Temporal Dominance of B.1.1.7 over B.1.354 SARS-CoV-2 Variant: A Hypothesis Based on Areas of Variant Co-Circulation
Abstract
1. Introduction
2. Materials and Methods
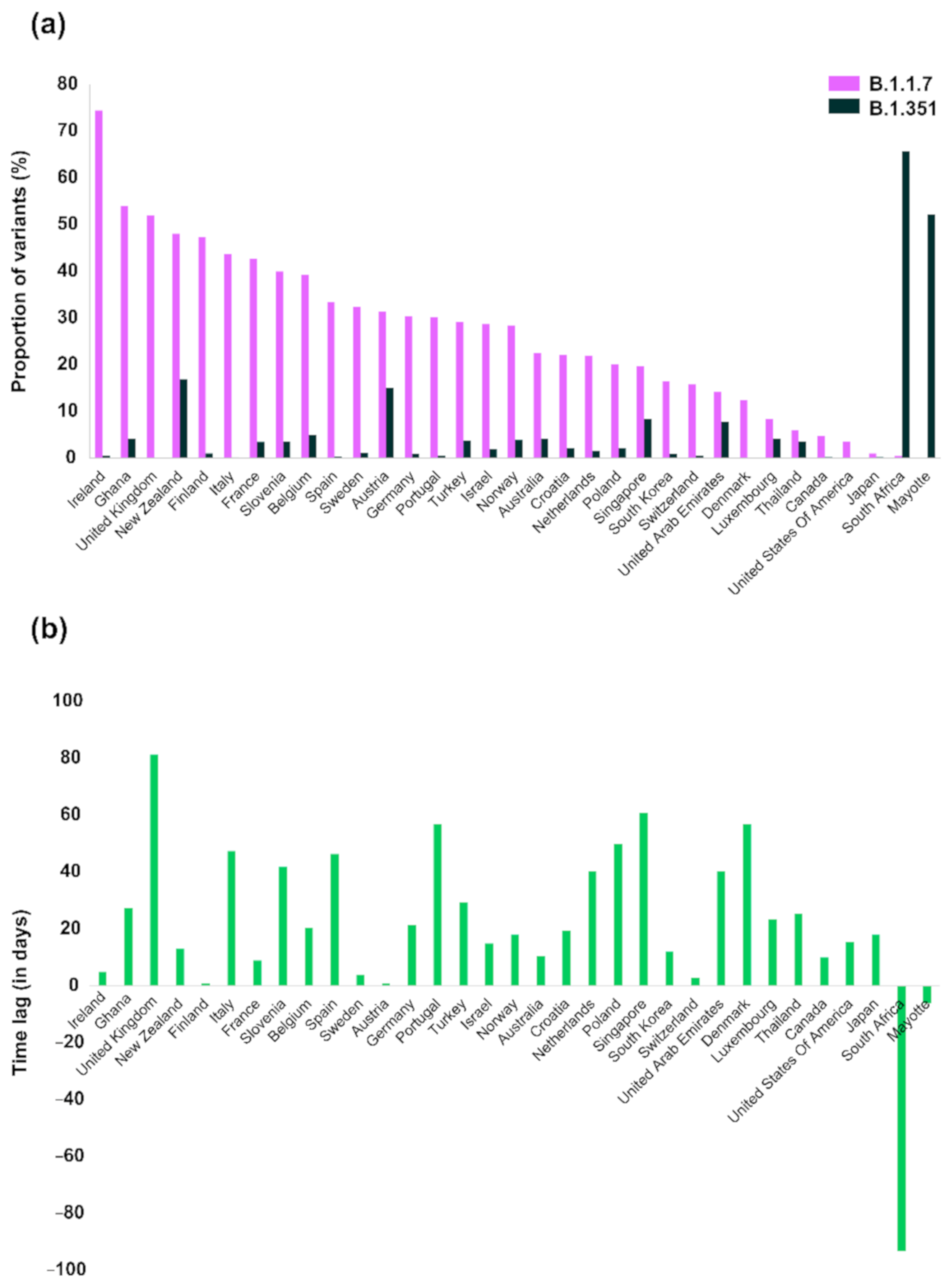
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Weekly Epidemiological Update—23 February 2021 Special Edition: Proposed Working Definitions of SARS-CoV-2 Variants of Interestand Variants of Concern; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Peng, H.; Quinlan, B.D.; Rangarajan, E.S.; Pan, A.; Vanderheiden, A.; Suthar, M.S.; et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020, 11, 6013. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Loman, N.; Pybus, O.; Barclay, W.; Barrett, J.; Carabelli, A.; Connor, T.; Peacock, T.; Robertson, D.L.; Volz, E. Preliminary Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in the UK Defined by a Novel Set of Spike Mutations. Available online: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (accessed on 2 April 2021).
