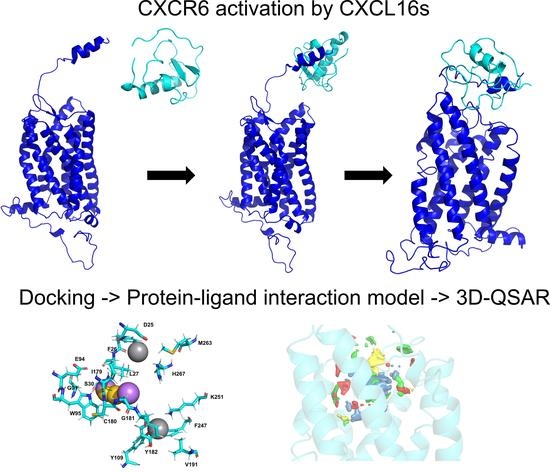
Behavior of Chemokine Receptor 6 (CXCR6) in Complex with CXCL16 Soluble form Chemokine by Molecular Dynamic Simulations: General Protein‒Ligand Interaction Model and 3D-QSAR Studies of Synthetic Antagonists
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Structures, Homology Modeling of CXCR6 and CXCL16s, and Relaxation by All Atom Molecular Dynamics (MD) Simulations
2.2. Protein‒Protein Docking
2.3. Coarse-Grained Molecular Dynamics (CG-MD) Simulations
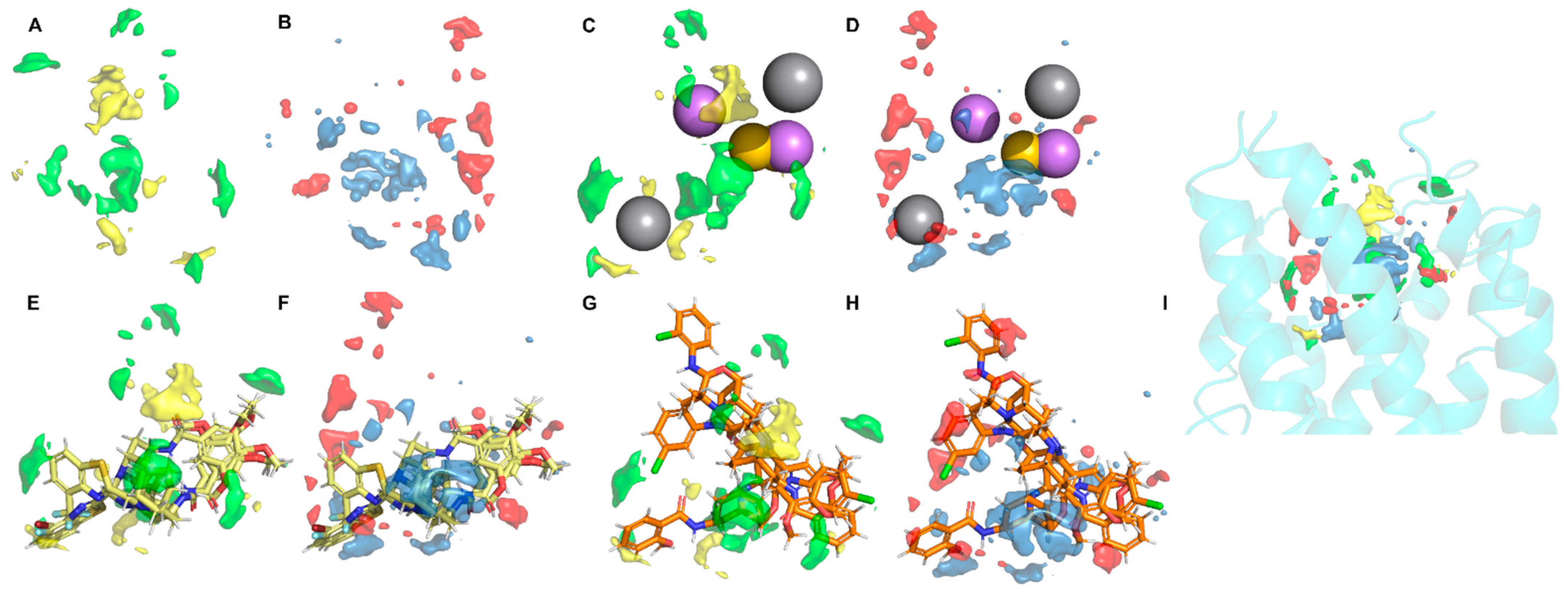
2.4. Ligand Docking, General Protein–Ligand Interaction Model, and Receptor-Based 3D-QSAR Model
2.5. Manipulations of Complexes and Figures
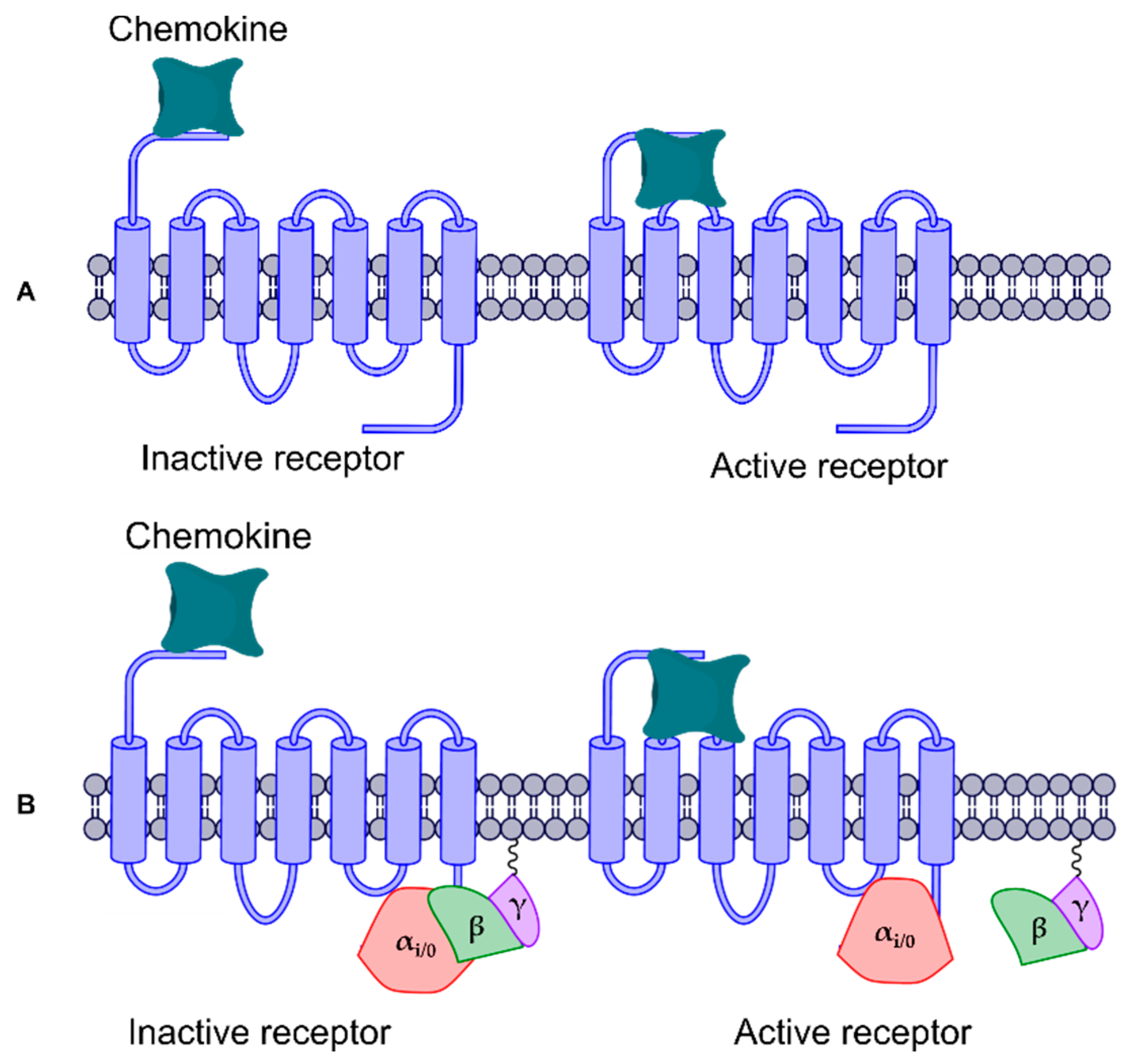
3. Results and Discussion
3.1. Protein Structures and Homology Modeling
3.2. Relaxation of CXCR6 and CXCL16s
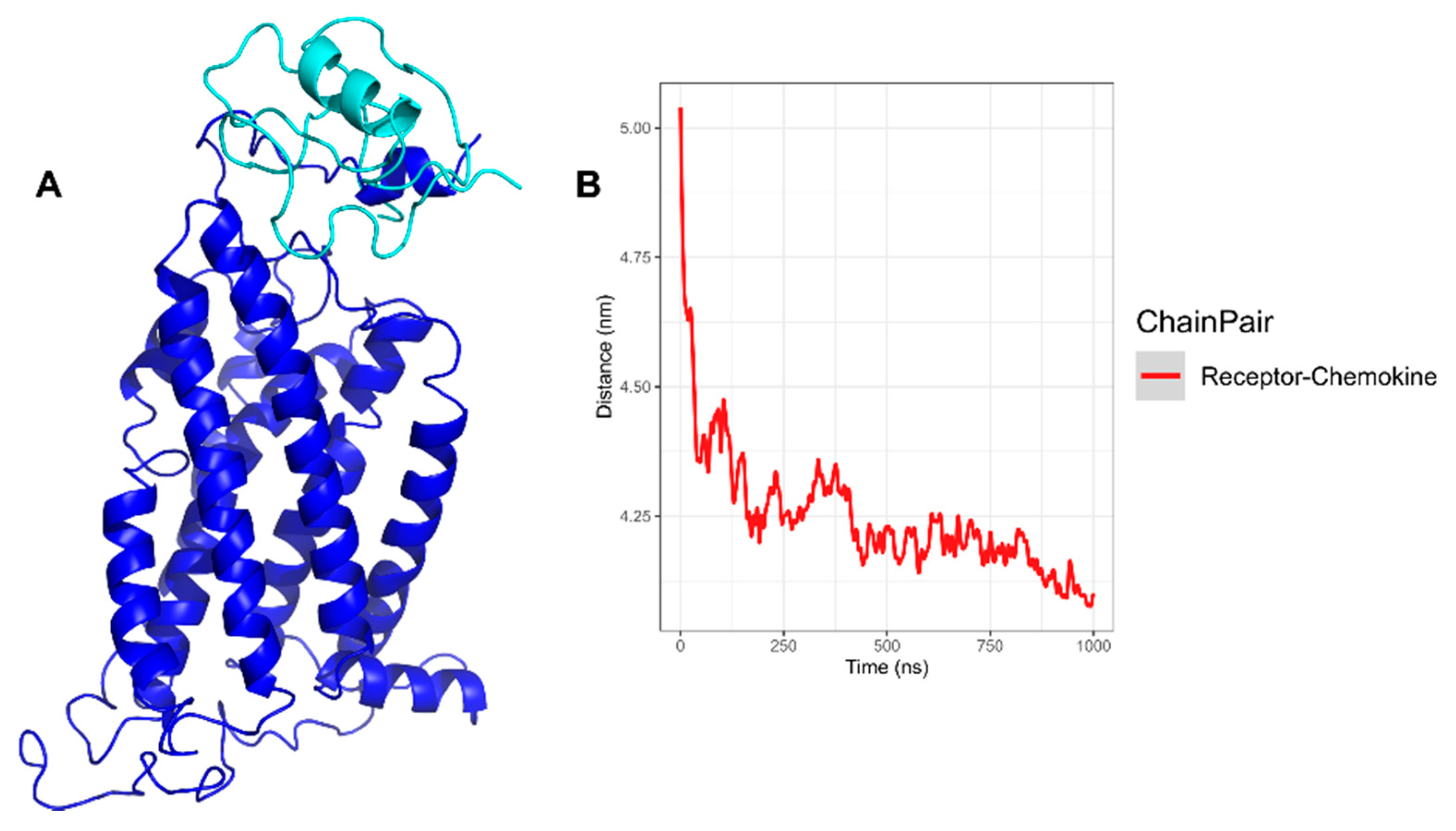
3.3. CXCR6/Chemokine Complex Building and CG-MD Simulations
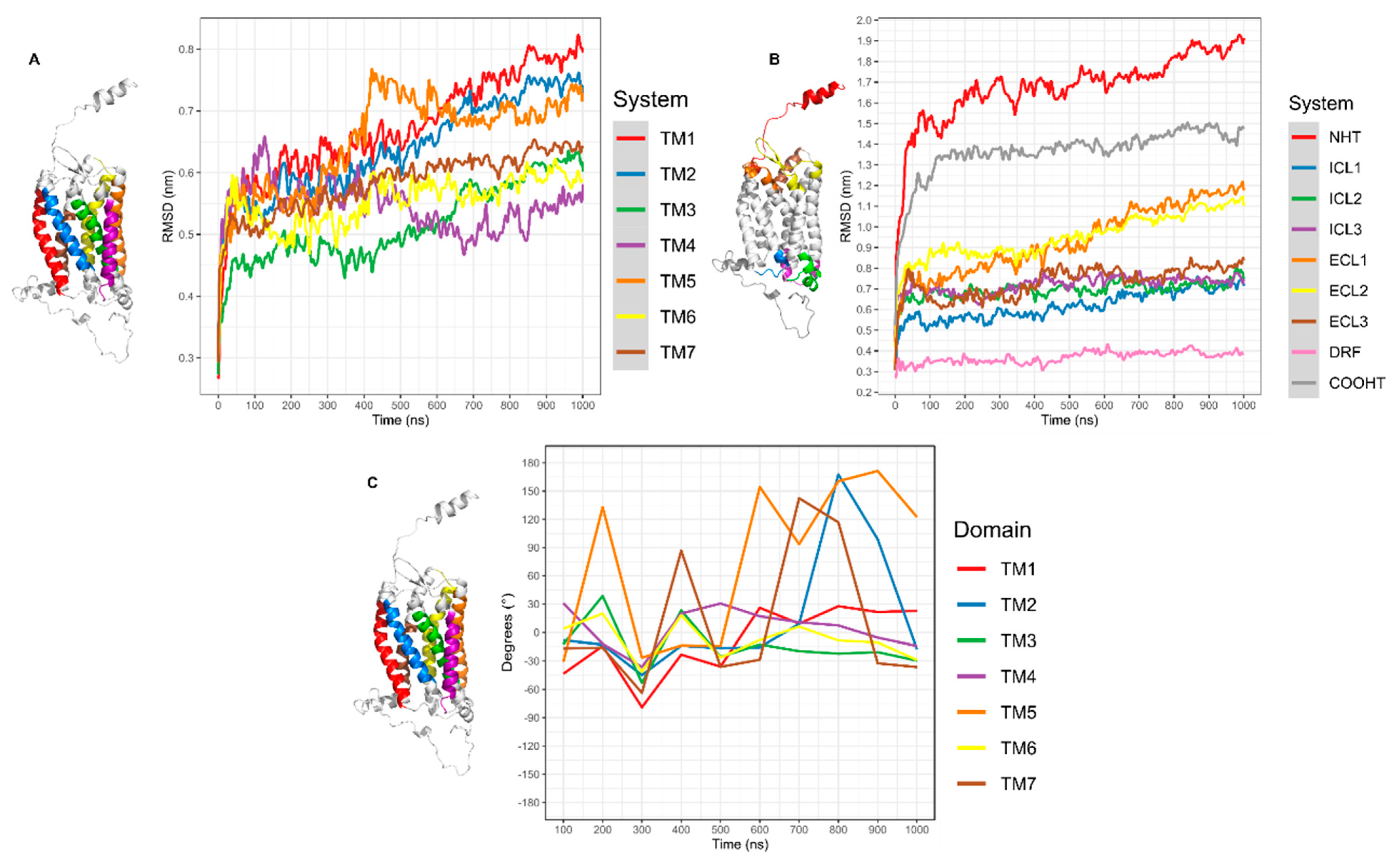
3.3.1. CXCR6 CG-MD Simulations
3.3.2. CXCR6‒CXCL16s CG-MD Simulations
3.4. General Protein‒Ligand Interaction Model and Receptor-Based 3D-QSAR Model of CXCR6 Antagonists
3.4.1. General Protein‒Ligand Interaction Model Based on pIC50
3.4.2. Receptor-Based 3D-QSAR Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Charo, I.F.; Ransohoff, R.M. The many roles of chemokines and chemokine receptors in inflammation. N. Engl. J. Med. 2006, 354, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Chea, S.; Possot, C.; Perchet, T.; Petit, M.; Cumano, A.; Golub, R. CXCR6 expression is important for retention and circulation of ILC precursors. Mediat. Inflamm. 2015, 2015. [Google Scholar] [CrossRef]
- Xiao, G.; Wang, X.; Wang, J.; Zu, L.; Cheng, G.; Hao, M.; Sun, X.; Xue, Y.; Lu, J.; Wang, J. CXCL16/CXCR6 chemokine signaling mediates breast cancer progression by pERK1/2-dependent mechanisms. Oncotarget 2015, 6, 14165–14178. [Google Scholar] [CrossRef] [PubMed]
