Priming by High Temperature Stress Induces MicroRNA Regulated Heat Shock Modules Indicating Their Involvement in Thermopriming Response in Rice
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
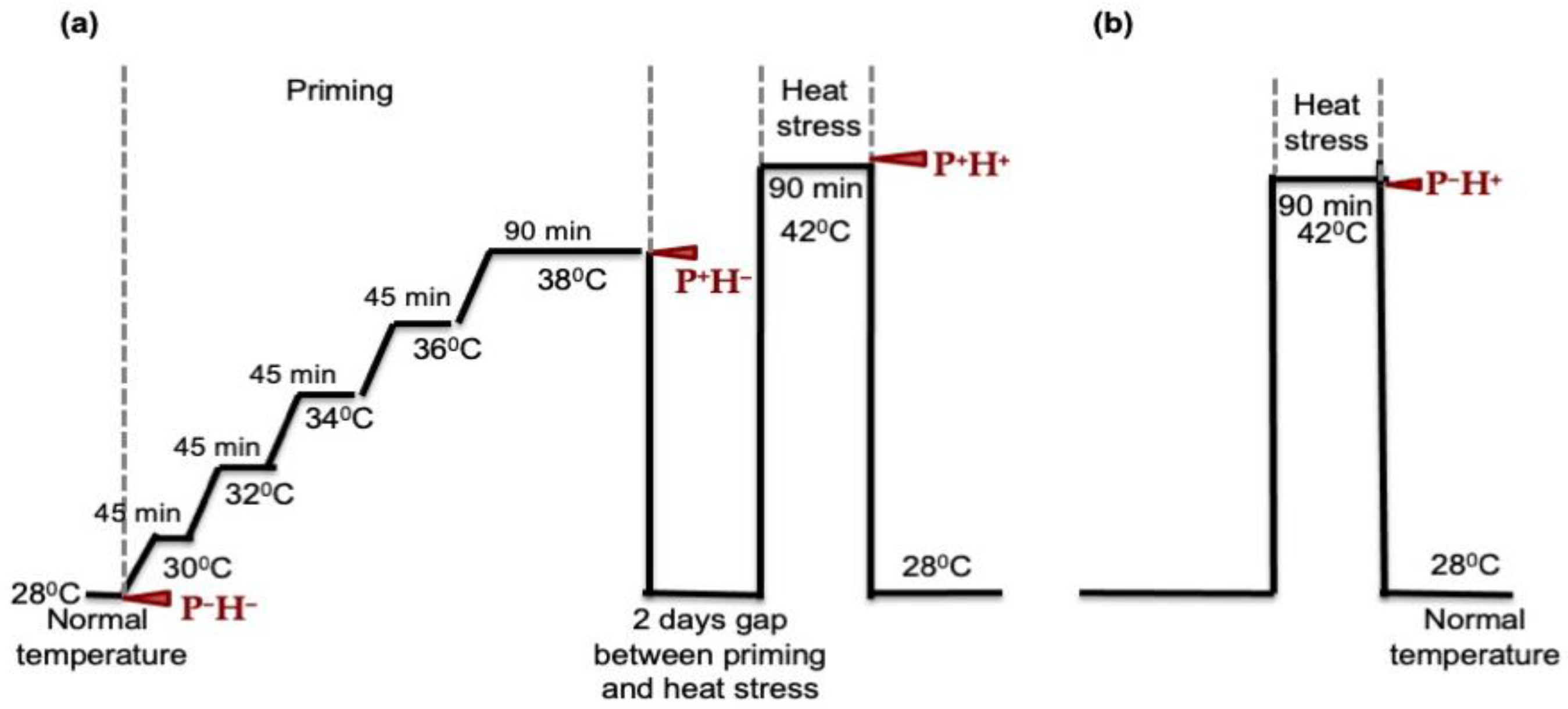
2.2. Heat Stress Priming and High Temperature Treatments
2.3. RNA-seq Data Analysis
2.4. Small RNA Data Analysis
2.5. Biological Pathways and Gene Ontology Enrichment Analysis
2.6. qRT-PCR Analysis
3. Results
3.1. Differential Expression Patterns Between Primed and Non-Primed Plants
3.1.1. Effect of High Temperature Stress on Primed Transcript Profiles
3.1.2. Effect of Priming on Transcript Profiles
3.1.3. Comparison of Priming Induced Transcript Profiles with Those in Stressed Tissues
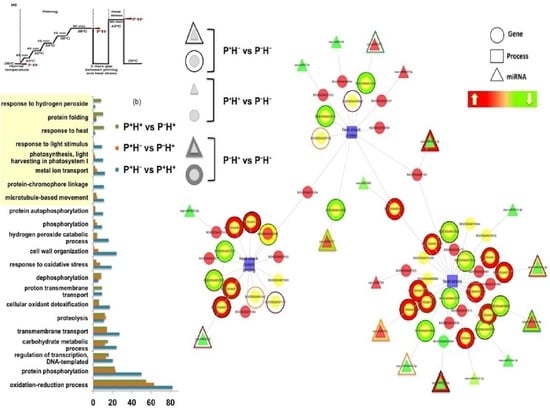
3.1.4. Functional Categorization of the Priming Induced Gene Expression
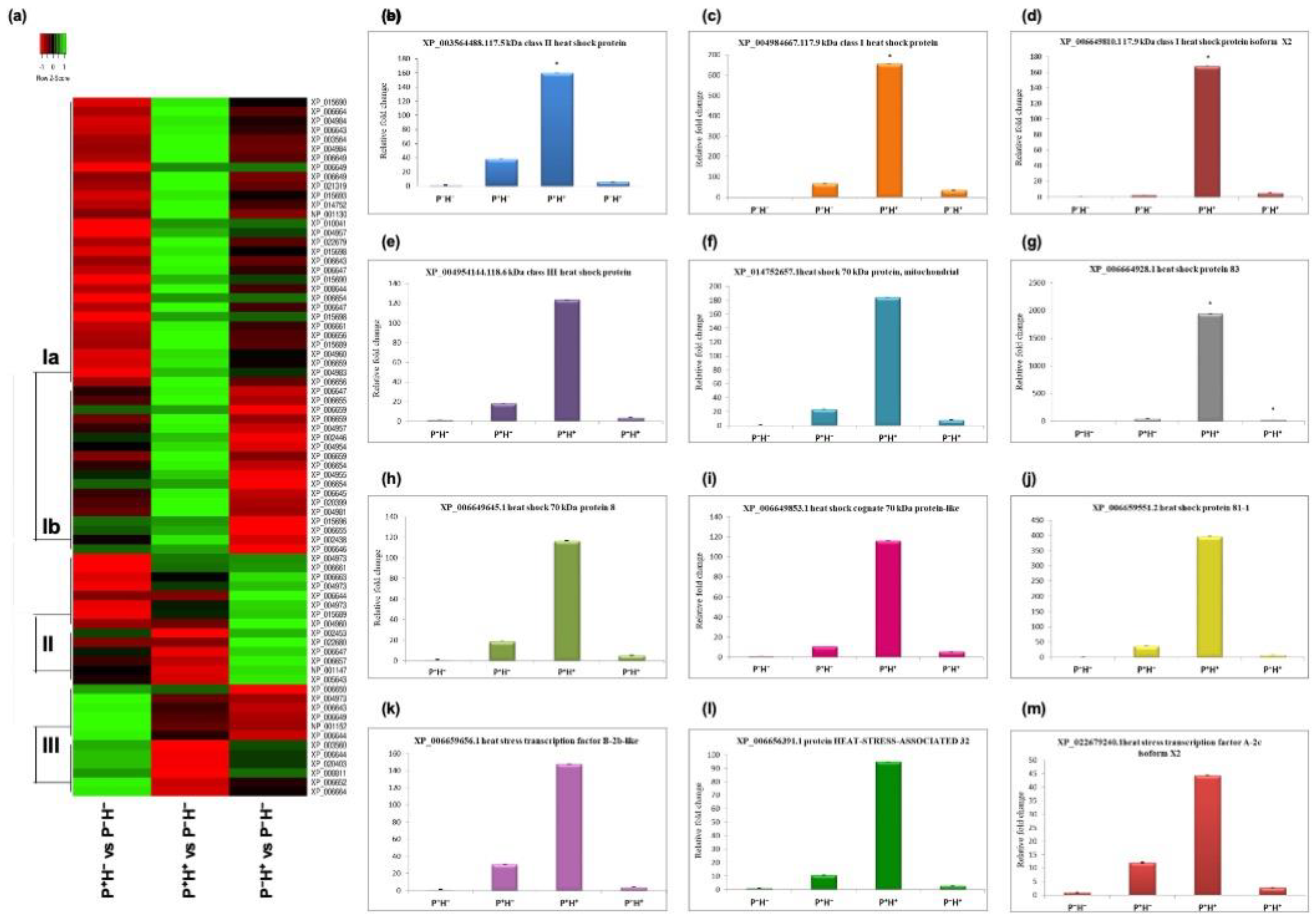
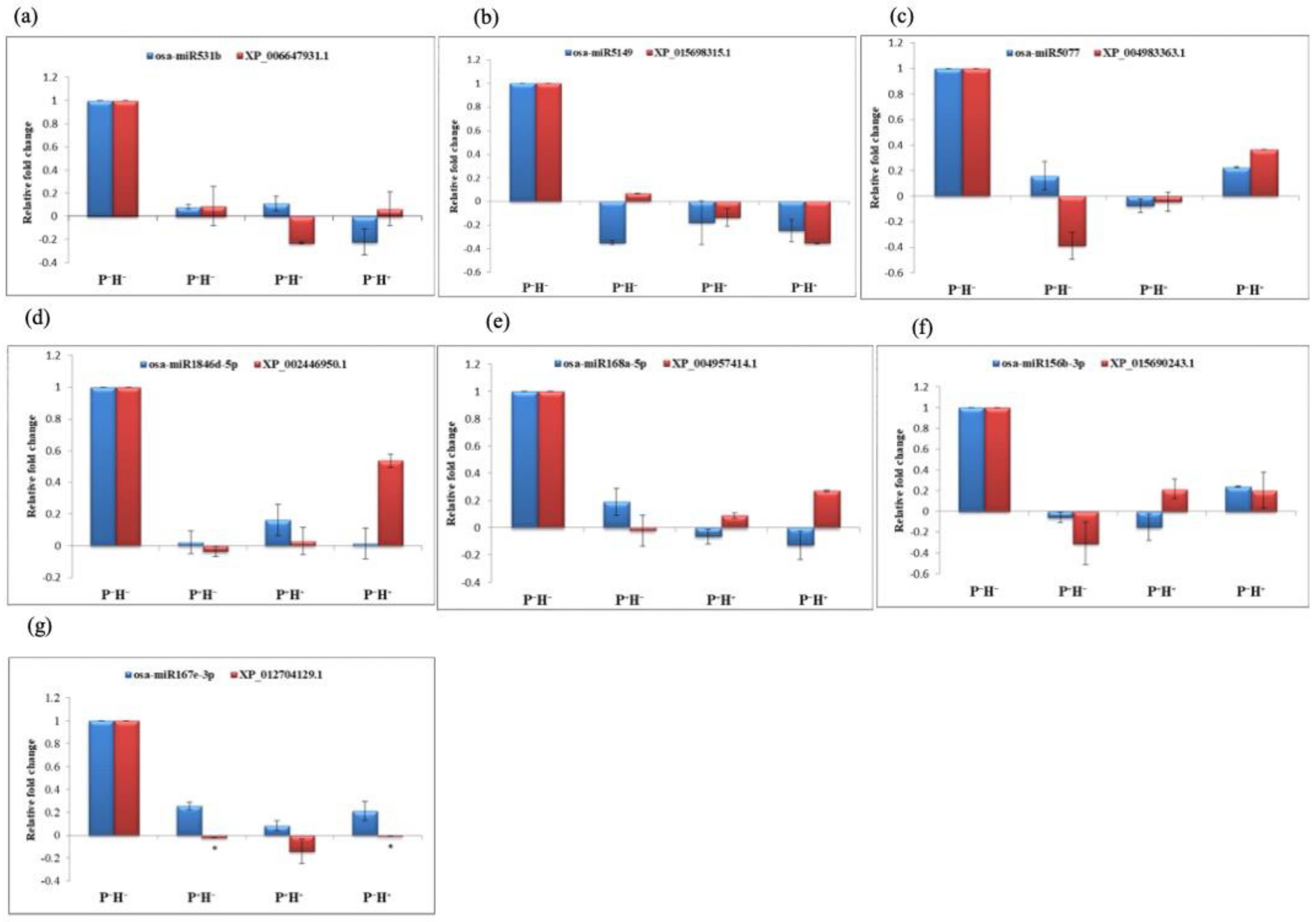
3.2. Validating the Priming Induced Gene Expression
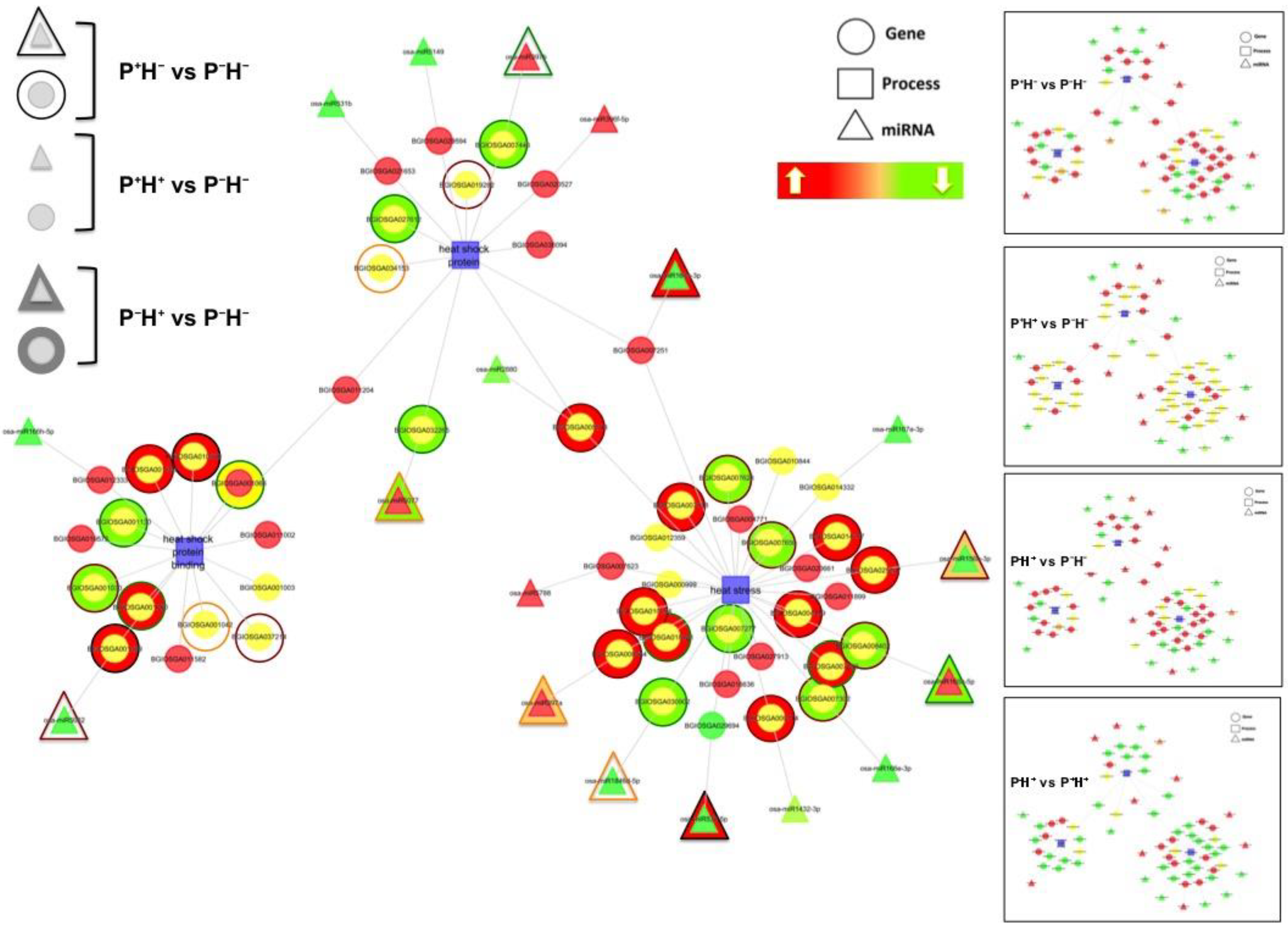
3.3. Capturing the miR Regulators in the Priming Response
Identifying the Windows of miR and Target Correlations
3.4. Validating the Role of HT Priming
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hasanuzzaman, M.; Nahar, K.; Fujita, M. Extreme temperature responses, oxidative stress and antioxidant defense in plants. In Abiotic Stress Plant Responses Appllications in Agriculture; Vahdati, K., Leslie, C., Eds.; IntechOpen: London, UK, 2013; pp. 169–205. [Google Scholar]
- Teixeira, E.I.; Fischer, G.; Van Velthuizen, H.; Walter, C.; Ewert, F. Global hot-spots of heat stress on agricultural crops due to climate change. Agric. For. Meteorol. 2013, 170, 206–215. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Zhu, Y.; Jones, A.; Rose, R.J.; Song, Y. Heat stress in legume seed setting: Effects, causes, and future prospects. Front. Plant Sci. 2019, 10, 938. [Google Scholar] [CrossRef]
- Wang, D.; Heckathorn, S.A.; Mainali, K.; Tripathee, R. Timing effects of heat-stress on plant ecophysiological characteristics and growth. Front. Plant Sci. 2016, 7, 1629. [Google Scholar] [CrossRef]
- Begcy, K.; Dresselhaus, T. Epigenetic responses to abiotic stresses during reproductive development in cereals. Plant Reprod. 2018, 31, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.K.; Bahuguna, R.N.; Shah, D.; Pal, M.; Jagadish, S.K. High temperature stress during flowering and grain filling offsets beneficial impact of elevated CO2 on assimilate partitioning and sink-strength in rice. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- De Storme, N.; Geelen, D. High temperatures alter cross-over distribution and induce male meiotic restitution in Arabidopsis thaliana. Commun. Biol. 2020, 3, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sedaghatmehr, M.; Thirumalaikumar, V.P.; Kamranfar, I.; Marmagne, A.; Masclaux-Daubresse, C.; Balazadeh, S. A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. 2019, 42, 1054–1064. [Google Scholar] [CrossRef]
