAF kinase inhibitor belvarafenib). Many of these therapeutics have been already approved by the US Food and Drug Administration (FDA) to treat patients with advanced melanoma [20]. The inherited or acquired resistance appears almost invariably, typically months after the beginning of the treatment. Additionally, tumor cells can adapt to the administered drug and even require it for further proliferation. In such a case, the removal of the drug from the therapy is beneficial. This “drug holiday” [8,21,22,23] improves treatment, and melanoma cells can become senescent or undergo cell death [8]. Furthermore, a therapy-induced secretome that stimulates tumor growth has been described [24]. Here we describe some of the main molecular mechanisms underlying the resistance to mutated BRAF and NRAS genes in melanomas. Overcoming resistance is crucial for effective treatment and durable remission.
2. General Mechanisms Underlying Targeted Drug Resistance
The mitogen-activated protein kinase (MAPK) pathway is frequently hyperactivated, signaling a cascade in cancers, including melanomas. It is an essential pathway required for melanoma progression and the prime target for therapeutic intervention [22,25,26,27,28]. The MAPK pathway is activated by a broad spectrum of extracellular signals, including mitogens, growth factors, and cytokines. Besides mutations of the BRAF and NRAS genes, activating mutations have also been found in the downstream kinases MEK and ERK. MEK1 mutations can be associated with either BRAF or NRAS mutations, while ERK mutations are less frequent (reviewed in [29]). The NRAS and RAF genes (A, B, C) are the main activators of the pathway [30]). Mutated CRAF was shown to be a rare driver oncogene overexpressed in BRAF or NRAS melanoma cells. These cells were sensitive to CRAF, but not BRAF knockdown [31]. Although BRAF is the most frequently mutated driver gene in melanoma, BRAF mutations alone are nevertheless not sufficient for melanoma development. They are already found in benign nevi (see above).
Numerous reports revealed additional genetic alterations in various oncogenes and tumor suppressors beyond the BRAF and NRAS genes. They modulate progression into an invasive state and include CDKN2A (encoding the tumor suppressor p16 protein), CDK4 (cyclin-dependent kinase 4), PTEN (phosphatase and tensin homolog), NF1 (neurofibromin 1), TP53, RAC1 (small GTPase 1), MEK, MITF (see below), MC1R (melanocortin 1 receptor), and many others [9,18] (reviewed also in [32,33,34]).
Although the main events leading to melanoma progression and drug resistance are genetic events (mainly BRAF and NRAS mutations), epigenetic mechanisms are also important. The early diversification of tumor micropopulations results in considerable heterogeneity of the whole tumoral population [35]. This results in different phenotypic features (some of the cells are BRAFwt, while some harbor the BRAFV600E mutation). Cancer heterogeneity is a common problem in oncology, hindering the more effective targeted therapy and considerably contributing to drug resistance [36,37]. Other factors, such as paracrine and autocrine cell communication and, e.g., the composition of stroma, can also promote tumor survival [38].
3. Resistance to Inhibitors of Mutated BRAF in Melanoma
3.1. BRAF and MAPK Cascade
Wild-type BRAF (Figure 1) activates MAPK route upon upstream activation from NRAS through the formation of homodimers or heterodimers with CRAF. Mutated BRAFV600E does not form dimers and is capable of activating MEK kinase as a monomeric protein. BRAF inhibitors are inactive toward BRAFwt, but they effectively block the activity of monomeric BRAFV600E [39,40,41]. Interestingly, in BRAFwt cells, the expression of CRAF has been shown to be higher. Vemurafenib stabilized BRAF–CRAF heterodimers, thus helping to keep the MAPK pathway active [42]. The BRAF gene can be also amplified in a small subset of melanomas [40]. The BRAFV600E fusion protein AGAP3-BRAF has been reported to cause BRAFi resistance in melanoma [43]. The alternatively spliced variant of BRAFV600E has been shown to enhance the interaction with MEK. This depends on increased phosphorylation on MEKS729 and causes resistance in a subset of melanomas [44]. Among other very scarce BRAF mutations, a vemurafenib resistance-conferring, MEK/ERK-activating mutation of BRAF (BRAFL505H) has also been reported [45].

Figure 1.
Some of the important mechanisms leading to resistance to mutated BRAF in melanoma. See text for a detailed explanation.
Resistance to BRAFV600E inhibitors logically comprises the biochemically closest downstream effectors in the MAPK pathway. Thus, MEK-activating mutations can bypass the block caused by inhibited mutated BRAF and reactivate the cascade [46,47]. Unfortunately, a resistance to the subsequently administered MEK inhibitor may also develop, causing the cells to become double-resistant. Reportedly, the ERK1/2 inhibitor SCH772984 efficiently suppressed the MAPK pathway and cell proliferation in BRAF, MEK, and combined BRAF/MEK inhibitor-resistant tumors [48,49]. However, long-term exposure of cells to the inhibitor also leads to acquired resistance through a mutation in ERK1 kinase [50].
3.2. Transcription Factors
Remarkably, resistance to the therapy is frequently mediated by mechanisms independent of ERK reactivation. A large number of mechanisms that may be very distant from the MAPK pathway can also cause BRAFV6000Ei resistance. Blocking these molecular events can release the sensitivity to the inhibitor. Alterations in other signaling pathways, transcription factors, oncogenes, and tumor suppressors are some examples of this kind of drug resistance.
Increased expression of the c-JUN oncogene, accompanied by the downregulation of SPROUTY4 and switching the phenotype to (EMT) epithelial-mesenchymal transition, was reported to be the reason behind BRAFi resistance in melanoma cells [51]. JNK/c-jun inhibitors synergy helped overcome BRAFi resistance [52]. Similar findings were reported by Titz et al., including co-targeting of BRAF and JUN which synergized in killing resistant cells [53]. In addition, JUN upregulation was a common outcome of BRAF inhibitor treatment in patients [53].
CRISPR–Cas9 knockout screen identified, besides ERK2 kinase, the transcription factors JUNB and FRA1 as essential factors responsible for the survival of BRAFi and combined BRAFi- and MEKi-resistant melanoma cells during drug holiday. The phenotype of resistant cells resembled the phenotype switch to EMT. The switch was ERK dependent, and the expression of the melanoma-specific transcription factor and oncogene MITF (melanoma-associated transcription factor) was decreased. Drug discontinuation synergized with the administration of dacarbazine, accompanied by decrease of MITF and its transcriptional target BCL-2 [8]. In another study, JUNB and FRA1 transcripts were induced upon MAPKi withdrawal, and the total FOS protein level was increased [23]. Drug holiday, thus, may select against BRAFi/MAPKi-resistant melanoma cells. The involvement of JUNB and FRA1 level decrease, therefore, seems to be crucial for sensitizing cells to the “drug holiday treatment.”
Overexpression of the transcription factor POU4F1 (POU Class 4 Homeobox 1) contributed to the resistance to BRAFi in vemurafenib-treated melanoma cells by increasing MEK expression and activating MEK/ERK signaling. POU4F1 also increased the expression of MITF, further supporting resistance [54].
Furthermore, elevated levels of protein p63 were described in melanoma cells exposed to BRAF and MEK inhibitors. This was caused by its reduced degradation by the E3 ubiquitin ligase FBXW (F-box and WD repeat domain-containing), whose degradation is regulated by MDM2. FBXW was found to be downregulated in BRAF/MEKi-resistant cells. The MDM2 inhibitor Nutlin-3A restored FBXW7 expression and p63 degradation and resensitized resistant cells to the BRAFi inhibitor, thus underpinning the role of p63 in BRAFi melanoma resistance [12].
