Stachybotrys musae sp. nov., S. microsporus, and Memnoniella levispora (Stachybotryaceae, Hypocreales) Found on Bananas in China and Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Morphological Studies, and Isolation
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Sequence Alignment
2.4. Phylogenetic Analyses
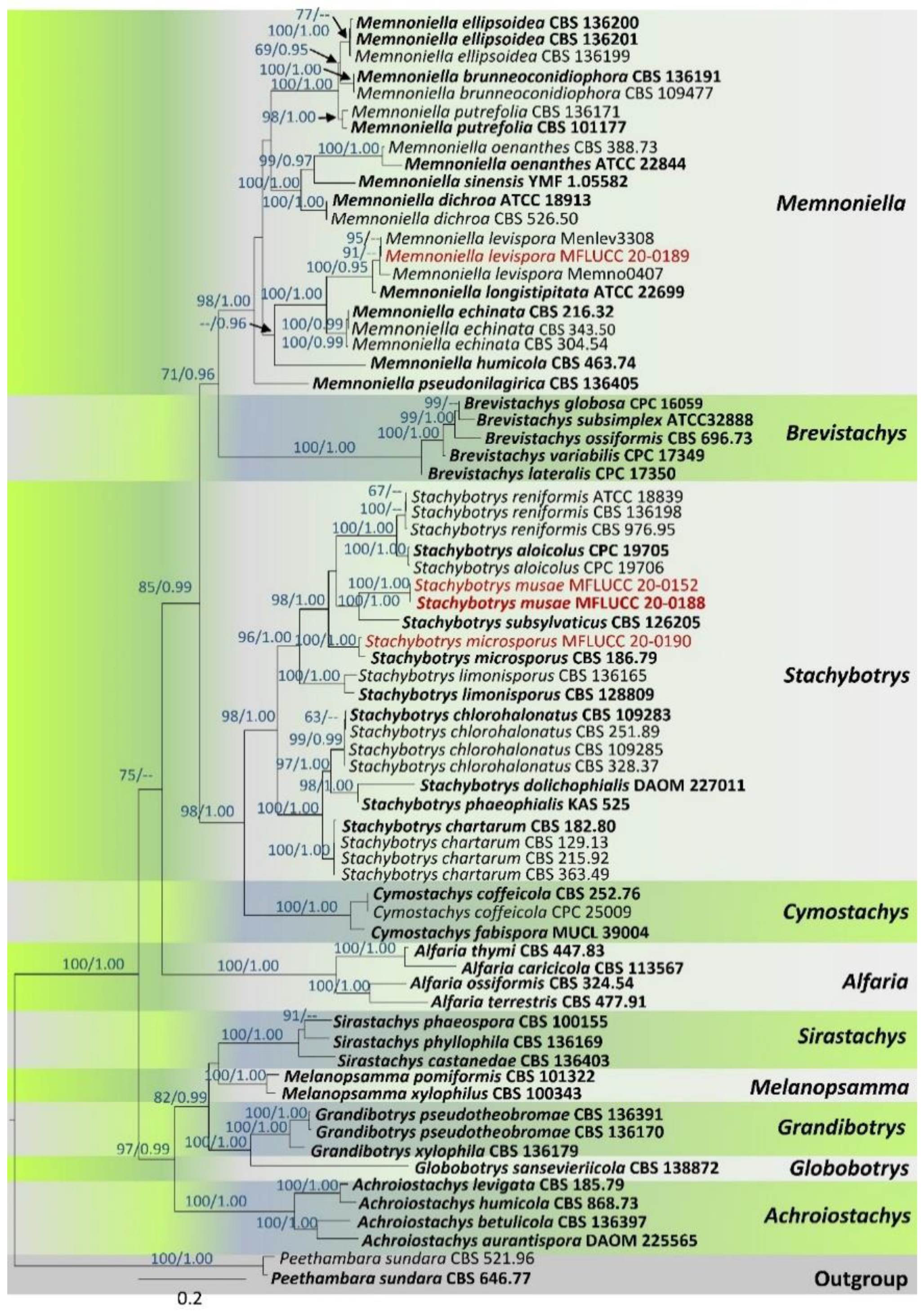
3. Results
3.1. Phylogenetic Analyses
3.2. Taxonomy
3.2.1. Stachybotrys musae Samarakoon & Chomnunti, sp. nov.
- MycoBank No.—MB 838529; FoF Number—FoF 09574.