- Soriani, A.; Stabile, H.; Gismondi, A.; Santoni, A.; Bernardini, G. Chemokine regulation of innate lymphoid cell tissue distribution and function. Cytokine Growth Factor Rev. 2018, 42, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yin, L.; Dai, Y. Modification of SR-PSOX functions by multi-point mutations of basic amino acid residues. Biochimie 2013, 95, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.U. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Scholz, F.; Schulte, A.; Adamski, F.; Hundhausen, C.; Mittag, J.; Schwarz, A.; Kruse, M.L.; Proksch, E.; Ludwig, A. Constitutive expression and regulated release of the transmembrane chemokine CXCL16 in human and murine skin. J. Investig. Dermatol. 2007, 127, 1444–1455. [Google Scholar] [CrossRef]
- Koenen, A.; Babendreyer, A.; Schumacher, J.; Pasqualon, T.; Schwarz, N.; Seifert, A.; Deupi, X.; Ludwig, A.; Dreymueller, D. The DRF motif of CXCR6 as chemokine receptor adaptation to adhesion. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Ludwig, A.; Hundhausen, C.; Lambert, M.; Broadway, N.; Andrews, R.; Bickett, D.; Leesnitzer, M.; Becherer, J. Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Comb. Chem. High Throughput Screen. 2005, 8, 161–171. [Google Scholar] [CrossRef]
- Reiss, K.; Ludwig, A.; Saftig, P. Breaking up the tie: Disintegrin-like metalloproteinases as regulators of cell migration in inflammation and invasion. Pharmacol. Ther. 2006, 111, 985–1006. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, A.; Weber, C. Transmembrane chemokines: Versatile “special agents” in vascular inflammation. Thromb. Haemost. 2007, 97, 694–703. [Google Scholar] [CrossRef] [PubMed]
- Gough, P.J.; Garton, K.J.; Wille, P.T.; Rychlewski, M.; Dempsey, P.J.; Raines, E.W. A disintegrin and metalloproteinase 10-mediated cleavage and shedding regulates the cell surface expression of CXC chemokine ligand 16. J. Immunol. 2004, 172, 3678–3685. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Unutmaz, D.; KewalRamani, V.N.; Littman, D.R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 1997, 388, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Loetscher, M.; Amara, A.; Oberlin, E.; Brass, N.; Legler, D.F.; Loetscher, P.; D’Apuzzo, M.; Meese, E.; Rousset, D.; Virelizier, J.L.; et al. TYMSTR, a putative chemokine receptor selectively expressed in activated T cells, exhibits HIV-1 coreceptor function. Curr. Biol. 1997, 7, 652–660. [Google Scholar] [CrossRef]