Zinc finger E-box-binding homeobox 1 (ZEB1) is a transcription factor known to induce phenotype switching, plasticity, and invasiveness. Consistently, high levels of ZEB1 expression were described to be associated with resistance to MEKi/BRAFi in BRAFV600E-melanoma cells. Conversely, ZEB1 inhibition promoted differentiation. It also inhibited tumor growth in vivo and sensitized melanoma cells to MEK/BRAF inhibitors, ultimately inducing cell death in resistant cells [55].
3.3. Protein Kinases
Many oncogenic protein kinases that act distantly from the MEK/ERK signaling have been demonstrated to cause resistance in BRAFi-treated melanoma cells. More pathways have cross-talks with MEK/ERK, and thus the hyperactivation of other oncogenic signaling cascades can often cause BRAFi resistance [56]. Studies have shown that the PI3K/AKT-mTOR pathway plays a role in the development of BRAFi resistance by triggering a surrogate survival signaling, leading to resistant BRAFi-treated melanoma cells [57,58]. The PI3K/AKT pathway has been recently identified as a key factor in stress-induced mutagenesis and stress-sensing mediating resistance in many types of cancer [59]. Inhibition of the PI3K/AKT may reactivate the sensitivity of melanoma cells to a BRAF inhibitor. Full activation of the PI3K/AKT pathway depends on the alterations and inactivity of its inhibitor and the tumor suppressor PTEN, which often regulates both acquired and intrinsic drug resistance. It was further shown that the oncogenic kinase AXL, together with AKT, mediates resistance to BRAFi, depending on the PTEN status. PTEN-inactive melanoma cells required only the active ERK pathway. However, AXL was shown to be an upstream effector of the AKT pathway-associated resistance to BRAFi in melanoma with wild-type PTEN [60]. Moreover, mutation of AKT kinase could alone trigger the adaptive resistance to BRAF inhibition [61].
Previous investigation also showed that the cell cycle regulator cyclin D1, often overexpressed in tumors, increased resistance concurrently with mutated or overexpressed CDK4 in BRAFi melanoma cells [62]. CDK2 also appeared to be a driver of resistance to inhibitors of BRAF and Hsp90 (XL888). Dinaciclib (a CDK2 inhibitor) attenuated resistance to both classes of inhibitors [63].
Oncogenic anaplastic lymphoma kinase (ALK) is an oncogenic driver in non-small-cell lung cancer and anaplastic large-cell lymphomas. Alterations in the ALK gene are gene fusions EML4-ALK, amplifications, or activating mutations. ALK was identified as another kinase conferring BRAFi resistance in melanomas. The PI3K/AKT signaling pathway, downstream of ALK, was required for ALK activity, and the ALK inhibitor ceritinib abrogated the BRAFi resistance in melanoma cells [64].
ABL kinase is frequently hyperactivated in tumors, e.g., in leukemias as a fusion gene with BCR. The expression of constitutively active ABL1/2 in melanomas has been described to be sufficient to promote resistance to vemurafenib, dabrafenib, MEK inhibitors, and their combination by inducing reactivation of MEK/ERK/MYC signaling. Targeting ABL kinases by the inhibitor nilotinib reversed the resistance and resensitized cells resistant to BRAF inhibitors. This indicates the role of ABL kinases as important factors supporting the active MEK/ERK pathway during the evolution of drug resistance in melanomas [65].
The P21-activated kinase (PAK) is a serine/threonine kinase mediating signaling in multiple pathways that are involved in numerous cell functions, including cell survival. This kinase has also been identified as one of the culprits causing BRAFi and combined inhibitor resistance in melanomas. Whereas the resistance to BRAFi was mediated classically through the MEK/ERK pathway reactivation (via the phosphorylation of CRAF and MEK), the resistance to combined treatment was independent of MEK/ERK activity. This second mechanisms involved-more complex mechanisms: regulation of JNK (c-Jun N-terminal kinase), β-catenin, and AKT/mTOR pathways. However, all types of resistant cells were sensitive to the PAK inhibitor PF-3758309. Thus, inhibiting PAK may also help treat patients whose tumors progress through initial inhibitor resistance in targeted therapies [66].
Aurora kinase B is overexpressed in melanoma. A study showed that the inhibitor of this kinase, HI-511, which also inhibits BRAFV600E, can suppress the growth of both vemurafenib-sensitive and vemurafenib-resistant melanoma cells. After inhibitor administration, both the MEK/ERK and PI3K/AKT pathways were inhibited. Aurora kinase B is thus another potential target in BRAFi-resistant cells [67].
Receptor tyrosine kinases from the MET and ERBB family have been reported to be involved in melanoma progression and metastasis, as well as in the development of therapy resistance. The epidermal growth factor receptor (EGFR), hepatocyte growth factor receptor (MET), and erb-b2 receptor tyrosine kinase 3 (ERBB3) were grossly overexpressed in BRAFi cells. Inhibition of these kinases with the combination of the drugs afatinib (an ERBB family inhibitor) and crizotinib (an MET inhibitor) proved to have deleterious and synergistic effects on melanomas. Interestingly, this effect was independent of their BRAF/NRAS mutational status [68].
3.4. MicroRNAs
Several microRNAs were described to be the cause of BRAFi resistance in melanomas. BRAFV600E-positive A375 melanoma cells were made resistant to vemurafenib, and two miRNAs, miR-204-5p and miR-211-5p, occurred overexpressed in these cells. Ectopic expression of both miRNA in parental and BRAFi-treated cells conferred vemurafenib resistance. Silencing their expression in BRAFi-resistant cells inhibited cell growth. The study thus shows that the miRNAs miR-204-5p and miR-211-5p enable resistance to BRAF inhibitors [69]. The miRNA miR-579-3p was shown to be a negative prognostic factor correlating with shorter survival, and its expression increased gradually during melanoma progression to later stages. The ectopic miR-579-3p maintained drug-resistant human melanoma cells, and its level sharply dropped in patients treated with targeted BRAFi therapies [70]. Conversely, the microRNA miR-524-5p, which behaves as a tumor suppressor in cancers, was indeed inhibitory for BRAFi-resistant melanoma cells. Overexpression of miR-524-5p reduced the proliferation and the migratory and invasive properties of BRAFi-resistant melanoma cells. In addition, the MEK/ERK signaling pathway was attenuated after treatment with miR-524-5p. This miRNA can therefore be considered a tumor suppressor for BRAFi-resistant melanomas [71].
3.5. Other Diverse Mechanisms
Additionally, very different mechanisms can contribute to BRAFi resistance. In earlier studies, the loss of the tumor suppressor NF1 has been observed in melanomas. Alterations of NF1 occurred in BRAF/NRASwt tumors, but also frequently occurred jointly with RAS or BRAF mutations. These BRAFV600E cells resulted in elevated resistance to RAF inhibitors. The loss of NF1 in mutated BRAF cells was therefore sufficient to overcome the upstream negative feedback and conferred resistance to vemurafenib. Furthermore, the MEK inhibitor trametinib attenuated MEK phosphorylation and exhibited greater potency than another MEK inhibitor, PD0325901, in NF1-null melanoma cells. Together, the loss of NF1 is associated with MEK dependence and resistance to BRAF inhibition [72].
The antiapoptotic BCL2 family member BCL2A1, a transcriptional target of MITF, is amplified in about one-third of melanomas. BCL2A1 expression is associated with more dismal clinical responses to BRAF inhibitors in melanoma patients. Cotreatment of melanomas with BRAF inhibitors and obatoclax, an inhibitor BCL2 family member, overcomes resistance to BRAF inhibitors in BCL2A1-amplified cells. Pharmacological inactivation of BCL2A1 thus leads to improved response to therapy by BRAF inhibitors [73].