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020. [Google Scholar] [CrossRef]
- Faria, N.R.; Claro, I.M.; Candido, D.; Franco, L.A.M.; Andrade, P.S.; Coletti, T.M.; Silva, C.A.M.; Sales, F.C.; Manuli, E.R.; Aguiar, R.S.; et al. Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in Manaus: Preliminary Findings. Available online: https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 (accessed on 2 April 2021).
- Zhang, W.; Davis, B.D.; Chen, S.S.; Martinez, J.M.S.; Plummer, J.T.; Vail, E. Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA 2021, 325, 1324–1326. [Google Scholar] [CrossRef] [PubMed]
- Lasek-Nesselquist, E.; Lapierre, P.; Schneider, E.; George, K.S.; Pata, J. The localized rise of a B.1.526 SARS-CoV-2 variant containing an E484K mutation in New York State. medRxiv 2021. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef] [PubMed]
- Nelson, G.; Buzko, O.; Spilman, P.; Niazi, K.; Rabizadeh, S.; Soon-Shiong, P. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y.V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escape mutant. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kissler, S.; Fauver, J.R.; Mack, C.; Tai, C.G.; Breban, M.I.; Watkins, A.E.; Samant, R.M.; Anderson, D.J.; Ho, D.D.; Grubaugh, N.D.; et al. Densely sampled viral trajectories suggest longer duration of acute infection with B.1.1.7 variant relative to non-B.1.1.7 SARS-CoV-2. Harvard University’s DASH repository 2021. medRxiv 2021. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021, 2021, 1–6. [Google Scholar] [CrossRef]
- Cele, S.; Gazy, I.; Jackson, L.; Hwa, S.-H.; Tegally, H.; Lustig, G.; Giandhari, J.; Pillay, S.; Wilkinson, E.; Naidoo, Y.; et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature 2021, 2021, 1–9. [Google Scholar] [CrossRef]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; de Oliveira, T.; Vermeulen, M.; van der Berg, K.; et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021, 27, 622–625. [Google Scholar] [CrossRef] [PubMed]
- Wadman, M.; Cohen, J. Novavax vaccine delivers 89% efficacy against COVID-19 in U.K.—But is less potent in South Africa. Science 2021. [Google Scholar] [CrossRef]
- Callaway, E.; Mallapaty, S. Novavax offers first evidence that COVID vaccines protect people against variants. Nature 2021, 590, 17. [Google Scholar]
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021. [Google Scholar] [CrossRef] [PubMed]
- Baele, G.; Suchard, M.A.; Rambaut, A.; Lemey, P. Emerging Concepts of Data Integration in Pathogen Phylodynamics. Syst. Biol. 2017, 66, e47–e65. [Google Scholar] [CrossRef] [PubMed]
- Cobey, S. Pathogen evolution and the immunological niche. Ann. N. Y. Acad. Sci. 2014, 1320, 1–15. [Google Scholar] [CrossRef] [PubMed]

| VOC | B.1.1.7 | B.1.351 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Countries | Proportion (%) | Proportion of SGTF 1—Week 8 of 2021 (%) | Earliest Variant Sequence | Number of Variant Sequences | Number of Sequences 2 | Proportion (%) | Earliest Variant Sequence | Number of Variant Sequences | Number of Sequences 2 |
| Ireland | 74.50 | 88.6 | 17/12/2020 | 1966 | 2639 | 0.62 | 22/12/2020 | 16 | 2594 |
| United Kingdom | 51.99 | NA 3 | 20/09/2020 | 111,140 | 213,761 | 0.16 | 10/12/2020 | 212 | 135,667 |
| Ghana | 54.03 | NA | 10/12/2020 | 67 | 124 | 4.21 | 06/01/2021 | 4 | 95 |
| Finland | 47.43 | NA | 18/12/2020 | 268 | 565 | 1.07 | 19/12/2020 | 6 | 563 |
| Italy | 43.67 | NA | 14/12/2020 | 2244 | 5139 | 0.21 | 30/01/2021 | 8 | 3788 |