- Webster, J.M.; Darling, A.L.; Uversky, V.N.; Blair, L.J. Small heat shock proteins, big impact on protein aggregation in neurodegenerative disease. Front. Pharmacol. 2019, 10, 1047. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Hossain, M.A.; da Silva, J.A.T.; Fujita, M. Plant responses and tolerance to abiotic oxidative stress: Antioxidant defense is a key factor. In Crop Stress and Its Management: Perspectives and Strategies; Bandi, V., Shanker, A.K., Shanker, C., Mandapaka, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 261–316. [Google Scholar]
- Sharma, R.; De Vleesschauwer, D.; Sharma, M.K.; Ronald, P.C. Recent advances in dissecting stress-regulatory crosstalk in rice. Mol. Plant 2013, 6, 250–260. [Google Scholar] [CrossRef]
- Chen, S.; Li, H. Heat stress regulates the expression of genes at transcriptional and post-transcriptional levels, revealed by RNA-seq in Brachypodium distachyon. Front. Plant Sci. 2017, 7, 2067. [Google Scholar] [CrossRef]
- Gomez-Pastor, R.; Burchfiel, E.T.; Thiele, D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 4. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef]
- Haslbeck, M.; Vierling, E. A first line of stress defense: Small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Scharf, K.-D.; Rose, S.; Zott, W.; Schöffl, F.; Nover, L.; Schöff, F. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990, 9, 4495–4501. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-C.; Charng, Y.-Y. Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 2013, 163, 276–290. [Google Scholar] [CrossRef]
- Guo, M.; Lu, J.-P.; Zhai, Y.-F.; Chai, W.-G.; Gong, Z.-H.; Lu, M.-H. Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 2015, 15, 1–20. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, G.; Sutoh, K.; Zhu, J.-K.; Zhang, W. Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim. Biophys. Acta Gene Regul. Mech. 2008, 1779, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Barciszewska-Pacak, M.; Milanowska, K.; Knop, K.; Bielewicz, D.; Nuc, P.; Plewka, P.; Pacak, A.M.; Vazquez, F.; Karlowski, W.; Jarmolowski, A. Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 2015, 6, 410. [Google Scholar] [CrossRef]
- Liu, W.; Xu, L.; Wang, Y.; Shen, H.; Zhu, X.; Zhang, K.; Chen, Y.; Yu, R.; Limera, C.; Liu, L. Transcriptome-wide analysis of chromium-stress responsive microRNAs to explore miRNA-mediated regulatory networks in radish (Raphanus sativus L.). Sci. Rep. 2015, 5, 1–17. [Google Scholar] [CrossRef]
- Chen, L.; Ren, Y.; Zhang, Y.; Xu, J.; Sun, F.; Zhang, Z.; Wang, Y. Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in Populus tomentosa. Gene 2012, 504, 160–165. [Google Scholar] [CrossRef]