Other MEK/ERK-independent mechanisms participate in the acquired BRAFi melanoma cell resistance. For instance, mRNA translation can have a key role in melanoma drug resistance. Wobble tRNA modifications are required for specific codon decoding during translation. It was demonstrated that BRAFV600E melanoma cells are dependent on U34 enzymes for survival. The wobble uridine 34 (U34) tRNA plays a key role in translation rewiring. It is induced by BRAFV600E and promotes glycolysis in cells through the regulation of the translation of HIF1α mRNA. U34 tRNA thus has an impact on BRAFi resistance in melanomas [74].
Recently, it has been described that melanoma cells can adapt to targeted therapies through a mechano-signaling loop involving autocrine remodeling. Melanoma cells secrete specific proteins to an extracellular matrix. In BRAFi-resistant cells, matrix remodeling and tumor stiffening were observed, depending largely on the Yes-associated protein (YAP) and myocardin. Targeting this biomechanical adaptation by Yes inhibitors may improve the sensitivity of BRAFi-resistant cells to targeted drugs [75].
Tumor stroma is known to be an important player in cancer progression. It has been reported that a more fibrotic stroma can influence the BRAFi resistance in melanomas [76]. Some studies support the notion that MEK/ERK inhibitor treatment in BRAF-mutant melanoma promotes the stromal microenvironment of melanoma cells toward a more fibrotic phenotype. Through this, drug resistance and tumor progression appears. Molecules such as fibronectin or type I collagen accumulate in the fibrotic stroma. Thus, therapeutically, relieving fibrosis in stroma with collagen or myofibroblast-targeting drugs could be promising. Such drugs can be pirfenidone, blocking myofibroblast proliferation, or aminopropionitrile, a lysyl oxidase inhibitor.
Another mode for overcoming BRAFV600Ei resistance is targeting the mitochondrial oxidative phosphorylation. IACS-010759 is a complex I mitochondrial oxidative phosphorylation inhibitor that inhibits the activity of both MEK/ERK and mTOR pathways. However, its activity is not synergized with MEK/ERK inhibitors. IACS-010759 enforces glycolysis and decreases nucleotide and amino acid pools. Thus, mitochondrial stress can mediate MEKi resistance and could be potentially clinically useful for melanoma treatment [77].
The authors of another study identified the fatty acid transporter CD36 as upregulated in MAPK-inhibited cells. These cells displayed increased fatty acid oxidation. Increased fatty acid oxidation was required for BRAFV600E-mutant melanoma cells to survive under the MAPK inhibition prior to acquiring drug resistance. Remarkably, the upregulation of CD36 itself was not involved in changes in fatty acid oxidation. These data indicated a potential metabolic approach to overcoming MAPKi resistance in BRAFV600E melanomas [78].
It is known that low-MITF/high-AXL expression signature is typical for invasive subpopulations of melanoma cells. Phenotype switching to this poorer phenotype is supported during the resistance to BRAF inhibitor therapy. It was shown that during the response phase of BRAFi therapy, endothelin 1 expression was increased. This occurred through the MITF-induced transcription and conferred drug resistance via MEK/ERK pathway re-activation. Antagonists of the endothelin receptor diminished AXL-high-expressing population and sensitized cells to BRAF inhibition. This suggests that targeting endothelin 1 can improve the BRAFi response and help overcome resistance [79].
4. Resistance to Inhibitors of NRAS Mutations in Melanoma
About one-third of cancers harbor mutations of KRAS, HRAS, or NRAS genes, occurring in codons 12, 13, and 61. In melanomas, only NRAS is exclusively mutated in about 15–20% of patients. The mutations are located predominantly in codon 61 (major mutations: Q61L, Q61K, and Q61R), while rare mutations are found in codons 12 or 13 [61,80,81]. Whereas many BRAFV600E-efficient inhibitors were found, specific NRAS inhibition is rather a disappointing matter compared with BRAF. The reason is that only a limited number of potentially active drugs have been developed [82,83]. Due to this, NRAS has been regarded as an “undruggable” target, and therapies aimed directly at NRAS mutations still remain elusive. BRAFV600E resistance develops during treatment and, similarly, resistance also appears when NRAS-mutant melanomas are treated with targeted drugs. Several distant mechanisms may be a cause of the acquired NRAS resistance (Figure 2).
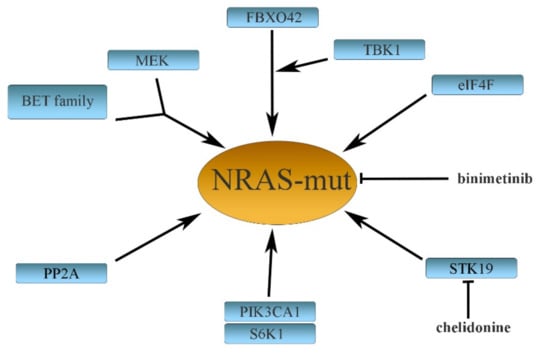
Figure 2.
Some mechanisms causing resistance to mutated NRAS in melanoma. See the text for a more detailed explanation.
Binimetinib (an MEK1/2 inhibitor) and the STK19 kinase inhibitor chelidonine were described to be relatively effective in NRAS-mutated melanomas [84,85], although controversies were raised against the efficacy of the STK19 inhibitor [86]. Like BRAF, NRAS also triggers the MEK/ERK signaling pathway. Therefore, binimetinib (used also in BRAFi-resistant melanomas) has been used for the treatment of NRAS-mutated melanomas either alone [85] or in combination with encorafenib [87]. Combining BET (bromodomain and extra-terminal domain) family and MEK inhibitors synergistically inhibited the growth of NRAS-mutant melanoma in mice not responding to MAPK inhibitors [82]. NRAS also stimulates the AKT/mTOR pathway, which is involved in mediating translational mechanisms. Thus, pharmacological targeting of the translation initiation complex component eIF4A, combined with an MEK inhibitor, seems to be an efficient treatment for NRAS-mutant melanomas [88].
Recently, Kelch domain-containing F-Box protein 42 (FBXO42) has been recognized as a factor causing the resistance of NRAS-mutant melanomas to trametinib. FBXO42 is involved in the transforming growth factor β-activated kinase 1 (TAK1) pathway, leading to the activation of p38 kinase. The TAK1 inhibitor takinib synergized with trametinib in eradicating NRAS-mutant melanoma cells [89].
A kinase TBK1 (TANK-binding kinase 1) is an atypical IκB kinase family member. It was demonstrated that NRAS overexpression increases TBK1 phosphorylation. Subsequent TBK1 downregulation inhibits cell migration and invasion, whereas its overexpression increases invasion of NRAS mutant melanoma cells [90,91]. TBK1 depletion cooperates with MEK inhibitors in the context of MEK-insensitive mutant NRAS, but not in wtNRAS. TKB1 thus appears to be a promising target in NRAS-mutant melanomas.
Another study revealed the E545K mutation in PIK3CA (a catalytic subunit of phosphatidylinositol 3 kinase PI3K) present in rare tumor subpopulations. These cells were nevertheless rapidly expanding during combined targeted therapy with MEK and CDK4 inhibitors. This represented a pivotal mechanism of resistance in NRAS-mutated melanomas. As PIK3CA-E545K increases S6K1 and S6 phosphorylation in the AKT/mTOR pathway, inhibition of the S6K1 kinase could be used to revert the resistance [92]. Growth inhibition of NRASQ61R melanomas (as well as BRAF-mutant melanomas) was also achieved by administering the protein phosphatase 2A (PP2A) inhibitor, a phendione derivative, or through depletion of PP2A catalytic subunits [26].
