A Super Stable Mutant of the Plant Protein Monellin Endowed with Enhanced Sweetness
Abstract
1. Introduction
2. Materials and Methods
2.1. Cloning, Expression and Purification of the Mutant
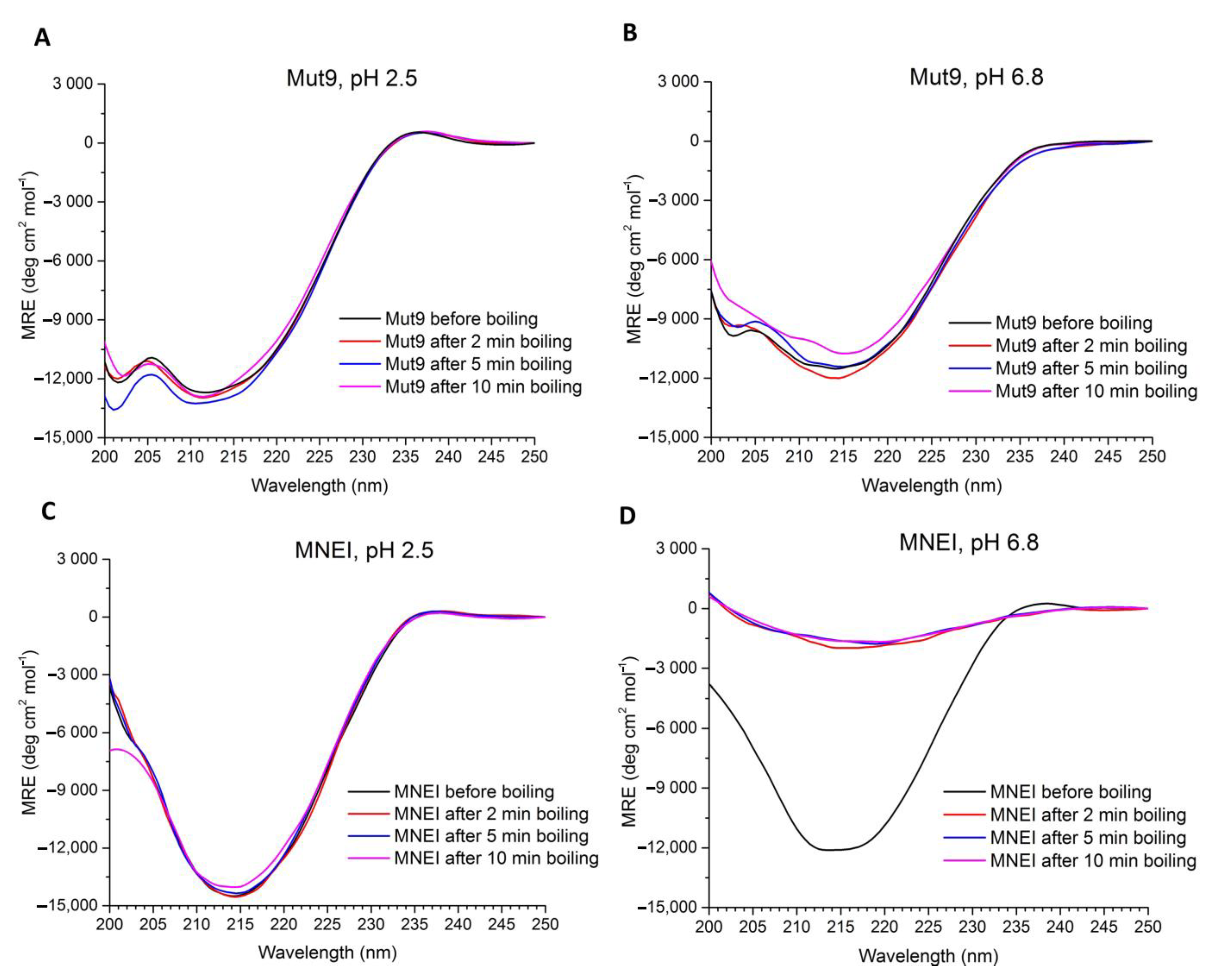
2.2. Circular Dichroism Spectroscopy (CD Spectroscopy)
2.3. Differential Scanning Calorimetry (DSC)
2.4. Sensory Analysis
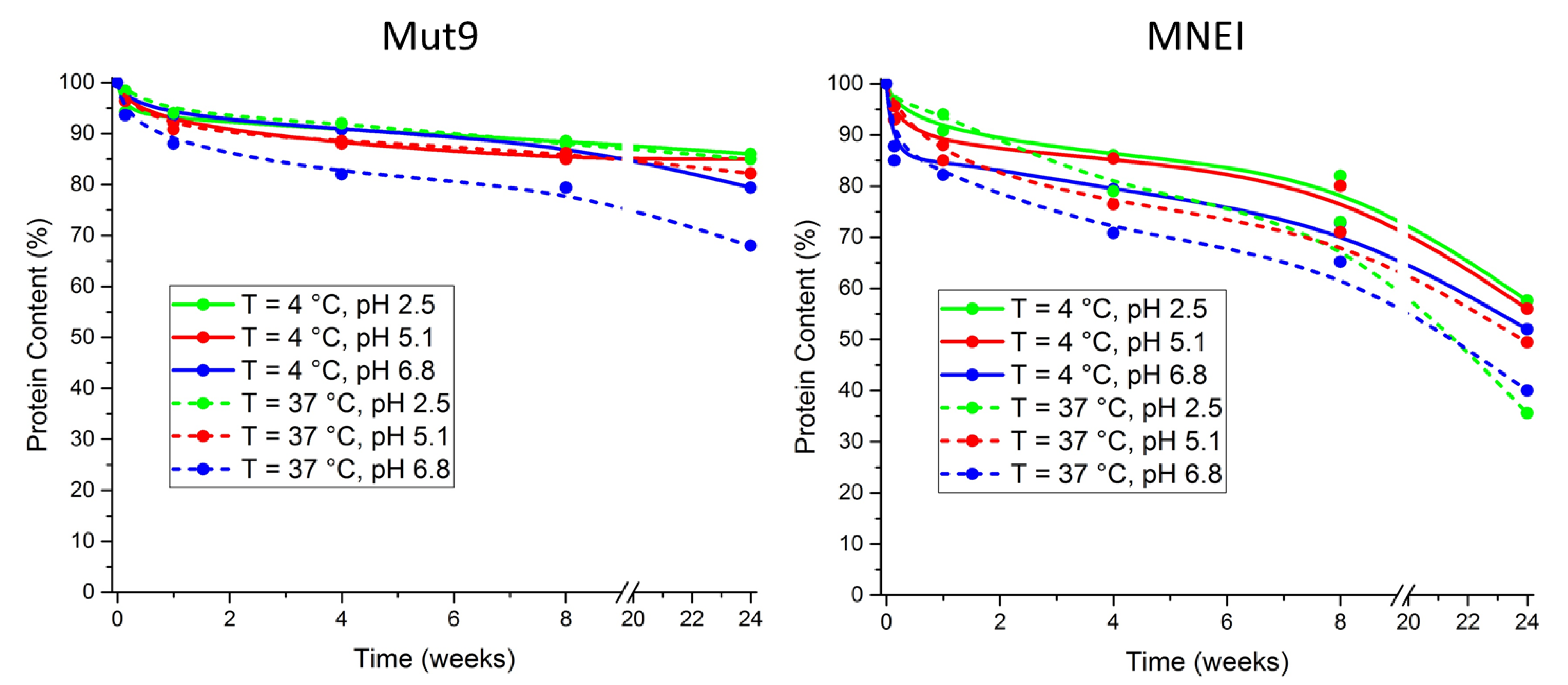
2.5. Shelf-Life Studies
3. Results