- Petit, S.J.; Chayen, N.E.; Pease, J.E. Site-directed mutagenesis of the chemokine receptor CXCR6 suggests a novel paradigm for interactions with the ligand CXCL16. Eur. J. Immunol. 2008, 38, 2337–2350. [Google Scholar] [CrossRef]
- Hydes, T.; Noll, A.; Salinas-Riester, G.; Abuhilal, M.; Armstrong, T.; Hamady, Z.; Primrose, J.; Takhar, A.; Walter, L.; Khakoo, S.I. IL-12 and IL-15 induce the expression of CXCR6 and CD49a on peripheral natural killer cells. Immun. Inflamm. Dis. 2018, 6, 34–46. [Google Scholar] [CrossRef]
- Booth, V.; Keizer, D.W.; Kamphuis, M.B.; Clark-Lewis, I.; Sykes, B.D. The CXCR3 binding chemokine IP-10/CXCL10: Structure and receptor interactions. Biochemistry 2002, 41, 10418–10425. [Google Scholar] [CrossRef]
- Benredjem, B.; Girard, M.; Rhainds, D.; St.-Onge, G.; Heveker, N. Mutational analysis of atypical chemokine receptor 3 (ACKR3/CXCR7) interaction with its chemokine ligands CXCL11 and CXCL12. J. Biol. Chem. 2017, 292, 31–42. [Google Scholar] [CrossRef]
- Sanchez, J.; Huma, Z.E.; Lane, J.R.; Liu, X.; Bridgford, J.L.; Payne, R.J.; Canals, M.; Stone, M.J. Evaluation and extension of the two-site, two-step model for binding and activation of the chemokine receptor CCR1. J. Biol. Chem. 2019, 294, 3464–3475. [Google Scholar] [CrossRef] [PubMed]
- Aguilera-Durán, G.; Romo-Mancillas, A. Computational study of C-X-C chemokine receptor (CXCR)3 binding with its natural agonists chemokine (C-X-C Motif) ligand (CXCL)9, 10 and 11 and with synthetic antagonists: Insights of receptor activation towards drug design for vitiligo. Molecules 2020, 25, 4413. [Google Scholar] [CrossRef]