5. Discussion
Melanoma is a resilient kind of cancer and the deadliest form of skin cancer. Melanoma tumors harbor driver mutations mainly in two genes, BRAF and NRAS. Whereas many inhibitors directed to mutated BRAF and MEK have been synthesized and used for treatment, more specific and efficient NRAS-directed agents are still lacking. Resistance develops almost invariably during treatment with one inhibitor. In such setting, micropopulations of cells resistant to a drug gain growth advantage over others. This leads to the overgrowth of these cell populations and tumor resistance. Intratumoral heterogeneity is a driver of treatment resistance, causing phenotype switching and tumor plasticity [93,94,95]. Switching between various phenotypes, e.g., high MITF (less invasive and proliferating) and low MITF (slowly proliferating but invasive, giving rise to metastatic cells) is typical for melanoma. The extremely plastic phenotypic feature definitely underlies the highly malignant and metastatic properties of a melanoma. The plasticity is not necessarily a genomic event, but a consequence of epigenetic and environmental changes. It reflects, e.g., cell metabolic limitations [96,97], a stress-induced response of melanoma cells to environmental cues [98] or hypoxia [99]. Tumor plasticity is a pivotal mechanism in tumor resistance, appears as an adaptive plasticity to various epigenetic stimuli, and results in transcriptional reprogramming [100]. In an earlier study, the sensitivity of MAPK signaling to low-molecular drug inhibitors has been recognized in BRAF-mutated melanomas [101]. Vemurafenib resistance has been described. Resistant cells showed growing disadvantage upon drug withdrawal, which demonstrated the drug holiday phenomenon [102]. A typical mechanism of pigmentation in melanocytes is cAMP-dependent signaling. It supports the pathway leading to MITF stimulation through CREB. It is also noteworthy that it has been recognized as a mechanism conferring melanoma resistance to MAPK route inhibition [103]. Collectively, despite enormous effort, the exact molecular mechanisms why drug resistance evolves in mutated BRAF or NRAS melanomas remain elusive.
6. Conclusions
A number of pathways and, very surprisingly, MAPK-distant events have been known to contribute to melanoma development and progression, including the MAPK signaling pathway. This pathway notoriously overcomes its pharmacological inhibition and develops a remarkable trick to exploit the initially used drug for the further fitness and growth of malignant cells. The drug holiday becomes then a treatment possibility [8,23].
Sometimes, a combination with a second drug may help, especially when the two drugs have a synthetic lethality. Combined therapy of BRAF or NRAS inhibitors with various other agents (e.g., MEK/ERK pathway inhibitors) could be beneficial. Drug resistance is caused by adopting the phenotype plasticity and phenotype switching in tumor cells. Thus, the biological behavior of the tumor is constituted on the basis how rapidly the resistant cells evolve and ultimately exploit the drug to promote growth. A solution to overcome the acquired resistance could be, e.g., the use of three drugs simultaneously from the beginning of the treatment. This approach would profoundly decrease the probability that tumor cells will acquire resistance to all drugs. Additionally, the probability of a faster rate of tumor cell eradication by three efficient agents simultaneously is much higher. These drugs should target independent pathways important for melanoma progression. Another option is the use of agents that do not target the BRAF, NRAS, or MEK/ERK pathway at all, but inhibit essential mechanisms (such as antiapoptosis) important for tumor growth [104]. This approach can also constitute the basis for a more durable eradication of the tumor cells. In addition, such treatment can have an additional advantage, in that both BRAF/NRAS-mutated and non-mutated cells could be killed. A suitable cocktail of two—or better, three—drugs should be used that are effective outside pathways that notoriously acquire drug resistance.
Author Contributions
J.V. and L.O. searched the literature and wrote the manuscript. J.V. supervised the manuscript preparation. Both authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the League Against Cancer Prague and by the institutional program PROGRESQ25 from the Charles University, Prague.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors apologize to all researchers whose work has not been cited owing to the enormous data collection in this field.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Steingrimsson, E.; Copeland, N.G.; Jenkins, N.A. Melanocytes and the Microphthalmia Transcription Factor Network. Annu. Rev. Genet. 2004, 38, 365–411. [Google Scholar] [CrossRef] [PubMed]
- Meacham, C.E.; Morrison, S.J. Tumour Heterogeneity and Cancer Cell Plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef]
- Bogenrieder, T.; Herlyn, M. The Molecular Pathology of Cutaneous Melanoma. Cancer Biomarks 2011, 9, 267–286. [Google Scholar] [CrossRef] [PubMed]
- Daud, A.; Bastian, B.C. Beyond BRAF in Melanoma. Curr. Top. Microbiol. Immunol. 2012, 355, 99–117. [Google Scholar] [PubMed]
- Belum, V.R.; Fischer, A.; Choi, J.N.; Lacouture, M.E. Dermatological Adverse Events From BRAF Inhibitors: A Growing Problem. Curr. Oncol. Rep. 2013, 15, 249–259. [Google Scholar] [CrossRef]
- Holderfield, M.; Deuker, M.M.; McCormick, F.; McMahon, M. Targeting RAF Kinases for Cancer Therapy: BRAF-Mutated Melanoma and Beyond. Nat. Rev. Cancer 2014, 14, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, M.I.; Widmer, D.S.; Narechania, A.; Eichhoff, O.; Freiberger, S.N.; Wenzina, J.; Cheng, P.F.; Mihic-Probst, D.; Desalle, R.; Dummer, R.; et al. Co-Existence of BRAF and NRAS Driver Mutations in the Same Melanoma Cells Results in Heterogeneity of Targeted Therapy Resistance. Oncotarget 2016, 7, 77163–77174. [Google Scholar] [CrossRef]