- Hivrale, V.; Zheng, Y.; Puli, C.O.R.; Jagadeeswaran, G.; Gowdu, K.; Kakani, V.G.; Barakat, A.; Sunkar, R. Characterization of drought-and heat-responsive microRNAs in switchgrass. Plant Sci. 2016, 242, 214–223. [Google Scholar] [CrossRef]
- Raghuram, B.; Sheikh, A.H.; Sinha, A.K. Regulation of MAP kinase signaling cascade by microRNAs in Oryza sativa. Plant Signal. Behav. 2014, 9, e972130. [Google Scholar] [CrossRef][Green Version]
- Yu, X.; Wang, H.; Lu, Y.; de Ruiter, M.; Cariaso, M.; Prins, M.; van Tunen, A.; He, Y. Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J. Exp. Bot. 2012, 63, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Jyothi, M.; Rai, D. Identification and characterization of high temperature stress responsive novel miRNAs in French bean (Phaseolus vulgaris). Appl. Biochem. Biotechnol. 2015, 176, 835–849. [Google Scholar] [CrossRef] [PubMed]
- Kruszka, K.; Pacak, A.; Swida-Barteczka, A.; Nuc, P.; Alaba, S.; Wroblewska, Z.; Karlowski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. J. Exp. Bot. 2014, 65, 6123–6135. [Google Scholar] [CrossRef]
- Pan, C.; Ye, L.; Zheng, Y.; Wang, Y.; Yang, D.; Liu, X.; Chen, L.; Zhang, Y.; Fei, Z.; Lu, G. Identification and expression profiling of microRNAs involved in the stigma exsertion under high-temperature stress in tomato. BMC Genom. 2017, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Swapna, M.; Kumar, S. MicroRNAs and their regulatory role in sugarcane. Front. Plant Sci. 2017, 8, 997. [Google Scholar] [CrossRef]
- Xie, F.; Frazier, T.P.; Zhang, B. Identification and characterization of microRNAs and their targets in the bioenergy plant switchgrass (Panicum virgatum). Planta 2010, 232, 417–434. [Google Scholar] [CrossRef]
- Yu, N.; Niu, Q.W.; Ng, K.H.; Chua, N.H. The role of miR156/SPL s modules in Arabidopsis lateral root development. Plant J. 2015, 83, 673–685. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Xu, M.; Willmann, M.R.; McCormick, K.; Hu, T.; Yang, L.; Starker, C.G.; Voytas, D.F.; Meyers, B.C.; Poethig, R.S. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Genet. 2018, 14, e1007337. [Google Scholar] [CrossRef]
- Guo, C.; Xu, Y.; Shi, M.; Lai, Y.; Wu, X.; Wang, H.; Zhu, Z.; Poethig, R.S.; Wu, G. Repression of miR156 by miR159 regulates the timing of the juvenile-to-adult transition in Arabidopsis. Plant Cell 2017, 29, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Ye, M.; Sang, M.; Wu, R. A regulatory network for miR156-SPL module in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 6166. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; MacPherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, N.-H. IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Wu, S.; Van Houten, J.; Wang, Y.; Ding, B.; Fei, Z.; Clarke, T.H.; Reed, J.W.; Van Der Knaap, E. Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J. Exp. Bot. 2014, 65, 2507–2520. [Google Scholar] [CrossRef]