- Etymology—Name reflects the host genus Musa, from which the novel taxon was originally isolated.
- Holotype—MFLU 20-0626.
3.2.2. Stachybotrys microsporus (B.L. Mathur & Sankhla) S.C. Jong & E.E. Davis
3.2.3. Memnoniella levispora Subram
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhilabutra, W.; McKenzie, E.H.C.; Hyde, K.D.; Lumyong, S. Fungi on the grasses, Thysanolaena latifolia and Saccharum spontaneum, in Northern Thailand. Mycosphere 2010, 1, 301–314. [Google Scholar]
- Hyde, K.D.; Fröhlich, J.; Taylor, J.E. Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia 1998, 50, 21–80. [Google Scholar]
- Konta, S.; Maharachchikumbura, S.S.N.; Senanayake, I.C.; McKenzie, E.H.C.; Stadler, M.; Boonmee, S.; Phookamsak, R.; Jayawardena, R.S.; Senwanna, C.; Hyde, K.D.; et al. A new genus Allodiatrype, five new species and a new host record of diatrypaceous fungi from palms (Arecaceae). Mycosphere 2020, 11, 239–268. [Google Scholar] [CrossRef]
- Thambugala, K.M.; Wanasinghe, D.N.; Phillips, A.J.L.; Camporesi, E.; Bulgakov, T.S.; Phukhamsakda, C.; Ariyawansa, H.A.; Goonasekara, I.D.; Phookamsak, R.; Dissanayake, A.; et al. Mycosphere notes 1-50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 2017, 8, 697–796. [Google Scholar] [CrossRef]
- Thongkantha, S.; Lumyong, S.; McKenzie, E.H.C.; Hyde, K.D. Fungal saprobes and pathogens occurring on tissues of Dracaena lourieri and Pandanus spp. in Thailand. Fungal. Divers. 2008, 30, 149–169. [Google Scholar]
- Tibpromma, S.; Hyde, K.D.; McKenzie, E.H.C.; Bhat, D.J.; Phillips, A.J.L.; Wanasinghe, D.N.; Samarakoon, M.C.; Jayawardena, R.S.; Dissanayake, A.J.; Tennakoon, D.S.; et al. Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal. Divers. 2018, 93, 1–160. [Google Scholar] [CrossRef]
- Wong, M.K.M.; Hyde, K.D. Diversity of fungi on six species of Gramineae and one species of Cyperaceae in Hong Kong. Mycol. Res. 2001, 105, 1485–1491. [Google Scholar] [CrossRef]
- Phookamsak, R.; Norphanphoun, C.; Tanaka, K.; Dai, D.Q.; Luo, Z.L.; Liu, J.K.; Su, H.Y.; Bhat, D.J.; Bahkali, A.H.; Mortimer, P.E.; et al. Towards a natural classification of Astrosphaeriella-like species; introducing Astrosphaeriellaceae and Pseudoastrosphaeriellaceae fam. nov. and Astrosphaeriellopsis, gen. nov. Fungal. Divers. 2015, 74, 143–197. [Google Scholar] [CrossRef]
- Liu, J.K.; Phookamsak, R.; Dai, D.Q.; Tanaka, K.; Jones, E.B.G.; Xu, J.C.; Chukeatirote, E.; Hyde, K.D. Roussoellaceae, a new pleosporalean family to accommodate the genera Neoroussoella gen. nov., Roussoella and Roussoellopsis. Phytotaxa 2014, 181, 1–33. [Google Scholar] [CrossRef]
- Dai, D.Q.; Phookamsak, R.; Wijayawardene, N.N.; Li, W.J.; Bhat, D.J.; Xu, J.C.; Taylor, J.E.; Hyde, K.D.; Chukeatirote, E. Bambusicolous fungi. Fungal. Divers. 2017, 82, 1–105. [Google Scholar] [CrossRef]
- Photita, W.; Lumyong, P.; McKenzie, E.H.C.; Hyde, K.D.; Lumyong, S. Memnoniella and Stachybotrys species from Musa acuminata. Cryptogam. Mycol. 2003, 24, 147–152. [Google Scholar]