- Kong, X.; Kuilman, T.; Shahrabi, A.; Boshuizen, J.; Kemper, K.; Song, J.Y.; Niessen, H.W.M.; Rozeman, E.A.; Geukes Foppen, M.H.; Blank, C.U.; et al. Cancer Drug Addiction Is Relayed by an ERK2-Dependent Phenotype Switch. Nature 2017, 550, 270–274. [Google Scholar] [CrossRef]
- Reddy, B.Y.; Miller, D.M.; Tsao, H. Somatic Driver Mutations in Melanoma. Cancer 2017, 123, 2104–2117. [Google Scholar] [CrossRef]
- Bugaj, L.J.; Sabnis, A.J.; Mitchell, A.; Garbarino, J.E.; Toettcher, J.E.; Bivona, T.G.; Lim, W.A. Cancer Mutations and Targeted Drugs Can Disrupt Dynamic Signal Encoding by the Ras-Erk Pathway. Science 2018, 361. [Google Scholar] [CrossRef]
- Proietti, I.; Skroza, N.; Bernardini, N.; Tolino, E.; Balduzzi, V.; Marchesiello, A.; Michelini, S.; Volpe, S.; Mambrin, A.; Mangino, G.; et al. Mechanisms of Acquired BRAF Inhibitor Resistance in Melanoma: A Systematic Review. Cancers 2020, 12, 2801. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.; Yacoub, N.; Mishra, R.; White, A.; Long, Y.; Alanazi, S.; Garrett, J.T. Current Advances in the Treatment of BRAF-Mutant Melanoma. Cancers 2020, 12, 1661. [Google Scholar] [CrossRef]
- Nassar, K.W.; Tan, A.C. The Mutational Landscape of Mucosal Melanoma. Semin. Cancer Biol. 2020, 61, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Michaloglou, C.; Vredeveld, L.C.; Soengas, M.S.; Denoyelle, C.; Kuilman, T.; Van der Horst, C.M.; Majoor, D.M.; Shay, J.W.; Mooi, W.J.; Peeper, D.S. BRAFE600-Associated Senescence-Like Cell Cycle Arrest of Human Naevi. Nature 2005, 436, 720–724. [Google Scholar] [CrossRef]
- Michaloglou, C.; Vredeveld, L.C.; Mooi, W.J.; Peeper, D.S. BRAF(E600) in Benign and Malignant Human Tumours. Oncogene 2008, 27, 877–895. [Google Scholar] [CrossRef] [PubMed]
- Salgia, R.; Kulkarni, P. The Genetic/Non-Genetic Duality of Drug ‘Resistance’ in Cancer. Trends Cancer 2018, 4, 110–118. [Google Scholar] [CrossRef]
- Khaliq, M.; Fallahi-Sichani, M. Epigenetic Mechanisms of Escape From BRAF Oncogene Dependency. Cancers 2019, 11, 1480. [Google Scholar] [CrossRef] [PubMed]
- Arozarena, I.; Wellbrock, C. Phenotype Plasticity As Enabler of Melanoma Progression and Therapy Resistance. Nat. Rev. Cancer 2019, 19, 377–391. [Google Scholar] [CrossRef]
- Rossi, A.; Roberto, M.; Panebianco, M.; Botticelli, A.; Mazzuca, F.; Marchetti, P. Drug Resistance of BRAF-Mutant Melanoma: Review of Up-to-Date Mechanisms of Action and Promising Targeted Agents. Eur. J. Pharmacol. 2019, 862, 172621. [Google Scholar] [CrossRef]
- Kakadia, S.; Yarlagadda, N.; Awad, R.; Kundranda, M.; Niu, J.; Naraev, B.; Mina, L.; Dragovich, T.; Gimbel, M.; Mahmoud, F. Mechanisms of Resistance to BRAF and MEK Inhibitors and Clinical Update of US Food and Drug Administration-Approved Targeted Therapy in Advanced Melanoma. Onco Targets. Ther. 2018, 11, 7095–7107. [Google Scholar] [CrossRef]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved Survival With Vemurafenib in Melanoma With BRAF V600E Mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, L.; Huang, S.; Heynen, G.J.; Prahallad, A.; Robert, C.; Haanen, J.; Blank, C.; Wesseling, J.; Willems, S.M.; et al. Reversible and Adaptive Resistance to BRAF(V600E) Inhibition in Melanoma. Nature 2014, 508, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Hong, A.; Moriceau, G.; Sun, L.; Lomeli, S.; Piva, M.; Damoiseaux, R.; Holmen, S.L.; Sharpless, N.E.; Hugo, W.; Lo, R.S. Exploiting Drug Addiction Mechanisms to Select Against MAPKi-Resistant Melanoma. Cancer Discov. 2018, 8, 74–93. [Google Scholar] [CrossRef]
- Obenauf, A.C.; Zou, Y.; Ji, A.L.; Vanharanta, S.; Shu, W.; Shi, H.; Kong, X.; Bosenberg, M.C.; Wiesner, T.; Rosen, N.; et al. Therapy-Induced Tumour Secretomes Promote Resistance and Tumour Progression. Nature 2015, 520, 368–372. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF Gene in Human Cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Yue, J.; Vendramin, R.; Liu, F.; Lopez, O.; Valencia, M.G.; Gomes Dos, S.H.; Gaidosh, G.; Beckedorff, F.; Blumenthal, E.; Speroni, L.; et al. Targeted Chemotherapy Overcomes Drug Resistance in Melanoma. Genes Dev. 2020, 34, 637–649. [Google Scholar] [CrossRef]
- Fallahi-Sichani, M.; Becker, V.; Izar, B.; Baker, G.J.; Lin, J.R.; Boswell, S.A.; Shah, P.; Rotem, A.; Garraway, L.A.; Sorger, P.K. Adaptive Resistance of Melanoma Cells to RAF Inhibition Via Reversible Induction of a Slowly Dividing De-Differentiated State. Mol. Syst. Biol. 2017, 13, 905. [Google Scholar] [CrossRef]
- Eskiocak, B.; McMillan, E.A.; Mendiratta, S.; Kollipara, R.K.; Zhang, H.; Humphries, C.G.; Wang, C.; Garcia-Rodriguez, J.; Ding, M.; Zaman, A.; et al. Biomarker Accessible and Chemically Addressable Mechanistic Subtypes of BRAF Melanoma. Cancer Discov. 2017, 7, 832–851. [Google Scholar] [CrossRef]
- Savoia, P.; Fava, P.; Casoni, F.; Cremona, O. Targeting the ERK Signaling Pathway in Melanoma. Int. J. Mol. Sci. 2019, 20, 1483. [Google Scholar] [CrossRef] [PubMed]
- Druillennec, S.; Pouponnot, C.; Eychene, A. NRAS-Driven Melanoma: A RAF Can Hide Another. Mol. Cell. Oncol. 2017, 4, e1344758. [Google Scholar] [CrossRef]
- Atefi, M.; Titz, B.; Tsoi, J.; Avramis, E.; Le, A.; Ng, C.; Lomova, A.; Lassen, A.; Friedman, M.; Chmielowski, B.; et al. CRAF R391W Is a Melanoma Driver Oncogene. Sci. Rep. 2016, 6, 27454. [Google Scholar] [CrossRef]
- Liu, D.; Liu, X.; Xing, M. Activities of Multiple Cancer-Related Pathways Are Associated With BRAF Mutation and Predict the Resistance to BRAF/MEK Inhibitors in Melanoma Cells. Cell Cycle 2014, 13, 208–219. [Google Scholar] [CrossRef]
- Read, J.; Wadt, K.A.; Hayward, N.K. Melanoma Genetics. J. Med. Genet. 2016, 53, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.J.; Johnson, D.B.; Sosman, J.A.; Chandra, S. Melanoma: What Do All the Mutations Mean? Cancer 2018, 124, 3490–3499. [Google Scholar] [CrossRef]
- Almeida, F.V.; Douglass, S.M.; Fane, M.E.; Weeraratna, A.T. Bad Company: Microenvironmentally Mediated Resistance to Targeted Therapy in Melanoma. Pigment Cell Melanoma Res. 2019, 32, 237–247. [Google Scholar] [CrossRef]