- Neumann, A.; Engel, V.; Mahardhika, A.B.; Schoeder, C.T.; Namasivayam, V.; Kieć-Kononowicz, K.; Müller, C.E. Computational investigations on the binding mode of ligands for the cannabinoid-activated G protein-coupled receptor GPR18. Biomolecules 2020, 10, 686. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Foley, J.F.; Zhang, H.H.; Hurt, D.E.; Richards, J.L.; Smith, C.S.; Liao, F.; Farber, J.M. Selectivity in the use of G i/o proteins is determined by the DRF motif in CXCR6 and is cell-type specific. Mol. Pharmacol. 2015, 88, 894–910. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Ernst, O.P.; Palczewski, K.; Hofmann, K.P. Activation of rhodopsin: New insights from structural and biochemical studies. Trends Biochem. Sci. 2001, 26, 318–324. [Google Scholar] [CrossRef]
- Hofmann, K.P.; Scheerer, P.; Hildebrand, P.W.; Choe, H.-W.; Park, J.H.; Heck, M.; Ernst, O.P. A G protein-coupled receptor at work: The rhodopsin model. Trends Biochem. Sci. 2009, 34, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Deupi, X. Relevance of rhodopsin studies for GPCR activation. Biochim. Biophys. Acta Bioenergy 2014, 1837, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Woods, K.N.; Pfeffer, J.; Dutta, A.; Klein-Seetharaman, J. Vibrational resonance, allostery, and activation in rhodopsin-like G protein-coupled receptors. Sci. Rep. 2016, 6, 37290. [Google Scholar] [CrossRef]
- Gao, Y.; Hu, H.; Ramachandran, S.; Erickson, J.W.; Cerione, R.A.; Skiniotis, G. Structures of the rhodopsin-transducin complex: Insights into G-protein activation. Mol. Cell 2019, 75, 781–790.e3. [Google Scholar] [CrossRef]
- Gooden, M.J.M.; Wiersma, V.R.; Boerma, A.; Leffers, N.; Boezen, H.M.; Ten Hoor, K.A.; Hollema, H.; Walenkamp, A.M.E.; Daemen, T.; Nijman, H.W.; et al. Elevated serum CXCL16 is an independent predictor of poor survival in ovarian cancer and may reflect pro-metastatic ADAM protease activity. Br. J. Cancer 2014, 110, 1535–1544. [Google Scholar] [CrossRef]
- Hu, W.; Liu, Y.; Zhou, W.; Si, L.; Ren, L. CXCL16 and CXCR6 are coexpressed in human lung cancer In Vivo and mediate the invasion of lung cancer cell lines In Vitro. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Li, J.; Ley, K. Lymphocyte migration into atherosclerotic plaque. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 40–49. [Google Scholar] [CrossRef]