- Photita, W.; Lumyong, P.; McKenzie, E.H.C.; Hyde, K.D.; Lumyong, S. Saprobic fungi on dead wild banana. Mycotaxon 2003, 85, 345–356. [Google Scholar]
- Photita, W.; Lumyong, S.; Lumyong, P.; Ho, W.H.; McKenzie, E.H.C.; Hyde, K.D. Fungi on Musa acuminata in Hong Kong. Fungal. Divers. 2001, 6, 99–106. [Google Scholar]
- Samarakoon, B.C.; Phookamsak, R.; Wanasinghe, D.N.; Chomnunti, P.; Hyde, K.D.; McKenzie, E.H.C.; Promputtha, I.; Xu, J.C.; Li, Y.J. Taxonomy and phylogenetic appraisal of Spegazzinia musae sp. nov. and S. deightonii (Didymosphaeriaceae, Pleosporales) on Musaceae from Thailand. Mycokeys 2020, 70, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Samarakoon, B.C.; Wanasinghe, D.N.; Samarakoon, M.C.; Phookamsak, R.; McKenzie, E.H.C.; Chomnunti, P.; Hyde, K.D.; Lumyong, S.; Karunarathna, S.C. Multi-gene phylogenetic evidence suggests Dictyoarthrinium belongs in Didymosphaeriaceae (Pleosporales, Dothideomycetes) and Dictyoarthrinium musae sp. nov. on Musa from Thailand. Mycokeys 2020, 71, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Somrithipol, S. A synnematous species of Dictyoarthrinium from Thailand. Mycologia 2007, 99, 792–796. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Bao, D.F.; Boonmee, S.; Calabon, M.S.; et al. Refined families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Wang, Y.; Hyde, K.D.; McKenzie, E.H.C.; Jiang, Y.L.; Li, D.W.; Zhao, D.G. Overview of Stachybotrys (Memnoniella) and current species status. Fungal. Divers. 2015, 71, 17–83. [Google Scholar] [CrossRef]
- Lombard, L.; Houbraken, J.; Decock, C.; Samson, R.A.; Meijer, M.; Reblova, M.; Groenewald, J.Z.; Crous, P.W. Generic hyper-diversity in Stachybotriaceae. Persoonia 2016, 36, 156–246. [Google Scholar] [CrossRef] [PubMed]
- Smith, G. Some new and interesting species of micro-fungi. Trans. Brit. Mycol. Soc. 1962, 45, 387–394. [Google Scholar] [CrossRef]
- Doilom, M.; Dissanayake, A.J.; Wanasinghe, D.N.; Boonmee, S.; Liu, J.K.; Bhat, D.J.; Taylor, J.E.; Bahkali, A.H.; McKenzie, E.H.C.; Hyde, K.D. Microfungi on Tectona grandis (Teak) in Northern Thailand. Fungal. Divers. 2017, 82, 107–182. [Google Scholar] [CrossRef]
- Lin, C.G.; McKenzie, E.H.C.; Bhat, D.J.; Ran, S.F.; Chen, Y.; Hyde, K.D.; Li, D.W.; Wang, Y. Stachybotrys-like taxa from karst areas and a checklist of Stachybotrys-like species from Thailand. Mycosphere 2016, 7, 1273–1291. [Google Scholar] [CrossRef]
- Mapook, A.; Hyde, K.D.; McKenzie, E.H.C.; Jones, E.B.G.; Bhat, D.J.; Jeewon, R.; Stadler, M.; Samarakoon, M.C.; Malaithong, M.; Tanunchai, B.; et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (siam weed). Fungal. Divers. 2020, 101, 1–175. [Google Scholar] [CrossRef]
- Index Fungorum. Available online: http://www.indexfungorum.org (accessed on 15 February 2021).
- Zheng, H.; Zhang, Z.; Liu, D.Z.; Yu, Z.F. Memnoniella sinensis sp. nov., a new species from China and a key to species of the genus. Int. J. Syst. Evol. Microbiol. 2019, 69, 3155–3163. [Google Scholar] [CrossRef] [PubMed]
- Species Fungorum. Available online: http://www.speciesfungorum.org/Names/Names.asp (accessed on 10 December 2020).