- Kemper, K.; De Goeje, P.L.; Peeper, D.S.; Van Amerongen, R. Phenotype Switching: Tumor Cell Plasticity As a Resistance Mechanism and Target for Therapy. Cancer Res. 2014, 74, 5937–5941. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Baselga, J.; Hyman, D.M. A View on Drug Resistance in Cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Swayden, M.; Chhouri, H.; Anouar, Y.; Grumolato, L. Tolerant/Persister Cancer Cells and the Path to Resistance to Targeted Therapy. Cells 2020, 9, 2601. [Google Scholar] [CrossRef]
- Poulikakos, P.I.; Persaud, Y.; Janakiraman, M.; Kong, X.; Ng, C.; Moriceau, G.; Shi, H.; Atefi, M.; Titz, B.; Gabay, M.T.; et al. RAF Inhibitor Resistance Is Mediated by Dimerization of Aberrantly Spliced BRAF(V600E). Nature 2011, 480, 387–390. [Google Scholar] [CrossRef]
- Tian, Y.; Guo, W. A Review of the Molecular Pathways Involved in Resistance to BRAF Inhibitors in Patients With Advanced-Stage Melanoma. Med. Sci. Monit. 2020, 26, e920957. [Google Scholar] [CrossRef]
- Czarnecka, A.M.; Bartnik, E.; Fiedorowicz, M.; Rutkowski, P. Targeted Therapy in Melanoma and Mechanisms of Resistance. Int. J. Mol. Sci. 2020, 21, 4576. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Cohen, M.S. The Discovery of Vemurafenib for the Treatment of BRAF-Mutated Metastatic Melanoma. Expert Opin. Drug Discov. 2016, 11, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.; Al Hraishawi, H.; Simhadri, S.; Hirshfield, K.M.; Chen, S.; Pine, S.; Jeyamohan, C.; Sokol, L.; Ali, S.; Teo, M.L.; et al. BRAF Fusion As a Novel Mechanism of Acquired Resistance to Vemurafenib in BRAF(V600E) Mutant Melanoma. Clin. Cancer Res. 2017, 23, 5631–5638. [Google Scholar] [CrossRef] [PubMed]
- Vido, M.J.; Le, K.; Hartsough, E.J.; Aplin, A.E. BRAF Splice Variant Resistance to RAF Inhibitor Requires Enhanced MEK Association. Cell Rep. 2018, 25, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Landrette, S.F.; Wang, T.; Evans, P.; Bacchiocchi, A.; Bjornson, R.; Cheng, E.; Stiegler, A.L.; Gathiaka, S.; Acevedo, O.; et al. Identification of PLX4032-Resistance Mechanisms and Implications for Novel RAF Inhibitors. Pigment Cell Melanoma Res. 2014, 27, 253–262. [Google Scholar] [CrossRef]
- Wagle, N.; Van Allen, E.M.; Treacy, D.J.; Frederick, D.T.; Cooper, Z.A.; Taylor-Weiner, A.; Rosenberg, M.; Goetz, E.M.; Sullivan, R.J.; Farlow, D.N.; et al. MAP Kinase Pathway Alterations in BRAF-Mutant Melanoma Patients With Acquired Resistance to Combined RAF/MEK Inhibition. Cancer Discov. 2014, 4, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Ng, W.H.; Tian, Z.; Yap, J.; Baccarini, M.; Chen, Z.; Hu, J. Activating Mutations in MEK1 Enhance Homodimerization and Promote Tumorigenesis. Sci. Signal. 2018, 11, eaar6795. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.J.; Jha, S.; Restaino, C.R.; Dayananth, P.; Zhu, H.; Cooper, A.; Carr, D.; Deng, Y.; Jin, W.; Black, S.; et al. Discovery of a Novel ERK Inhibitor With Activity in Models of Acquired Resistance to BRAF and MEK Inhibitors. Cancer Discov. 2013, 3, 742–750. [Google Scholar] [CrossRef]
- Welsh, S.J.; Rizos, H.; Scolyer, R.A.; Long, G.V. Resistance to Combination BRAF and MEK Inhibition in Metastatic Melanoma: Where to Next? Eur. J. Cancer 2016, 62, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Jha, S.; Morris, E.J.; Hruza, A.; Mansueto, M.S.; Schroeder, G.K.; Arbanas, J.; McMasters, D.; Restaino, C.R.; Dayananth, P.; Black, S.; et al. Dissecting Therapeutic Resistance to ERK Inhibition. Mol. Cancer Ther. 2016, 15, 548–559. [Google Scholar] [CrossRef] [PubMed]
- Ramsdale, R.; Jorissen, R.N.; Li, F.Z.; Al Obaidi, S.; Ward, T.; Sheppard, K.E.; Bukczynska, P.E.; Young, R.J.; Boyle, S.E.; Shackleton, M.; et al. The Transcription Cofactor C-JUN Mediates Phenotype Switching and BRAF Inhibitor Resistance in Melanoma. Sci. Signal. 2015, 8, ra82. [Google Scholar] [CrossRef] [PubMed]
- Fallahi-Sichani, M.; Moerke, N.J.; Niepel, M.; Zhang, T.; Gray, N.S.; Sorger, P.K. Systematic Analysis of BRAF(V600E) Melanomas Reveals a Role for JNK/c-Jun Pathway in Adaptive Resistance to Drug-Induced Apoptosis. Mol. Syst. Biol. 2015, 11, 797. [Google Scholar] [CrossRef] [PubMed]
- Titz, B.; Lomova, A.; Le, A.; Hugo, W.; Kong, X.; Ten Hoeve, J.; Friedman, M.; Shi, H.; Moriceau, G.; Song, C.; et al. JUN Dependency in Distinct Early and Late BRAF Inhibition Adaptation States of Melanoma. Cell Discov. 2016, 2, 16028. [Google Scholar] [CrossRef]
- Liu, L.; Yue, Q.; Ma, J.; Liu, Y.; Zhao, T.; Guo, W.; Zhu, G.; Guo, S.; Wang, S.; Gao, T.; et al. POU4F1 Promotes the Resistance of Melanoma to BRAF Inhibitors Through MEK/ERK Pathway Activation and MITF Up-Regulation. Cell Death Dis. 2020, 11, 451. [Google Scholar] [CrossRef]
- Richard, G.; Dalle, S.; Monet, M.A.; Ligier, M.; Boespflug, A.; Pommier, R.M.; De la Fouchardière, A.; Perier-Muzet, M.; Depaepe, L.; Barnault, R.; et al. ZEB1-Mediated Melanoma Cell Plasticity Enhances Resistance to MAPK Inhibitors. EMBO Mol. Med. 2016, 8, 1143–1161. [Google Scholar] [CrossRef]
- Chan, X.Y.; Singh, A.; Osman, N.; Piva, T.J. Role Played by Signalling Pathways in Overcoming BRAF Inhibitor Resistance in Melanoma. Int. J. Mol. Sci. 2017, 18, 1527. [Google Scholar] [CrossRef] [PubMed]
- Deuker, M.M.; Durban, V.M.; Phillips, W.A.; McMahon, M. PI3’-Kinase Inhibition Forestalls the Onset of MEK1/2 Inhibitor Resistance in BRAF-Mutated Melanoma. Cancer Discov. 2015, 5, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Caporali, S.; Alvino, E.; Lacal, P.M.; Levati, L.; Giurato, G.; Memoli, D.; Caprini, E.; Antonini Cappellini, G.C.; D’Atri, S. Targeting the PI3K/AKT/MTOR Pathway Overcomes the Stimulating Effect of Dabrafenib on the Invasive Behavior of Melanoma Cells With Acquired Resistance to the BRAF Inhibitor. Int. J. Oncol. 2016, 49, 1164–1174. [Google Scholar] [CrossRef] [PubMed]