- Wang, C.; Li, Q.; Zhen, J.; Xu, Y.; Sun, S. Simvastatin ameliorates renal lipidosis through the suppression of renal CXCL16 expression in mice with adriamycin-induced nephropathy. Int. J. Clin. Exp. Pathol. 2015, 8, 15696–15707. [Google Scholar] [PubMed]
- Fallahi, P.; Corrado, A.; Di Domenicantonio, A.; Frenzilli, G.; Antonelli, A.; Martina Ferrari, S. CXCR3, CXCR5, CXCR6, and CXCR7 in diabetes. Curr. Drug Targets 2016, 17, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Li, Y.; Li, H.; Lyu, M.; Zhang, D.; Fu, R.; Guan, Y.; Wang, S.; Sun, B.; Dou, X.; et al. Increased plasma sCXCL16 levels may have a relationship with Th1/Th2 imbalance in primary immune thrombocytopenia. Cytokine 2017, 99, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Elmaci, İ.; Altinoz, M.; Sari, R. Immune pathobiology of schwannomas: A concise review. J. Neurol. Surg. Part A Cent. Eur. Neurosurg. 2018, 79, 159–162. [Google Scholar] [CrossRef]
- Hu, W.; Zhen, X.; Xiong, B.; Wang, B.; Zhang, W.; Zhou, W. CXCR6 is expressed in human prostate cancer in vivo and is involved in the in vitro invasion of PC3 and LNCap cells. Cancer Sci. 2008, 99, 1362–1369. [Google Scholar] [CrossRef] [PubMed]
- Peddibhotla, S.; Hershberger, P.M.; Jason Kirby, R.; Sugarman, E.; Maloney, P.R.; Hampton Sessions, E.; Divlianska, D.; Morfa, C.J.; Terry, D.; Pinkerton, A.B.; et al. Discovery of small molecule antagonists of chemokine receptor CXCR6 that arrest tumor growth in SK-HEP-1 mouse xenografts as a model of hepatocellular carcinoma. Bioorg. Med. Chem. Lett. 2020, 30, 126899. [Google Scholar] [CrossRef] [PubMed]
- Floudas, C.A.; Fung, H.K.; McAllister, S.R.; Mönnigmann, M.; Rajgaria, R. Advances in protein structure prediction and de novo protein design: A review. Chem. Eng. Sci. 2006, 61, 966–988. [Google Scholar] [CrossRef]
- De Ruyck, J.; Brysbaert, G.; Blossey, R.; Lensink, M.F. Molecular docking as a popular tool in drug design, an in silico travel. Adv. Appl. Bioinform. Chem. 2016, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mitsutake, A.; Nagai, T.; Takano, H. Relaxation mode analysis for simulations of biomolecules. Seibutsu Butsuri 2013, 1637, 164102. [Google Scholar] [CrossRef]