- Fang, X.; Zhao, G.; Zhang, S.; Li, Y.; Gu, H.; Li, Y.; Zhao, Q.; Qi, Y. Chloroplast-to-nucleus signaling regulates microRNA biogenesis in Arabidopsis. Dev. Cell 2019, 48, 371–382.e374. [Google Scholar] [CrossRef]
- Ling, Y.; Serrano, N.; Gao, G.; Atia, M.; Mokhtar, M.; Woo, Y.H.; Bazin, J.; Veluchamy, A.; Benhamed, M.; Crespi, M. Thermopriming triggers splicing memory in Arabidopsis. J. Exp. Bot. 2018, 69, 2659–2675. [Google Scholar] [CrossRef]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought’train’transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sani, E.; Herzyk, P.; Perrella, G.; Colot, V.; Amtmann, A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013, 14, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Finka, A.; Goloubinoff, P. How do plants feel the heat? Trends Biochem. Sci. 2012, 37, 118–125. [Google Scholar] [CrossRef]
- Charng, Y.-Y.; Liu, H.-C.; Liu, N.-Y.; Chi, W.-T.; Wang, C.-N.; Chang, S.-H.; Wang, T.-T. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007, 143, 251–262. [Google Scholar] [CrossRef]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.-R.; Bäurle, I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef]
- Khan, A.; Goswami, K.; Sopory, S.K.; Sanan-Mishra, N. “Mirador” on the potential role of miRNAs in synergy of light and heat networks. Indian J. Plant Physiol. 2017, 22, 587–607. [Google Scholar] [CrossRef]
- Kushawaha, A.K.; Khan, A.; Sopory, S.K.; Sanan-Mishra, N. Light Regulated Osa-miR169e Is Implicated during Priming under High Temperature Stress in Rice. Am. J. Plant Sci. 2019, 10, 1662. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562. [Google Scholar] [CrossRef]
- De Hoon, M.J.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, A.J. Java Treeview—Extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef]
- Goel, S.; Goswami, K.; Pandey, V.K.; Pandey, M.; Sanan-Mishra, N. Identification of microRNA-target modules from rice variety Pusa Basmati-1 under high temperature and salt stress. Funct. Integr. Genom. 2019, 19, 867–888. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Eur. Mol. Biol. Netw. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ (–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71. [Google Scholar]
- Hatfield, J.L.; Prueger, J.H. Temperature extremes: Effect on plant growth and development. Weather Clim. Extrem. 2015, 10, 4–10. [Google Scholar] [CrossRef]
- Khan, Z.; Shahwar, D. Role of Heat Shock Proteins (HSPs) and Heat Stress Tolerance in Crop Plants. In Sustainable Agriculture in the Era of Climate Change; Roychowdhury, R., Choudhury, S., Hasanuzzaman, M., Srivastava, S., Eds.; Springer: Cham, Switzerland, 2020; pp. 211–234. [Google Scholar]