- Cipponi, A.; Goode, D.L.; Bedo, J.; McCabe, M.J.; Pajic, M.; Croucher, D.R.; Rajal, A.G.; Junankar, S.R.; Saunders, D.N.; Lobachevsky, P.; et al. MTOR Signaling Orchestrates Stress-Induced Mutagenesis, Facilitating Adaptive Evolution in Cancer. Science 2020, 368, 1127–1131. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Liu, J.; Huang, L.; Qin, Y.; Hawley, T.; Seo, C.; Merlino, G.; Yu, Y. AXL/AKT Axis Mediated-Resistance to BRAF Inhibitor Depends on PTEN Status in Melanoma. Oncogene 2018, 37, 3275–3289. [Google Scholar] [CrossRef]
- Shi, H.; Hong, A.; Kong, X.; Koya, R.C.; Song, C.; Moriceau, G.; Hugo, W.; Yu, C.C.; Ng, C.; Chodon, T.; et al. A Novel AKT1 Mutant Amplifies an Adaptive Melanoma Response to BRAF Inhibition. Cancer Discov. 2014, 4, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Smalley, K.S.; Lioni, M.; Dalla, P.M.; Xiao, M.; Desai, B.; Egyhazi, S.; Hansson, J.; Wu, H.; King, A.J.; Van Belle, P.; et al. Increased Cyclin D1 Expression Can Mediate BRAF Inhibitor Resistance in BRAF V600E-Mutated Melanomas. Mol. Cancer Ther. 2008, 7, 2876–2883. [Google Scholar] [CrossRef]
- Azimi, A.; Caramuta, S.; Seashore-Ludlow, B.; Bostrom, J.; Robinson, J.L.; Edfors, F.; Tuominen, R.; Kemper, K.; Krijgsman, O.; Peeper, D.S.; et al. Targeting CDK2 Overcomes Melanoma Resistance Against BRAF and Hsp90 Inhibitors. Mol. Syst. Biol. 2018, 14, e7858. [Google Scholar] [CrossRef]
- Janostiak, R.; Malvi, P.; Wajapeyee, N. Anaplastic Lymphoma Kinase Confers Resistance to BRAF Kinase Inhibitors in Melanoma. Iscience 2019, 16, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.; Liu, Z.; Jain, A.; Lyon, A.; Meeks, C.; Richards, D.; Liu, J.; He, D.; Wang, C.; Nespi, M.; et al. Combating Acquired Resistance to MAPK Inhibitors in Melanoma by Targeting Abl1/2-Mediated Reactivation of MEK/ERK/MYC Signaling. Nat. Commun. 2020, 11, 5463. [Google Scholar] [CrossRef]
- Lu, H.; Liu, S.; Zhang, G.; Bin, W.; Zhu, Y.; Frederick, D.T.; Hu, Y.; Zhong, W.; Randell, S.; Sadek, N.; et al. PAK Signalling Drives Acquired Drug Resistance to MAPK Inhibitors in BRAF-Mutant Melanomas. Nature 2017, 550, 133–136. [Google Scholar] [CrossRef]
- Chang, X.; Zhang, T.; Wang, Q.; Rathore, M.G.; Reddy, K.; Chen, H.; Shin, S.H.; Ma, W.Y.; Bode, A.M.; Dong, Z. HI-511 Overcomes Melanoma Drug Resistance Via Targeting AURKB and BRAF V600E. Theranostics 2020, 10, 9721–9740. [Google Scholar] [CrossRef] [PubMed]
- Das, I.; Wilhelm, M.; Hoiom, V.; Franco, M.R.; Costa, S.F.; Hansson, J.; Tuominen, R.; Egyhazi, B.S. Combining ERBB Family and MET Inhibitors Is an Effective Therapeutic Strategy in Cutaneous Malignant Melanoma Independent of BRAF/NRAS Mutation Status. Cell Death Dis. 2019, 10, 663. [Google Scholar] [CrossRef]
- Díaz-Martínez, M.; Benito-Jardón, L.; Alonso, L.; Koetz-Ploch, L.; Hernando, E.; Teixidó, J. MiR-204-5p and MiR-211-5p Contribute to BRAF Inhibitor Resistance in Melanoma. Cancer Res. 2018, 78, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Fattore, L.; Mancini, R.; Acunzo, M.; Romano, G.; Lagana, A.; Pisanu, M.E.; Malpicci, D.; Madonna, G.; Mallardo, D.; Capone, M.; et al. MiR-579-3p Controls Melanoma Progression and Resistance to Target Therapy. Proc. Natl. Acad. Sci. USA 2016, 113, E5005–E5013. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Lin, C.H.; Liu, S.M.; Miyashita, A.; Ihn, H.; Lin, H.; Ng, C.H.; Tsai, J.C.; Chen, M.H.; Tsai, M.S.; et al. MiR-524-5p Reduces the Progression of the BRAF Inhibitor-Resistant Melanoma. Neoplasia 2020, 22, 789–799. [Google Scholar] [CrossRef]
- Nissan, M.H.; Pratilas, C.A.; Jones, A.M.; Ramirez, R.; Won, H.; Liu, C.; Tiwari, S.; Kong, L.; Hanrahan, A.J.; Yao, Z.; et al. Loss of NF1 in Cutaneous Melanoma Is Associated With RAS Activation and MEK Dependence. Cancer Res. 2014, 74, 2340–2350. [Google Scholar] [CrossRef] [PubMed]
- Haq, R.; Yokoyama, S.; Hawryluk, E.B.; Jonsson, G.B.; Frederick, D.T.; McHenry, K.; Porter, D.; Tran, T.N.; Love, K.T.; Langer, R.; et al. BCL2A1 Is a Lineage-Specific Antiapoptotic Melanoma Oncogene That Confers Resistance to BRAF Inhibition. Proc. Natl. Acad. Sci. USA 2013, 110, 4321–4326. [Google Scholar] [CrossRef]
- Rapino, F.; Delaunay, S.; Rambow, F.; Zhou, Z.; Tharun, L.; De Tullio, P.; Sin, O.; Shostak, K.; Schmitz, S.; Piepers, J.; et al. Codon-Specific Translation Reprogramming Promotes Resistance to Targeted Therapy. Nature 2018, 558, 605–609. [Google Scholar] [CrossRef]
- Girard, C.A.; Lecacheur, M.; Ben Jouira, R.; Berestjuk, I.; Diazzi, S.; Prod’homme, V.; Mallavialle, A.; Larbret, F.; Gesson, M.; Schaub, S.; et al. A Feed-Forward Mechanosignaling Loop Confers Resistance to Therapies Targeting the MAPK Pathway in BRAF-Mutant Melanoma. Cancer Res. 2020, 80, 1927–1941. [Google Scholar] [CrossRef]
- Diazzi, S.; Tartare-Deckert, S.; Deckert, M. Bad Neighborhood: Fibrotic Stroma As a New Player in Melanoma Resistance to Targeted Therapies. Cancers 2020, 12, 1364. [Google Scholar] [CrossRef]
- Vashisht Gopal, Y.N.; Gammon, S.; Prasad, R.; Knighton, B.; Pisaneschi, F.; Roszik, J.; Feng, N.; Johnson, S.; Pramanik, S.; Sudderth, J.; et al. A Novel Mitochondrial Inhibitor Blocks MAPK Pathway and Overcomes MAPK Inhibitor Resistance in Melanoma. Clin. Cancer Res. 2019, 25, 6429–6442. [Google Scholar] [CrossRef] [PubMed]
- Aloia, A.; Mullhaupt, D.; Chabbert, C.D.; Eberhart, T.; Flueckiger, S.; Vukolic, A.; Eichhoff, O.M.; Irmisch, A.; Alexander, L.T.; Scibona, E.; et al. A Fatty Acid Oxidation-Dependent Metabolic Shift Regulates the Adaptation of BRAF-Mutated Melanoma to MAPK Inhibitors. Clin. Cancer Res. 2019, 25, 6852–6867. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.P.; Rowling, E.J.; Miskolczi, Z.; Ferguson, J.; Spoerri, L.; Haass, N.K.; Sloss, O.; McEntegart, S.; Arozarena, I.; Von Kriegsheim, A.; et al. Targeting Endothelin Receptor Signalling Overcomes Heterogeneity Driven Therapy Failure. EMBO Mol. Med. 2017, 9, 1011–1029. [Google Scholar] [CrossRef]
- Burd, C.E.; Liu, W.; Huynh, M.V.; Waqas, M.A.; Gillahan, J.E.; Clark, K.S.; Fu, K.; Martin, B.L.; Jeck, W.R.; Souroullas, G.P.; et al. Mutation-Specific RAS Oncogenicity Explains NRAS Codon 61 Selection in Melanoma. Cancer Discov. 2014, 4, 1418–1429. [Google Scholar] [CrossRef]