- Ellis, M.B. Dematiaceous Hyphomycetes; Commonwealth Mycological Institute, Kew: London, UK, 1971. [Google Scholar]
- Seifert, K.A.; Morgan-Jones, G.; Gams, W.; Kendrick, B. The Genera of Hyphomycetes; CBS Biodiversity Series: Utrecht, The Netherlands, 2011; Volume 9, p. 997. [Google Scholar]
- Gond, A.K.; Ramanuj, P.; Sanjay, P.; Jamaluddin; Pandy, A.K. New record of Memnoniella levispora subram on Ficus carica L. from India. J. New Biol. Rep. 2013, 2, 272–274. [Google Scholar]
- Ellis, M.B. More Dematiaceous hyphomycetes; Commonwealth Mycological Institute, Kew: Surrey, UK, 1976. [Google Scholar]
- Izabel, T.D.S.S.; Cruz, A.C.R.D.; Barbosa, F.R.; Ferreira, S.M.L.; Marques, M.F.O.; Gusmão, L.F.P. The genus Stachybotrys (anamorphic fungi) in the semi-arid region of Brazil. Rev. Bras. Bot. 2010, 33, 479–487. [Google Scholar] [CrossRef]
- Jie, C.Y.; Geng, K.; Jiang, Y.L.; Xu, J.J.; Hyde, K.D.; McKenzie, E.H.C.; Zhang, T.Y.; Bahkali, A.H.; Li, D.W.; Wang, Y. Stachybotrys from soil in China, identified by morphology and molecular phylogeny. Mycol. Prog. 2013, 12, 693–698. [Google Scholar] [CrossRef]
- Tang, A.M.C.; Hyde, K.D.; Corlett, R.T. Diversity of fungi on wild fruits in Hong Kong. Fungal. Divers. 2003, 14, 165–185. [Google Scholar]
- Whitton, S.R.; McKenzie, E.H.C.; Hyde, K.D. Microfungi on the Pandanaceae: Stachybotrys, with three new species. N. Z. J. Bot. 2001, 39, 489–499. [Google Scholar] [CrossRef]
- Li, Q.R.; Jiang, Y.L. Stachybotrys subreniformis, new from soil in China. Mycotaxon 2011, 115, 171–173. [Google Scholar] [CrossRef]
- Pathak, P.D.; Chauhan, R.K.S. Leaf spot disease of Crotolaria juncea L. caused by Stachybotrys atra Corda. Curr. Sci. 1976, 45, 567. [Google Scholar]
- Zhao, G.H.; Li, D.W.; Jiang, J.H.; Peng, J. First report of Stachybotrys chartarum causing leaf blight of Tillandsia tenuifolia in China. Plant Dis. 2010, 94, 1166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, M.H.; Zhuo, F.F.; Gao, N.; Cheng, X.B.; Wang, X.B.; Pei, Y.H.; Kong, L.Y. Seven new cytotoxic phenylspirodrimane derivatives from the endophytic fungus Stachybotrys chartarum. RSCAdv. 2019, 9, 3520–3531. [Google Scholar] [CrossRef]
- Busby, P.E.; Ridout, M.; Newcombe, G. Fungal endophytes: Modifiers of plant disease. Plant Mol. Biol. 2016, 90, 645–655. [Google Scholar] [CrossRef]
- Cao, R.; Liu, X.; Gao, K.; Mendgen, K.; Kang, Z.; Gao, J.; Dai, Y.; Wang, X. Mycoparasitism of endophytic fungi isolated from reed on soilborne phytopathogenic fungi and production of cell wall-degrading enzymes in vitro. Curr. Microbiol. 2009, 59, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, A.K.H.; Newcombe, G. The contribution of foliar endophytes to quantitative resistance to Melampsora rust. New Phytol. 2013, 197, 909–918. [Google Scholar] [CrossRef]
- Yang, X.; Jin, H.; Xu, L.; Cui, H.; Xin, A.; Liu, H.; Qin, B. Diversity and functions of endophytic fungi associated with roots and leaves of Stipa purpurea in an Alpine Steppe at Tibet plateau. J. Microbiol. Biotechnol. 2020, 30, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Mason, C.D.; Rand, T.G.; Oulton, M.; MacDonald, J.M.; Scott, J.E. Effects of Stachybotrys chartarum (atra) conidia and isolated toxin on lung surfactant production and homeostasis. Nat. Toxins 1998, 6, 27–33. [Google Scholar] [CrossRef]
- Hossain, M.A.; Ahmed, M.S.; Ghannoum, M.A. Attributes of Stachybotrys chartarum and its association with human disease. J. Allergy Clin. Immunol. 2004, 113, 200–208. [Google Scholar] [CrossRef]
- Lichtenstein, J.H.R.; Molina, R.M.; Donaghey, T.C.; Amuzie, C.J.; Pestka, J.J.; Coull, B.A.; Brain, J.D. Pulmonary responses to Stachybotrys chartarum and its toxins: Mouse strain affects clearance and macrophage cytotoxicity. Toxicol. Sci. 2010, 116, 113–121. [Google Scholar] [CrossRef][Green Version]