| France | 42.75 | 65.8 | 13/12/2020 | 1847 | 4320 | 3.48 | 22/12/2020 | 144 | 4137 |
| Slovenia | 40.00 | NA | 29/12/2020 | 46 | 115 | 3.64 | 09/02/2021 | 2 | 55 |
| Belgium | 39.33 | 46.3 | 30/11/2020 | 2064 | 5248 | 5.02 | 20/12/2020 | 244 | 4857 |
| Spain | 33.53 | 25–30 | 08/11/2020 | 1323 | 3946 | 0.34 | 24/12/2020 | 10 | 2930 |
| Sweden | 32.39 | 41.5 4 | 20/12/2020 | 458 | 1414 | 1.22 | 24/12/2020 | 16 | 1310 |
| New Zealand | 48.09 | NA | 16/12/2020 | 63 | 131 | 16.96 | 29/12/2020 | 19 | 112 |
| Portugal | 30.17 | 50.5 | 09/11/2020 | 746 | 2473 | 0.51 | 04/01/2021 | 9 | 1782 |
| Germany | 30.44 | 54.5 | 30/11/2020 | 4427 | 14,543 | 1.03 | 21/12/2020 | 143 | 13,817 |
| Israel | 28.74 | ~90 5 | 16/12/2020 | 434 | 1510 | 1.92 | 31/12/2020 | 16 | 833 |
| Turkey | 29.15 | NA | 24/12/2020 | 479 | 1643 | 3.78 | 22/01/2021 | 54 | 1428 |
| Norway | 28.49 | 72.5 | 09/12/2020 | 337 | 1183 | 3.92 | 27/12/2020 | 40 | 1021 |
| Netherlands | 22.00 | 64.3 5 | 12/11/2020 | 1697 | 7715 | 1.63 | 22/12/2020 | 99 | 6091 |
| Croatia | 22.06 | NA | 20/01/2021 | 60 | 272 | 2.13 | 09/02/2021 | 2 | 94 |
| Australia | 22.52 | NA | 30/11/2020 | 134 | 595 | 4.14 | 10/12/2020 | 23 | 556 |
| Poland | 20.08 | 9 | 22/12/2020 | 151 | 752 | 2.11 | 10/02/2021 | 2 | 95 |
| Austria | 31.49 | 63.2 | 22/12/2020 | 336 | 1067 | 15.09 | 23/12/2020 | 159 | 1054 |
| South Korea | 16.45 | NA | 14/12/2020 | 90 | 547 | 0.97 | 26/12/2020 | 4 | 413 |
| Switzerland | 15.80 | 40.5 5 | 09/11/2020 | 1983 | 12,550 | 0.63 | 12/11/2020 | 77 | 12,313 |
| Denmark | 12.47 | 76.5 | 09/11/2020 | 4889 | 39,191 | 0.06 | 04/01/2021 | 12 | 19,655 |
| Singapore | 19.70 | NA | 08/12/2020 | 66 | 335 | 8.45 | 07/02/2021 | 6 | 71 |
| United Arab Emirates | 14.19 | NA | 16/11/2020 | 21 | 148 | 7.81 | 26/12/2020 | 5 | 64 |
| Canada | 4.83 | NA | 15/12/2020 | 54 | 1117 | 0.26 | 25/12/2020 | 2 | 761 |
| Luxembourg | 8.40 | 65.5 | 24/12/2020 | 32 | 381 | 4.17 | 16/01/2021 | 2 | 48 |
| United States of America | 3.58 | 26.2 | 17/12/2020 | 2652 | 74,129 | 0.06 | 01/01/2021 | 38 | 62,390 |
| Thailand | 6.06 | NA | 08/01/2021 | 8 | 132 | 3.61 | 03/02/2021 | 3 | 83 |
| Japan | 1.07 | NA | 01/12/2020 | 59 | 5527 | 0.26 | 19/12/2020 | 9 | 3441 |
| Mayotte | 0.19 | NA | 13/01/2021 | 1 | 518 | 52.24 | 07/01/2021 | 338 | 647 |
| South Africa | 0.47 | NA | 09/01/2021 | 1 | 213 | 65.74 | 08/10/2020 | 1086 | 1652 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostaki, E.G.; Tseti, I.; Tsiodras, S.; Pavlakis, G.N.; Sfikakis, P.P.; Paraskevis, D. Temporal Dominance of B.1.1.7 over B.1.354 SARS-CoV-2 Variant: A Hypothesis Based on Areas of Variant Co-Circulation. Life 2021, 11, 375. https://doi.org/10.3390/life11050375
Kostaki EG, Tseti I, Tsiodras S, Pavlakis GN, Sfikakis PP, Paraskevis D. Temporal Dominance of B.1.1.7 over B.1.354 SARS-CoV-2 Variant: A Hypothesis Based on Areas of Variant Co-Circulation. Life. 2021; 11(5):375. https://doi.org/10.3390/life11050375
Chicago/Turabian StyleKostaki, Evangelia Georgia, Ioulia Tseti, Sotirios Tsiodras, George N. Pavlakis, Petros P. Sfikakis, and Dimitrios Paraskevis. 2021. "Temporal Dominance of B.1.1.7 over B.1.354 SARS-CoV-2 Variant: A Hypothesis Based on Areas of Variant Co-Circulation" Life 11, no. 5: 375. https://doi.org/10.3390/life11050375
APA StyleKostaki, E. G., Tseti, I., Tsiodras, S., Pavlakis, G. N., Sfikakis, P. P., & Paraskevis, D. (2021). Temporal Dominance of B.1.1.7 over B.1.354 SARS-CoV-2 Variant: A Hypothesis Based on Areas of Variant Co-Circulation. Life, 11(5), 375. https://doi.org/10.3390/life11050375