- Dämgen, M.A.; Biggin, P.C. Computational methods to examine conformational changes and ligand-binding properties: Examples in neurobiology. Neurosci. Lett. 2019, 700, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Legler, D.F.; Thelen, M. New insights in chemokine signaling. F1000 Res. 2018, 7, 95. [Google Scholar] [CrossRef]
- Wasilko, D.J.; Johnson, Z.L.; Ammirati, M.; Che, Y.; Griffor, M.C.; Han, S.; Wu, H. Structural basis for chemokine receptor CCR6 activation by the endogenous protein ligand CCL20. Nat. Commun. 2020, 11, 3031. [Google Scholar] [CrossRef] [PubMed]
- Medina-Ruiz, D.; Erreguin-Luna, B.; Luna-Vázquez, F.J.; Romo-Mancillas, A.; Rojas-Molina, A.; Ibarra-Alvarado, C. Vasodilation elicited by isoxsuprine, identified by high-throughput virtual screening of compound libraries, involves activation of the NO/cGMP and H2S/KATP pathways and blockade of α1-adrenoceptors and calcium channels. Molecules 2019, 24, 987. [Google Scholar] [CrossRef]
- The Uniprot Consortium Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2014, 42, D191–D198. [CrossRef] [PubMed]
- Zhang, J.; Yang, J.; Jang, R.; Zhang, Y. GPCR-I-TASSER: A hybrid approach to g protein-coupled receptor structure modeling and the application to the human genome. Structure 2015, 23, 1538–1549. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2014, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Zhu, Y.; Li, J.; Chen, Z.; Han, G.W.; Kufareva, I.; Li, T.; Ma, L.; Fenalti, G.; Li, J.; et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science 2013, 341, 1387–1390. [Google Scholar] [CrossRef]
- Chan, D.I.; Hunter, H.N.; Tack, B.F.; Vogel, H.J. Human macrophage inflammatory protein 3α: Protein and peptide nuclear magnetic resonance solution structures, dimerization, dynamics, and anti-infective properties. Antimicrob. Agents Chemother. 2008, 52, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins Struct. Funct. Bioinform. 2013, 81, 2159–2166. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Vajda, S.; Yueh, C.