- Hyde, K.D.; Xu, J.C.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal. Divers. 2019, 97, 1–136. [Google Scholar] [CrossRef]
- Jarvis, B.B.; Salemme, J.; Morals, A. Stachybotrys toxins. 1. Nat. Toxins 1995, 3, 10–16. [Google Scholar] [CrossRef]
- Pan, H.; Kong, J.; Xu, J.; Zhang, Y.; Hang, T. Four new species of Stachybotrys and a key to species of the genus known from soil in China. Mycosystema 2014, 33, 785–792. [Google Scholar]
- Wu, Y.M.; Zhang, T.Y. Two new species of Stachybotrys from soil. Mycotaxon 2009, 109, 461–464. [Google Scholar] [CrossRef]
- Wang, H.; Kong, J.; Wu, Y.; Zhang, T. Notes on soil dematiaceous hyphomycetes from the Qaidam basin, Qinghai province, China I. Mycosystema 2009, 28, 20–24. [Google Scholar]
- Kong, H.Z. Stachybotrys yunnanensis sp. nov. And Neosartorya delicata sp. nov. isolated from Yunnan, China. Mycotaxon 1997, 62, 427–433. [Google Scholar]
- Liu, H.; Zhang, T. A preliminary report of soil dematiaceous hyphomycetes from the Yellow river delta I. Mycosystema 2004, 23, 338–344. [Google Scholar]
- Farr, D.F.; Rossman, A.Y. Fungal Databases, Systematic Mycology and Microbiology Laboratory, ARS, USDA. Available online: http://nt.ars-grin.gov/fungaldatabases/ (accessed on 15 December 2020).
- Castellani, E.; Ciferri, R. Prodromus Mycoflorae Africae Orientalis Italicae; Istituto Agricolo Coloniale Italiano: Florence, Italy, 1937; pp. 121–161. [Google Scholar]
- Misra, P.C.; Srivastava, S.K. Two undescribed Stachybotrys species from India. Trans. Brit. Mycol. Soc. 1982, 78, 556–559. [Google Scholar] [CrossRef]
- Matsushima, T. Icones Microfungorum: A Matsushima Lectorum; Nippon printing Publishing Co.: Osaka, Japan, 1975. [Google Scholar]
- McGuire, J.U.; Crandall, B.S. Survey of Insect Pests and Plant Diseases of Selected Food Crops of Mexico, Central America and Panama; US Department of Agriculture: Washinghton, DC, USA, 1967.
- Samarakoon, S.M.B.C.; Samarakoon, M.C.; Aluthmuhandiram, J.V.S. The first report of Daldinia eschscholtzii as an endophyte from leaves of Musa sp.(musaceae) in Thailand. Asian J. Mycol. 2019, 2, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The faces of fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal. Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Bhunjun, C.S.; Maharachchikumbura, S.S.M.; Liu, J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 2020, 11, 2652–2676. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Groenewald, J.Z.; Nakashima, C.; Nishikawa, J.; Shin, H.D.; Park, J.H.; Jama, A.N.; Groenewald, M.; Braun, U.; Crous, P.W. Species concepts in Cercospora: Spotting the weeds among the roses. Stud. Mycol. 2013, 75, 115–170. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Liu, Y.J.J.; Whelen, S.; Benjamin, D.H. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. Mafft online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Stamatakis, A. Raxml version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the cipres science gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; IEEE: New Orleans, LA, USA, 2010; pp. 1–8. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Rannala, B.; Yang, Z.H. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Zhaxybayeva, O.; Gogarten, J.P. Bootstrap, bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genom. 2002, 3, 1–15. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree Tree Figure Drawing Tool Version 131, Institute of Evolutionary 623 Biology, University of Edinburgh. Available online: http://treebioedacuk/software/figtree/ (accessed on 24 May 2020).