- Hong, S.-W.; Vierling, E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc. Natl. Acad. Sci. USA 2000, 97, 4392–4397. [Google Scholar] [CrossRef]
- Maestri, E.; Klueva, N.; Perrotta, C.; Gulli, M.; Nguyen, H.T.; Marmiroli, N. Molecular genetics of heat tolerance and heat shock proteins in cereals. Plant Mol. Biol. 2002, 48, 667–681. [Google Scholar] [CrossRef]
- Huang, B.; Xu, C. Identification and characterization of proteins associated with plant tolerance to heat stress. J. Integr. Plant Biol. 2008, 50, 1230–1237. [Google Scholar] [CrossRef]
- Chandel, G.; Dubey, M.; Meena, R. Differential expression of heat shock proteins and heat stress transcription factor genes in rice exposed to different levels of heat stress. J. Plant Biochem. Biotechnol. 2013, 22, 277–285. [Google Scholar] [CrossRef]
- Guo, J.; Wu, J.; Ji, Q.; Wang, C.; Luo, L.; Yuan, Y.; Wang, Y.; Wang, J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008, 35, 105–118. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Q.; Shou, H.-X. Identification and expression analysis of OsHsfs in rice. J. Zhejiang Univ. Sci. B 2009, 10, 291–300. [Google Scholar] [CrossRef]
- Scharf, K.-D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Li, M.; Berendzen, K.W.; Schöffl, F. Promoter specificity and interactions between early and late Arabidopsis heat shock factors. Plant Mol. Biol. 2010, 73, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Schramm, F.; Ganguli, A.; Kiehlmann, E.; Englich, G.; Walch, D.; von Koskull-Döring, P. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 2006, 60, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-D.; Wang, X.-L.; Cai, Y.-M.; Wu, J.-H.; Mo, B.-T.; Yu, E.-R. Arabidopsis heat stress transcription factors A2 (HSFA2) and A3 (HSFA3) function in the same heat regulation pathway. Acta Physiol. Plant. 2017, 39, 67. [Google Scholar] [CrossRef]
- Sato, H.; Mizoi, J.; Tanaka, H.; Maruyama, K.; Qin, F.; Osakabe, Y.; Morimoto, K.; Ohori, T.; Kusakabe, K.; Nagata, M. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 2014, 26, 4954–4973. [Google Scholar] [CrossRef]
- Qu, A.-L.; Ding, Y.-F.; Jiang, Q.; Zhu, C. Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 2013, 432, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 2011, 157, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.-P.; Sadat, S.; Drenth, J.; McIntyre, C.L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 2014, 65, 539–557. [Google Scholar] [CrossRef]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Döring, P.; Vierling, E.; Scharf, K.-D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef]
- Usman, M.G.; Rafii, M.; Ismail, M.; Malek, M.; Latif, M.A.; Oladosu, Y. Heat shock proteins: Functions and response against heat stress in plants. Int. J. Sci. Technol. Res. 2014, 3, 204–218. [Google Scholar]