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Kozakov, D. New additions to the ClusPro server motivated by CAPRI. Proteins Struct. Funct. Bioinform. 2017, 85, 435–444. [Google Scholar] [CrossRef]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and its limits in rigid body protein-protein docking. Structure 2020, 28, 1071–1081.e3. [Google Scholar] [CrossRef]
- Xia, B.; Vajda, S.; Kozakov, D. Accounting for pairwise distance restraints in FFT-based protein-protein docking. Bioinformatics 2016, 32, 3342–3344. [Google Scholar] [CrossRef] [PubMed]
- Yershova, A.; Jain, S.; LaValle, S.M.; Mitchell, J.C. Generating uniform incremental grids on SO (3) using the hopf fibration. Int. J. Rob. Res. 2010, 29, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. Ligplot: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Ingólfsson, H.I.; Cheng, X.; Lee, J.; Marrink, S.J.; Im, W. CHARMM-gui martini maker for coarse-grained simulations with the martini force field. J. Chem. Theory Comput. 2015, 11, 4486–4494. [Google Scholar] [CrossRef]
- Schrödinger Release 2019-3; Maestro, Schrödinger, LLC: New York, NY, USA, 2019.
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 12 August 2020).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Santos-Martins, D.; Solis-Vasquez, L.; Tillack, A.F.; Sanner, M.F.; Koch, A.; Forli, S. Accelerating AutoDock4 with GPUs and gradient-based local search. J. Chem. Theory Comput. 2021, 17, 1060–1073. [Google Scholar] [CrossRef] [PubMed]
- Koes, D.R.; Camacho, C.J. Pharmer: Efficient and exact pharmacophore search. J. Chem. Inf. Model. 2011, 51, 1307–1314. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, A.P.; Richards, G.; de la Iglesia, B.; Rayward-Smith, V.J. Clustering rules: A comparison of partitioning and hierarchical clustering algorithms. J. Math. Model. Algorithms 2006, 5, 475–504. [Google Scholar] [CrossRef]
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K. Cluster: Cluster Analysis Basics and Extensions. R Package Version 2.1.0. Available online: https://cran.r-project.org/package=cluster (accessed on 12 August 2020).