- Crous, P.W.; Shivas, R.G.; Quaedvlieg, W.; van der Bank, M.; Zhang, Y.; Summerell, B.A.; Guarro, J.; Wingfield, M.J.; Wood, A.R.; Alfenas, A.C.; et al. Fungal planet description sheets: 214–280. Persoonia 2014, 32, 184–306. [Google Scholar] [CrossRef] [PubMed]
- Johnston-Monje, D.; Loewen, S.; Lazarovits, G. Mycobiomes of tomato plants with vine decline. Canad. J. Plant Pathol. 2017, 39, 184–200. [Google Scholar] [CrossRef]
- Santos, T.A.B. Relações Filogenéticas dos Complexos Stachybotrys-Memmoniella e Phalangispora-Speiropsis-Wiesneriomyces; Universidade Federal de Pernambuco: Pernambuco, Brazil, 2015. [Google Scholar]
- Arnold, G.R.W. Lista de Hongos Fitopatogenos de Cuba; Ministerio de Cultura, Editorial Cientifico-Tecnica Cuba: Havana, Cuba, 1986. [Google Scholar]
- Barrios, L.M.; Pérez, I.O. Nuevos registros de hongos en semillas de Oryza sativa en cuba. Manejo Integr. Plagas Agroecol. 2005, 75, 64–67. [Google Scholar]
- Munjal, R.L.; Kapoor, J.N. Some hyphomycetes from Himalayas. Mycopathol. Mycol. Appl. 1969, 39, 121–128. [Google Scholar] [CrossRef]
- Rifai, M.A. Stachybotrys bambusicola sp. nov. Trans. Brit. Mycol. Soc. 1964, 47, 269–272. [Google Scholar] [CrossRef]
- McKenzie, E.C. Dematiaceous hyphomycetes on Freycinetia (Pandanaceae). I., Stachybotrys. Mycotaxon 1991, 41, 179–188. [Google Scholar]



| Taxa | Culture Collection | cmdA | ITS | rpb2 | TUB2 | tef1 |
|---|---|---|---|---|---|---|
| Achroiostachys aurantispora | DAOMC 225565 T | KU845784 | KU845804 | KU845840 | NA | KU845859 |
| Ac. betulicola | CBS 136397 T | KU845772 | KU845792 | KU845831 | KU845753 | KU845848 |
| Ac. humicola | CBS 868.73 T | KU845779 | KU845799 | KU845837 | KU845760 | KU845854 |
| Ac. levigata | CBS 185.79 T | KU845785 | KU845805 | KU845841 | KU845765 | KU845860 |
| Alfaria caricicola | CBS 113567 T | KU845976 | KU845983 | KU846001 | KU846014 | KU846008 |
| Al. ossiformis | CBS 324.54 T | KU845977 | KU845984 | KU846002 | KU846015 | KU846009 |
| Al. terrestris | CBS 477.91 T | KU845979 | KU845988 | KU846006 | KU846019 | KU846011 |
| Al. thymi | CBS 447.83 T | KU845981 | KU845990 | NA | KU846021 | KU846013 |
| Brevistachys globosa | CBS 141056 T | KU846024 | KU846038 | NA | KU846101 | KU846085 |
| Br. lateralis | CBS 141058 T | KU846027 | KU846043 | KU846074 | KU846106 | KU846090 |
| Br. ossiformis | CBS 696.73T | NA | KU846044 | NA | KU846107 | NA |
| Br. subsimplex | ATCC 32888 T | NA | AF205439 | NA | NA | NA |
| Br. variabilis | CBS 141057 | KU846030 | KU846047 | KU846076 | KU846110 | KU846093 |
| Cymostachys coffeicola | CPC 25009 | NA | KU846053 | NA | NA | NA |
| Cy. coffeicola | CBS 252.76 T | KU846035 | KU846052 | KU846081 | KU846113 | KU846097 |
| Cy. fabispora | CBS 136180 T | KU846036 | KU846054 | KU846082 | KU846114 | KU846098 |
| Globobotrys sansevieriicola | CBS 138872 T | NA | KR476717 | NA | KR476794 | KR476793 |