- Sung, D.Y.; Kaplan, F.; Guy, C.L. Plant Hsp70 molecular chaperones: Protein structure, gene family, expression and function. Physiol. Plant. 2001, 113, 443–451. [Google Scholar] [CrossRef]
- Sarkar, N.K.; Kim, Y.-K.; Grover, A. Rice sHsp genes: Genomic organization and expression profiling under stress and development. BMC Genom. 2009, 10, 1–18. [Google Scholar] [CrossRef]
- Malik, M.K.; Slovin, J.P.; Hwang, C.H.; Zimmerman, J.L. Modified expression of a carrot small heat shock protein gene, Hsp17. 7, results in increased or decreased thermotolerance. Plant J. 1999, 20, 89–99. [Google Scholar] [CrossRef]
- Murakami, T.; Matsuba, S.; Funatsuki, H.; Kawaguchi, K.; Saruyama, H.; Tanida, M.; Sato, Y. Over-expression of a small heat shock protein, sHSP17. 7, confers both heat tolerance and UV-B resistance to rice plants. Mol. Breed. 2004, 13, 165–175. [Google Scholar] [CrossRef]
- Bäurle, I. Plant heat adaptation: Priming in response to heat stress. F1000Research 2016, 5, 694. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, R.P.; Misra, H.S.; Saini, A. Heat-stress priming and alternative splicing-linked memory. J. Exp. Bot. 2018, 69, 2431–2434. [Google Scholar] [CrossRef] [PubMed]
- Hilker, M.; Schmülling, T. Stress priming, memory, and signalling in plants. Plant Cell Environ. 2019, 42, 753–761. [Google Scholar] [CrossRef]
- Serrano, N.; Ling, Y.; Bahieldin, A.; Mahfouz, M.M. Thermopriming reprograms metabolic homeostasis to confer heat tolerance. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.-D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K.-D. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002, 16, 1555–1567. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa-Yokoi, A.; Nosaka, R.; Hayashi, H.; Tainaka, H.; Maruta, T.; Tamoi, M.; Ikeda, M.; Ohme-Takagi, M.; Yoshimura, K.; Yabuta, Y. HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 2011, 52, 933–945. [Google Scholar] [CrossRef]
- Song, X.; Liu, G.; Duan, W.; Liu, T.; Huang, Z.; Ren, J.; Li, Y.; Hou, X. Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in Chinese cabbage. Mol. Genet. Genom. 2014, 289, 541–551. [Google Scholar] [CrossRef]
- Ding, X.; Guo, Q.; Li, Q.; Gai, J.; Yang, S. Comparative Transcriptomics Analysis and Functional Study Reveal Important Role of High-Temperature Stress Response Gene GmHSFA2 During Flower Bud Development of CMS-Based F1 in Soybean. Front. Plant Sci. 2020, 11, 600217. [Google Scholar] [CrossRef]
- Suzuki, N.; Sejima, H.; Tam, R.; Schlauch, K.; Mittler, R. Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J. 2011, 66, 844–851. [Google Scholar] [CrossRef]
- Bruce, T.J.; Matthes, M.C.; Napier, J.A.; Pickett, J.A. Stressful “memories” of plants: Evidence and possible mechanisms. Plant Sci. 2007, 173, 603–608. [Google Scholar] [CrossRef]