- Tosco, P.; Balle, T. Open3DQSAR: A new open-source software aimed at high-throughput chemometric analysis of molecular interaction fields. J. Mol. Model. 2011, 17, 201–208. [Google Scholar] [CrossRef]
- Cramer, R.D.; Patterson, D.E.; Bunce, J.D. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. [Google Scholar] [CrossRef] [PubMed]
- Naceiri Mrabti, N.; Elhallaoui, M. QSAR study and molecular docking of benzimidazole derivatives as potent activators of AMP-activated protein kinase. J. Taibah Univ. Sci. 2017, 11, 18–39. [Google Scholar] [CrossRef][Green Version]
- Schrödinger, L. The PyMOL Molecular Graphics System; Version 2.0; Schrödinger Inc.: New York, NY, USA, 2015. [Google Scholar]
- Hillisch, A.; Pineda, L.F.; Hilgenfeld, R. Utility of homology models in the drug discovery process. Drug Discov. Today 2004, 9, 659–669. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Chaturvedi, S.; Swastika Pal, S.; Jain, N.; Mishra, A.K. Improvising 5-HT 7 R homology model for design of high affinity ligands: Model validation with docking, embrace minimization, MM-GBSA, and molecular dynamic simulations. J. Biomol. Struct. Dyn. 2018, 36, 2475–2494. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- States, D.J.; Gish, W. QGB: Combined use of sequence similarity and codon bias for coding region identification. J. Comput. Biol. 1994, 1, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Sobolev, O.V.; Afonine, P.V.; Moriarty, N.W.; Hekkelman, M.L.; Joosten, R.P.; Perrakis, A.; Adams, P.D. A global ramachandran score identifies protein structures with unlikely stereochemistry. Structure 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Clark-Lewis, I.; Mattioli, I.; Gong, J.-H.; Loetscher, P. Structure-function relationship between the human chemokine receptor CXCR3 and its ligands. J. Biol. Chem. 2003, 278, 289–295. [Google Scholar] [CrossRef] [PubMed]





| ID | C-Score | TM-Score | Ramachandran Favored | * z-Score |
|---|---|---|---|---|
| CXCR6-M1 | −0.46 | 0.65 ± 0.13 | 85.29% | −3.03 ± 0.38 |
| CXCR6 | - | - | 84.71% | −3.08 ± 0.37 |
| CXCR6 ** | - | - | 91.76% | −2.05 ± 0.39 |
| CXCL16s-M1 | −1.51 | 0.53 ± 0.15 | 67.57% | −6.24 ± 0.71 |
| CXCL16s-C1 | - | - | 89.19% | −2.50 ± 0.81 |
| TM1 | TM2 | TM3 * | TM4 | TM5 | TM6 | TM7 |
|---|---|---|---|---|---|---|
| 33–59 | 69–89 | 104–128 | 144–164 | 188–215 | 232–259 | 276–293 |
| Pharmacophoric Features | Residues Around 5 Å |
|---|---|
| HAc | L27, S30, G91, E94, W95, I179, C180, G181, Y182 |
| HDn | D25, F26, L27, Y109, Y182, V191, F247, K251, M263, H267 |
| Hph | L27, S30, W95, I179, C180 |
| Models | q2 | R2 | F Test | Test Group |
|---|---|---|---|---|
| Model 7 | 0.6397 | 0.8731 | 72.8899 | 18, 74, 38, 2, 72, 80, 28, 52, 16, 15, 12, 73, 70, 26, 47, 13, 32, 54, 65, 62, 59, 79, 56 |
| Model 1 | 0.6329 | 0.8639 | 67.2740 | 33, 30, 39, 22, 61, 28, 36, 53, 9, 66, 56, 59, 24, 48, 72, 18, 2, 52, 13, 41, 62, 46, 75 |
| Model 4 | 0.4906 | 0.8718 | 72.0680 | 27, 50, 25, 9, 2, 23, 14, 67, 41, 70, 33, 28, 39, 79, 66, 36, 13, 1, 71, 55, 82, 3, 76 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguilera-Durán, G.; Romo-Mancillas, A. Behavior of Chemokine Receptor 6 (CXCR6) in Complex with CXCL16 Soluble form Chemokine by Molecular Dynamic Simulations: General Protein‒Ligand Interaction Model and 3D-QSAR Studies of Synthetic Antagonists. Life 2021, 11, 346. https://doi.org/10.3390/life11040346
Aguilera-Durán G, Romo-Mancillas A. Behavior of Chemokine Receptor 6 (CXCR6) in Complex with CXCL16 Soluble form Chemokine by Molecular Dynamic Simulations: General Protein‒Ligand Interaction Model and 3D-QSAR Studies of Synthetic Antagonists. Life. 2021; 11(4):346. https://doi.org/10.3390/life11040346
Chicago/Turabian StyleAguilera-Durán, Giovanny, and Antonio Romo-Mancillas. 2021. "Behavior of Chemokine Receptor 6 (CXCR6) in Complex with CXCL16 Soluble form Chemokine by Molecular Dynamic Simulations: General Protein‒Ligand Interaction Model and 3D-QSAR Studies of Synthetic Antagonists" Life 11, no. 4: 346. https://doi.org/10.3390/life11040346
APA StyleAguilera-Durán, G., & Romo-Mancillas, A. (2021). Behavior of Chemokine Receptor 6 (CXCR6) in Complex with CXCL16 Soluble form Chemokine by Molecular Dynamic Simulations: General Protein‒Ligand Interaction Model and 3D-QSAR Studies of Synthetic Antagonists. Life, 11(4), 346. https://doi.org/10.3390/life11040346