| Grandibotrys pseudotheobromae | CBS 136391 | NA | KU846136 | KU846189 | KU846242 | KU846215 |
| Gr. pseudotheobromae | CBS 136170 T | NA | KU846135 | KU846188 | KU846241 | KU846216 |
| Gr. xylophilus | CBS 136179 T | KU846115 | KU846137 | KU846190 | NA | KU846217 |
| Melanopsamma pomiformis | CBS 101322 T | KU846032 | KU846049 | KU846078 | NA | NA |
| Me. xylophila | CBS 100343 T | KU846034 | KU846051 | KU846080 | NA | KU846096 |
| Memnoniella brunneoconidiophora | CBS 109477 | NA | KU846138 | KU846192 | KU846243 | KU846218 |
| M. brunneoconidiophora | CBS 136191 T | KU846116 | KU846139 | KU846193 | KU846244 | KU846219 |
| M. dichroa | CBS 526.50 | KU846117 | KU846140 | KU846194 | NA | KU846220 |
| M. dichroa | ATCC 18913 T | NA | AF081472 | NA | NA | NA |
| M. echinata | CBS 304.54 | KU846120 | KU846143 | KU846197 | NA | NA |
| M. echinata | CBS 343.50 | KU846121 | KU846144 | KU846198 | KU846246 | NA |
| M. echinata | CBS 216.32 T | KU846119 | KU846142 | KU846196 | KU846245 | KU846222 |
| M. ellipsoidea | CBS 136199 | KU846127 | KU846150 | KU846204 | KU846252 | KU846230 |
| M. ellipsoidea | CBS 136200 | KU846128 | KU846151 | KU846205 | KU846253 | KU846231 |
| M. ellipsoidea | CBS 136201 T | KU846129 | KU846152 | KU846206 | KU846254 | KU846232 |
| M. humicola | CBS 463.74 T | KU846130 | KU846154 | KU846208 | NA | KU846234 |
| M. levispora | Menlev3308 | NA | KF626495 | NA | NA | NA |
| M. levispora | Memno0407 | NA | KF626494 | NA | NA | NA |
| M. levispora | MFLUCC 20-0189 | NA | MW477993 | NA | MW480236 | NA |
| M. longistipitata | ATCC 22699 T | NA | AF081471 | NA | NA | NA |
| M. oenanthes | CBS 388.73 | NA | KU846156 | KU846210 | NA | NA |
| M. oenanthes | ATCC 22844 T | NA | AF081473 | NA | NA | KU846236 |
| M. pseudonilagirica | CBS 136405 T | KU846132 | KU846157 | KU846211 | KU846257 | NA |
| M. putrefolia | CBS 136171 | KU846133 | KU846159 | KU846213 | KU846259 | KU846238 |
| M. putrefolia | CBS 101177 T | NA | KU846158 | KU846212 | KU846258 | KU846239 |
| M. sinensis | YMF 1.05582 T | MK772065 | MK773576 | MK773575 | MK773574 | NA |
| Peethambara sundara | CBS 521.96 | NA | KU846470 | KU846508 | KU846550 | KU846530 |
| Pe. sundara | CBS 646.77 T | NA | KU846471 | KU846509 | KU846551 | KU846531 |
| Sirastachys castanedae | CBS 136403T | KU846555 | KU846660 | KU846887 | KU847096 | KU846992 |
| Si. phaeospora | CBS 100155 T | KU846560 | KU846666 | KU846891 | KU847102 | KU846995 |
| Si. phyllophila | CBS 136169 T | KU846566 | KU846672 | KU846897 | KU847108 | KU846999 |
| Stachybotrys aloicolus | CBS 137941 | KU846571 | KJ817889 | KU846902 | KJ817887 | NA |
| S. aloicolus | CBS 137940 T | KU846570 | KJ817888 | KU846901 | KJ817886 | NA |
| S. chartarum | CBS 129.13 | NA | KM231858 | KM232434 | KM232127 | KM231994 |
| S. chartarum | CBS 215.92 | NA | KU846680 | KU846905 | KU847116 | KU847003 |
| S. chartarum | CBS 363.49 | NA | KU846681 | KU846906 | KU847117 | KU847004 |