- González-Schain, N.; Dreni, L.; Lawas, L.M.; Galbiati, M.; Colombo, L.; Heuer, S.; Jagadish, K.S.; Kater, M.M. Genome-wide transcriptome analysis during anthesis reveals new insights into the molecular basis of heat stress responses in tolerant and sensitive rice varieties. Plant Cell Physiol. 2016, 57, 57–68. [Google Scholar] [CrossRef]
- Hilker, M.; Schwachtje, J.; Baier, M.; Balazadeh, S.; Bäurle, I.; Geiselhardt, S.; Hincha, D.K.; Kunze, R.; Mueller-Roeber, B.; Rillig, M.C. Priming and memory of stress responses in organisms lacking a nervous system. Biol. Rev. 2016, 91, 1118–1133. [Google Scholar] [CrossRef]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, T.; Faivre, L.; Bäurle, I.; Schubert, D. Chromatin-based mechanisms of temperature memory in plants. Plant Cell Environ. 2019, 42, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Turgut-Kara, N.; Arikan, B.; Celik, H. Epigenetic memory and priming in plants. Genetica 2020, 148, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhou, X.; Zheng, Y.; Zhang, W.; Zhu, J.-K. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Wang, Y.; Yao, Y.; Song, N.; Hu, Z.; Qin, D.; Xie, C.; Peng, H.; Ni, Z.; Sun, Q. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 2011, 11, 1–13. [Google Scholar] [CrossRef]
- Gao, R.; Wang, Y.; Gruber, M.Y.; Hannoufa, A. miR156/SPL10 modulates lateral root development, branching and leaf morphology in Arabidopsis by silencing AGAMOUS-LIKE 79. Front. Plant Sci. 2018, 8, 2226. [Google Scholar] [CrossRef]
- Guan, Q.; Lu, X.; Zeng, H.; Zhang, Y.; Zhu, J. Heat stress induction of mi R 398 triggers a regulatory loop that is critical for thermotolerance in A rabidopsis. Plant J. 2013, 74, 840–851. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, H.; Zhang, Q.; Zhang, J.; Ni, F.; Liu, C.; Qi, Y. DNA methylation mediated by a microRNA pathway. Mol. Cell 2010, 38, 465–475. [Google Scholar] [CrossRef]
- Sanchez, D.H.; Paszkowski, J. Heat-induced release of epigenetic silencing reveals the concealed role of an imprinted plant gene. PLoS Genet. 2014, 10, e1004806. [Google Scholar] [CrossRef]
- Naydenov, M.; Baev, V.; Apostolova, E.; Gospodinova, N.; Sablok, G.; Gozmanova, M.; Yahubyan, G. High-temperature effect on genes engaged in DNA methylation and affected by DNA methylation in Arabidopsis. Plant Physiol. Biochem. 2015, 87, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Giacomelli, J.I.; Weigel, D.; Chan, R.L.; Manavella, P.A. Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol. 2012, 195, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, F.; Cao, H.; Peng, H.; Ni, Z.; Sun, Q.; Yao, Y. TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 2012, 7, e48445. [Google Scholar] [CrossRef] [PubMed]



| P−H− | P+H− | P+H+ | P−H+ | |
|---|---|---|---|---|
| RNA-seq Data | ||||
| Total number of raw reads (in millions) | 25.82 | 25.57 | 25.29 | 25.24 |