- Sarkisian, S.; Davar, D. MEK Inhibitors for the Treatment of NRAS Mutant Melanoma. Drug Des. Dev. Ther. 2018, 12, 2553–2565. [Google Scholar] [CrossRef] [PubMed]
- Echevarria-Vargas, I.M.; Reyes-Uribe, P.I.; Guterres, A.N.; Yin, X.; Kossenkov, A.V.; Liu, Q.; Zhang, G.; Krepler, C.; Cheng, C.; Wei, Z.; et al. Co-Targeting BET and MEK As Salvage Therapy for MAPK and Checkpoint Inhibitor-Resistant Melanoma. EMBO Mol. Med. 2018, 10, e8446. [Google Scholar] [CrossRef] [PubMed]
- Delyon, J.; Lebbe, C.; Dumaz, N. Targeted Therapies in Melanoma Beyond BRAF: Targeting NRAS-Mutated and KIT-Mutated Melanoma. Curr. Opin. Oncol. 2020, 32, 79–84. [Google Scholar] [CrossRef]
- Yin, C.; Zhu, B.; Zhang, T.; Liu, T.; Chen, S.; Liu, Y.; Li, X.; Miao, X.; Li, S.; Mi, X.; et al. Pharmacological Targeting of STK19 Inhibits Oncogenic NRAS-Driven Melanomagenesis. Cell 2019, 176, 1113–1127. [Google Scholar] [CrossRef] [PubMed]
- Matter, A.V.; Micaletto, S.; Urner-Bloch, U.; Dummer, R.; Goldinger, S.M. Long-Term Response to Intermittent Binimetinib in Patients With NRAS-Mutant Melanoma. Oncologist 2020, 25, e1593–e1597. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, M.; Boissiére, T.; Noe, G.M.; Litchfield, K.; Mitter, R.; Walker, J.; Kjœr, S.; Ismail, M.; Downward, J.; Swanton, C.; et al. Evidence That STK19 Is Not an NRAS-Dependent Melanoma Driver. Cell 2020, 181, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Tran, B.; Cohen, M.S. The Discovery and Development of Binimetinib for the Treatment of Melanoma. Expert Opin. Drug Discov. 2020, 15, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Malka-Mahieu, H.; Girault, I.; Rubington, M.; Leriche, M.; Welsch, C.; Kamsu-Kom, N.; Zhao, Q.; Desaubry, L.; Vagner, S.; Robert, C. Synergistic Effects of EIF4A and MEK Inhibitors on Proliferation of NRAS-Mutant Melanoma Cell Lines. Cell Cycle 2016, 15, 2405–2409. [Google Scholar] [CrossRef]
- Nagler, A.; Vredevoogd, D.W.; Alon, M.; Cheng, P.F.; Trabish, S.; Kalaora, S.; Arafeh, R.; Goldin, V.; Levesque, M.P.; Peeper, D.S.; et al. A Genome-Wide CRISPR Screen Identifies FBXO42 Involvement in Resistance Toward MEK Inhibition in NRAS-Mutant Melanoma. Pigment Cell Melanoma Res. 2020, 33, 334–344. [Google Scholar] [CrossRef]
- Vu, H.L.; Aplin, A.E. Targeting TBK1 Inhibits Migration and Resistance to MEK Inhibitors in Mutant NRAS Melanoma. Mol. Cancer Res. 2014, 12, 1509–1519. [Google Scholar] [CrossRef]
- Vu, H.L.; Aplin, A.E. Targeting Mutant NRAS Signaling Pathways in Melanoma. Pharmacol. Res. 2016, 107, 111–116. [Google Scholar] [CrossRef]
- Romano, G.; Chen, P.L.; Song, P.; McQuade, J.L.; Liang, R.J.; Liu, M.; Roh, W.; Duose, D.Y.; Carapeto, F.C.L.; Li, J.; et al. A Preexisting Rare PIK3CA(E545K) Subpopulation Confers Clinical Resistance to MEK Plus CDK4/6 Inhibition in NRAS Melanoma and Is Dependent on S6K1 Signaling. Cancer Discov. 2018, 8, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Roesch, A. Tumor Heterogeneity and Plasticity As Elusive Drivers for Resistance to MAPK Pathway Inhibition in Melanoma. Oncogene 2015, 34, 2951–2957. [Google Scholar] [CrossRef]
- Rambow, F.; Marine, J.C.; Goding, C.R. Melanoma Plasticity and Phenotypic Diversity: Therapeutic Barriers and Opportunities. Genes Dev. 2019, 33, 1295–1318. [Google Scholar] [CrossRef]
- Ahmed, F.; Haass, N.K. Microenvironment-Driven Dynamic Heterogeneity and Phenotypic Plasticity As a Mechanism of Melanoma Therapy Resistance. Front. Oncol. 2018, 8, 173. [Google Scholar] [CrossRef] [PubMed]
- Falletta, P.; Sanchez-Del-Campo, L.; Chauhan, J.; Effern, M.; Kenyon, A.; Kershaw, C.J.; Siddaway, R.; Lisle, R.; Freter, R.; Daniels, M.J.; et al. Translation Reprogramming Is an Evolutionarily Conserved Driver of Phenotypic Plasticity and Therapeutic Resistance in Melanoma. Genes Dev. 2017, 31, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Avagliano, A.; Fiume, G.; Pelagalli, A.; SanitÃ, G.; Ruocco, M.R.; Montagnani, S.; Arcucci, A. Metabolic Plasticity of Melanoma Cells and Their Crosstalk With Tumor Microenvironment. Front. Oncol. 2020, 10, 722. [Google Scholar] [CrossRef]
- Menon, D.R.; Das, S.; Krepler, C.; Vultur, A.; Rinner, B.; Schauer, S.; Kashofer, K.; Wagner, K.; Zhang, G.; Rad, E.B.; et al. A Stress-Induced Early Innate Response Causes Multidrug Tolerance in Melanoma. Oncogene 2015, 34, 4545. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.P.; Marchbank, K.; Webster, M.R.; Valiga, A.A.; Kaur, A.; Vultur, A.; Li, L.; Herlyn, M.; Villanueva, J.; Liu, Q.; et al. Hypoxia Induces Phenotypic Plasticity and Therapy Resistance in Melanoma Via the Tyrosine Kinase Receptors ROR1 and ROR2. Cancer Discov. 2013, 3, 1378–1393. [Google Scholar] [CrossRef]
- Roesch, A.; Paschen, A.; Landsberg, J.; Helfrich, I.; Becker, J.C.; Schadendorf, D. Phenotypic Tumour Cell Plasticity As a Resistance Mechanism and Therapeutic Target in Melanoma. Eur. J. Cancer 2016, 59, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Solit, D.B.; Garraway, L.A.; Pratilas, C.A.; Sawai, A.; Getz, G.; Basso, A.; Ye, Q.; Lobo, J.M.; She, Y.; Osman, I.; et al. BRAF Mutation Predicts Sensitivity to MEK Inhibition. Nature 2006, 439, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Das, T.M.; Salangsang, F.; Landman, A.S.; Sellers, W.R.; Pryer, N.K.; Levesque, M.P.; Dummer, R.; McMahon, M.; Stuart, D.D. Modelling Vemurafenib Resistance in Melanoma Reveals a Strategy to Forestall Drug Resistance. Nature 2013, 494, 251–255. [Google Scholar]
- Johannessen, C.M.; Johnson, L.A.; Piccioni, F.; Townes, A.; Frederick, D.T.; Donahue, M.K.; Narayan, R.; Flaherty, K.T.; Wargo, J.A.; Root, D.E.; et al. A Melanocyte Lineage Program Confers Resistance to MAP Kinase Pathway Inhibition. Nature 2013, 504, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Vlčková, K.; Réda, J.; Ondrušová, L.; Krayem, M.; Ghanem, G.; Vachtenheim, J. GLI Inhibitor GANT61 Kills Melanoma Cells and Acts in Synergy With Obatoclax. Int. J. Oncol. 2016, 49, 953–960. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).