| S. chartarum | CBS 182.80 T | NA | KU846679 | KU846904 | KU847115 | KU847005 |
| S. chlorohalonatus | CBS 328.37 | KU846619 | KU846725 | KU846950 | KU847160 | KU847048 |
| S. chlorohalonatus | CBS 109283 | KU846622 | KU846728 | KU846953 | KU847163 | KU847049 |
| S. chlorohalonatus | CBS 251.89 | KU846618 | KU846724 | KU846949 | KU847159 | KU847052 |
| S. chlorohalonatus | CBS 109285 T | KU846623 | KU846729 | KU846954 | KU847164 | KU847053 |
| S. dolichophialis | DAOMC 227011 | KU846628 | KU846734 | KU846958 | KU847169 | NA |
| S. limonisporus | CBS 136165 | KU846630 | KU846736 | KU846960 | KU847171 | KU847058 |
| S. limonisporus | CBS 128809 T | KU846629 | KU846735 | KU846959 | KU847170 | KU847059 |
| S. microsporus | CBS 186.79 | KU846631 | KU846737 | DQ676580 | KU847172 | NA |
| S. microsporus | ATCC 18852 T | NA | AF081475 | NA | NA | NA |
| S. microsporus | MFLUCC 20-0190 | NA | MW477992 | NA | MW480235 | MW480237 |
| S. musae | MFLUCC 20-0152 | MW480231 | MW477991 | MW480229 | MW480233 | NA |
| S. musae | MFLUCC 20-0188T | MW480232 | MW477990 | MW480230 | MW480234 | NA |
| S. phaeophialis | KAS 525 T | KU846632 | KU846738 | KU846962 | KU847173 | NA |
| S. reniformis | ATCC 18839 | NA | AF081476 | NA | NA | NA |
| S. reniformis | CBS 136198 | NA | KU846740 | NA | NA | KU847063 |
| S. reniformis | CBS 976.95 | KU846633 | KU846739 | KU846963 | KU847174 | KU847064 |
| S. subsylvaticus | CBS 126205T | KU846634 | KU846741 | KU846964 | KU847175 | KU847076 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samarakoon, B.C.; Wanasinghe, D.N.; Phookamsak, R.; Bhat, J.; Chomnunti, P.; Karunarathna, S.C.; Lumyong, S. Stachybotrys musae sp. nov., S. microsporus, and Memnoniella levispora (Stachybotryaceae, Hypocreales) Found on Bananas in China and Thailand. Life 2021, 11, 323. https://doi.org/10.3390/life11040323
Samarakoon BC, Wanasinghe DN, Phookamsak R, Bhat J, Chomnunti P, Karunarathna SC, Lumyong S. Stachybotrys musae sp. nov., S. microsporus, and Memnoniella levispora (Stachybotryaceae, Hypocreales) Found on Bananas in China and Thailand. Life. 2021; 11(4):323. https://doi.org/10.3390/life11040323
Chicago/Turabian StyleSamarakoon, Binu C., Dhanushka N. Wanasinghe, Rungtiwa Phookamsak, Jayarama Bhat, Putarak Chomnunti, Samantha C. Karunarathna, and Saisamorn Lumyong. 2021. "Stachybotrys musae sp. nov., S. microsporus, and Memnoniella levispora (Stachybotryaceae, Hypocreales) Found on Bananas in China and Thailand" Life 11, no. 4: 323. https://doi.org/10.3390/life11040323
APA StyleSamarakoon, B. C., Wanasinghe, D. N., Phookamsak, R., Bhat, J., Chomnunti, P., Karunarathna, S. C., & Lumyong, S. (2021). Stachybotrys musae sp. nov., S. microsporus, and Memnoniella levispora (Stachybotryaceae, Hypocreales) Found on Bananas in China and Thailand. Life, 11(4), 323. https://doi.org/10.3390/life11040323