| Total high quality reads (in millions) | 24.40 | 24.24 | 23.89 | 23.78 |
| Reads aligned to the genome (in millions) | 21.97 | 21.87 | 21.69 | 21.20 |
| Percentage alignment | 90.04 | 90.23 | 90.8 | 89.14 |
| Number of transcripts | 22,417 | 22,297 | 22,440 | 22,412 |
| Total number of differentially expressed transcripts (DETs) | 935 # | 2467 # | 1734 # | |
| Up regulated DETs (FC ≥ 2, p ≤ 0.05) | 546 # | 1026 # | 751 # | |
| Down regulated DETs (FC ≤ −2, p ≤ 0.05) | 389 # | 1441 # | 983 # | |
| Small RNA-seq Data | ||||
| Total number of raw reads (in millions) | 27.34 | 24.15 | 25.17 | 24.96 |
| Total high quality reads (in millions) | 25.51 | 22.54 | 23.4 | 23.29 |
| Number of unique Tags (in millions) | 2.18 | 1.71 | 2.31 | 2.23 |
| Number of miRNAs identified | 296 | 294 | 296 | 296 |
| Number of DE miRs (FC ≥ 2) | 5 # | 10 # | 5 # | |
| Number of DE miRs (FC ≥ 1.5) | 21 # | 26 # | 15 # | |
| Traits | Control | PAH | PBH | NAH | NBH |
|---|---|---|---|---|---|
| 100-grains weight (g) | 1.90 ± 0.065 | 2.10 ± 0.11 | 2.09 ± 0.068 * | 1.78 ± 0.049 | 1.74 ± 0.057 * |
| Panicle length (cm) | 32.2 ± 2.75 | 25.8 ± 5.99 | 29.32 ± 0.59 | 30.37 ± 2.71 | 31.65 ± 2.63 |
| Grains per panicle (Filled) | 73.5 ± 13.77 | 48.25 ± 39.16 | 49.0 ± 13.19 * | 21.25 ± 10.21 ** | 46 ± 18.01 * |
| Grains per panicle (Empty) | 10.5 ± 21.98 | 21.25 ± 5.90 ** | 21.75 ± 10.71 ** | 86.25 ± 21.56 | 50.5 ± 19.75 ** |
| Seed length (mm) | 11.12 ± 0.54 | 10.96 ± 0.56 | 11.03 ± 0.60 | 10.98 ± 0.54 | 10.84 ± 0.51 ** |
| Seed width (mm) | 2.26 ± 0.12 | 2.20 ± 0.20 | 2.63 ± 0.28 ** | 2.53 ± 0.22 ** | 2.59 ± 0.21 ** |
| Ratio of seed length to seed width | 4.90 | 4.84 | 4.19 | 4.33 | 4.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kushawaha, A.K.; Khan, A.; Sopory, S.K.; Sanan-Mishra, N. Priming by High Temperature Stress Induces MicroRNA Regulated Heat Shock Modules Indicating Their Involvement in Thermopriming Response in Rice. Life 2021, 11, 291. https://doi.org/10.3390/life11040291
Kushawaha AK, Khan A, Sopory SK, Sanan-Mishra N. Priming by High Temperature Stress Induces MicroRNA Regulated Heat Shock Modules Indicating Their Involvement in Thermopriming Response in Rice. Life. 2021; 11(4):291. https://doi.org/10.3390/life11040291
Chicago/Turabian StyleKushawaha, Akhilesh Kumar, Ambreen Khan, Sudhir Kumar Sopory, and Neeti Sanan-Mishra. 2021. "Priming by High Temperature Stress Induces MicroRNA Regulated Heat Shock Modules Indicating Their Involvement in Thermopriming Response in Rice" Life 11, no. 4: 291. https://doi.org/10.3390/life11040291
APA StyleKushawaha, A. K., Khan, A., Sopory, S. K., & Sanan-Mishra, N. (2021). Priming by High Temperature Stress Induces MicroRNA Regulated Heat Shock Modules Indicating Their Involvement in Thermopriming Response in Rice. Life, 11(4), 291. https://doi.org/10.3390/life11040291