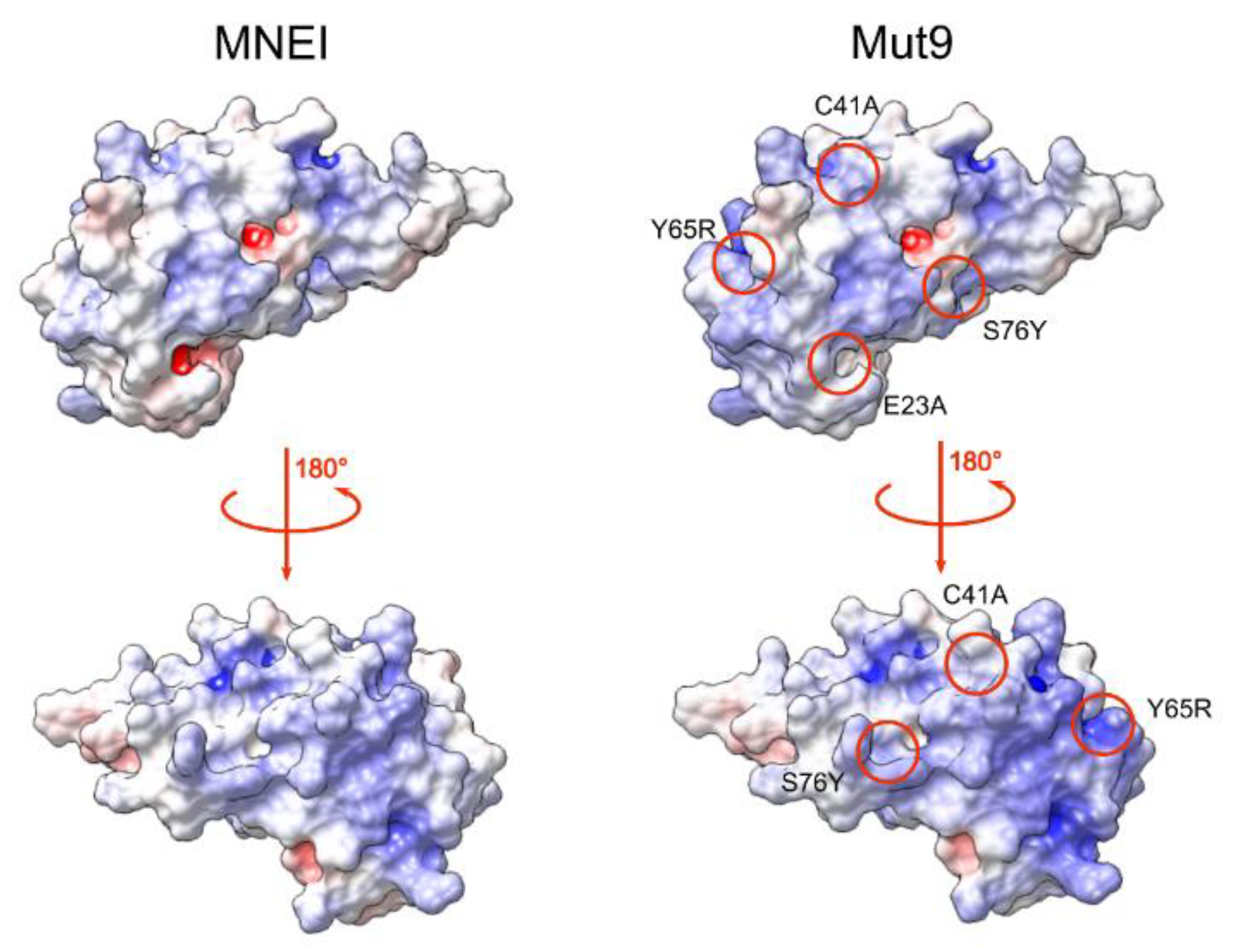
3.1. Protein Design and Production
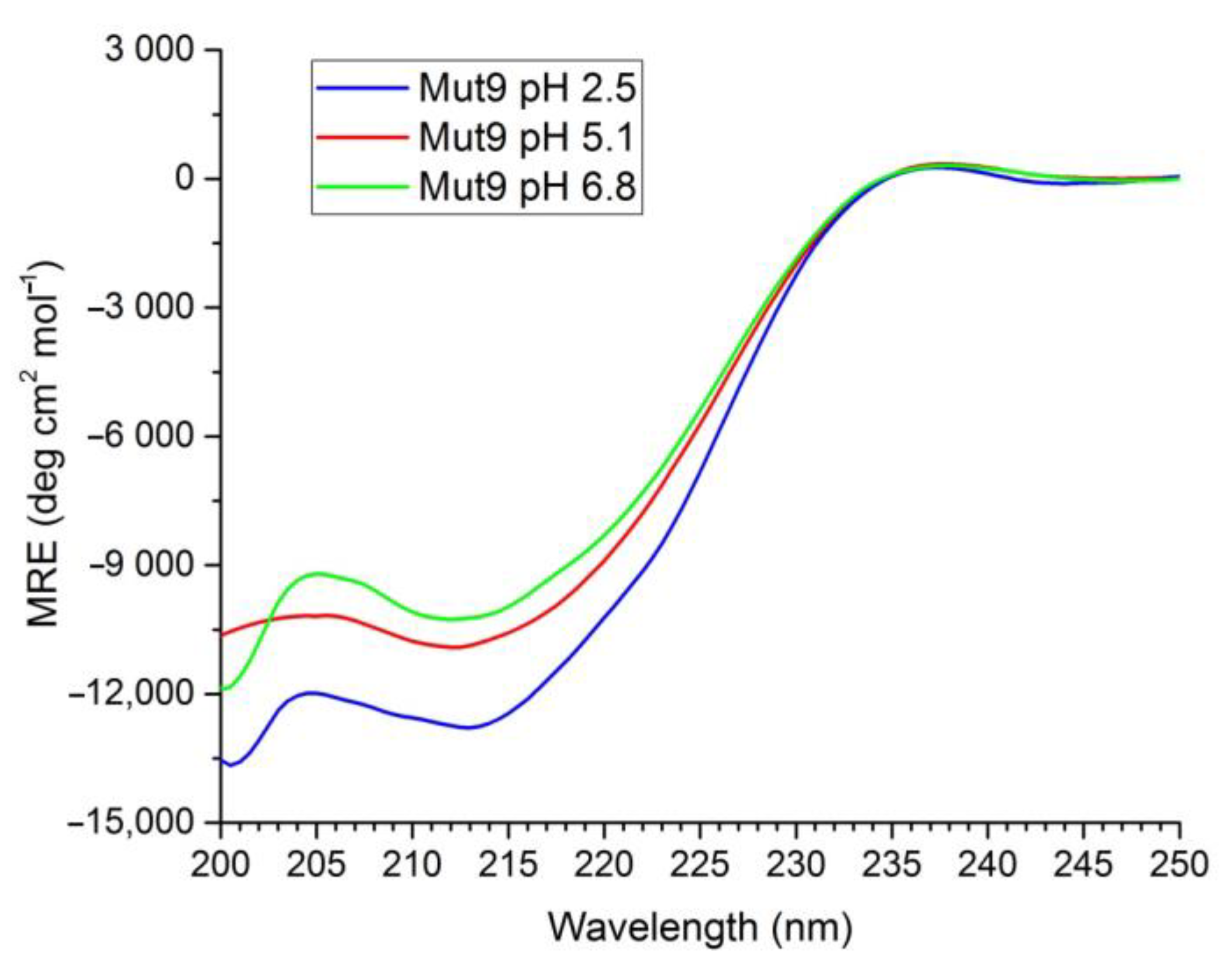
3.2. Secondary Structure Assessment
3.3. Sensory Analysis
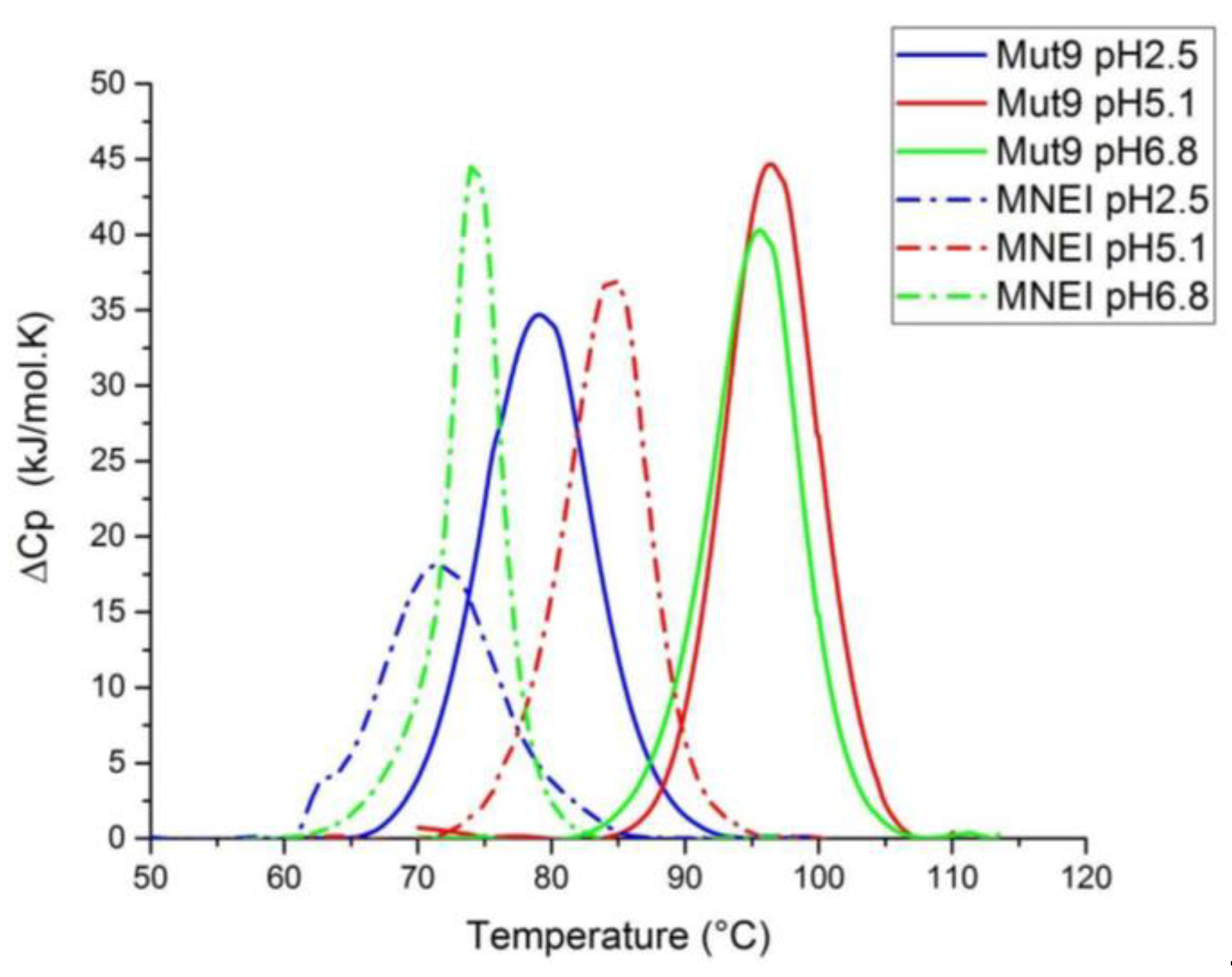
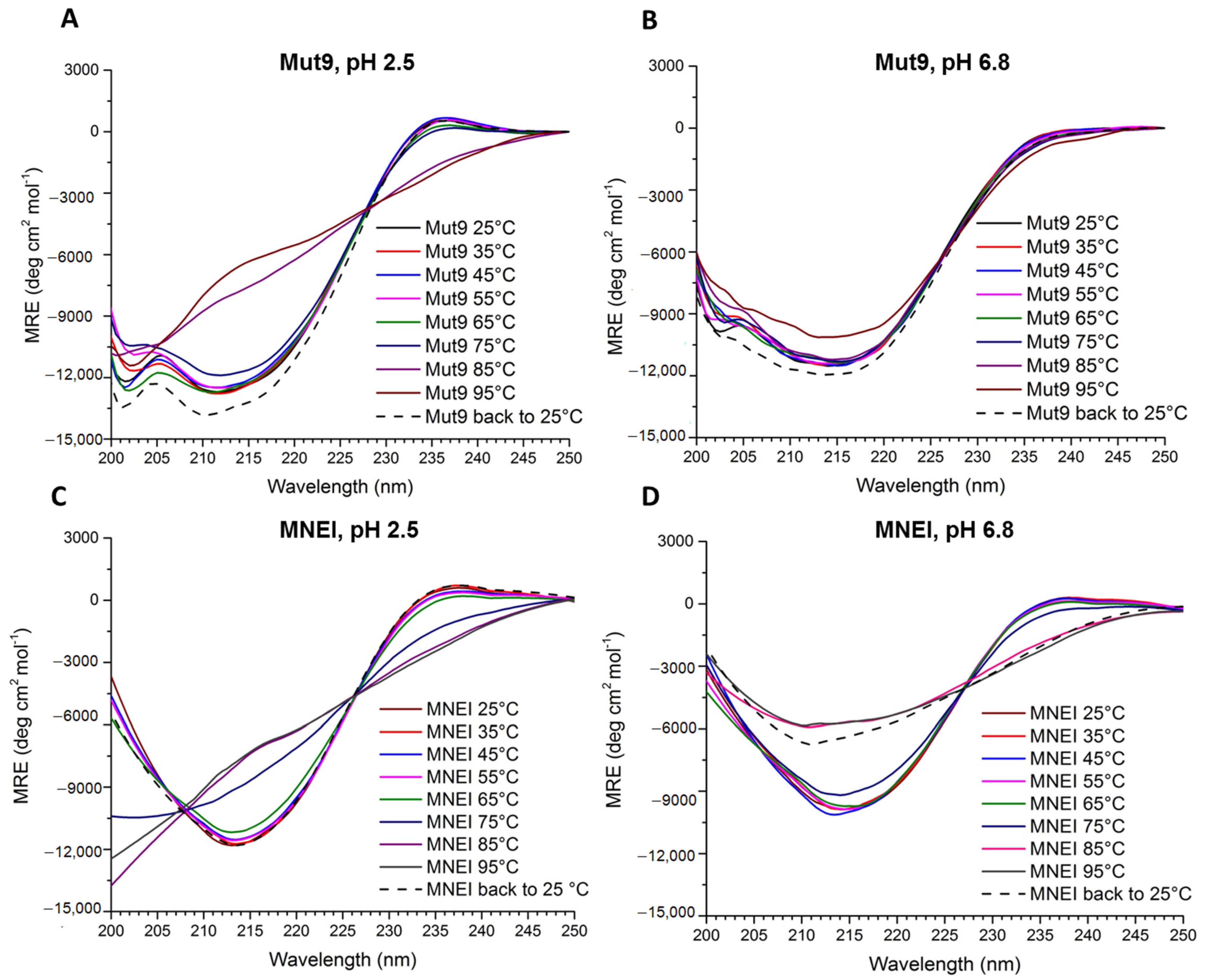
3.4. Thermal Stability Assessment
3.5. Shelf-Life Assessment
4. Discussion
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Temussi, P.A. Natural sweet macromolecules: How sweet proteins work. Cell. Mol. Life Sci. C 2006, 63, 1876–1888. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.A.; Cagan, R.H. Purification of monellin, the sweet principle of Dioscoreophyllum cumminsii. Biochim. Biophys. Acta Gen. Subj. 1972, 261, 114–122. [Google Scholar] [CrossRef]
- Van der Wel, H.; Loeve, K. Isolation and characterization of thaumatin I and II, the sweet-tasting proteins from Thaumatococcus daniellii Benth. Eur. J. Biochem. 1972, 31, 221–225. [Google Scholar]
- Ming, D.; Hellekant, G. Brazzein, a new high-potency thermostable sweet protein from Pentadiplandra brazzeana B. FEBS Lett. 1994, 355, 106–108. [Google Scholar] [CrossRef]
- Liu, X.; Maeda, S.; Hu, Z.; Aiuchi, T.; Nakaya, K.; Kurihara, Y. Purification, complete amino acid sequence and structural characterization of the heat-stable sweet protein, mabinlin II. Eur. J. Biochem. 1993, 211, 281–287. [Google Scholar] [CrossRef]
- Theerasilp, S.; Hitotsuya, H.; Nakajo, S.; Nakaya, K.; Nakamura, Y.; Kurihara, Y. Complete amino acid sequence and structure characterization of the taste-modifying protein, miraculin. J. Biol. Chem. 1989, 264, 6655–6659. [Google Scholar] [CrossRef]
- Yamashita, H.; Theerasilp, S.; Aiuchi, T.; Nakaya, K.; Nakamura, Y.; Kurihara, Y. Purification and complete amino acid sequence of a new type of sweet protein taste-modifying activity, curculin. J. Biol. Chem. 1990, 265, 15770–15775. [Google Scholar] [CrossRef]
- Morris, J.A. Sweetening agents from natural sources. Lloydia 1976, 39, 25–38. [Google Scholar]
- Kurihara, K.; Beidler, L.M. Taste-modifying protein from miracle fruit. Science 1968, 161, 1241–1243. [Google Scholar] [CrossRef]
- Yamashita, H.; Akabane, T.; Kurihara, Y. Activity and stability of a new sweet protein with taste-modifying action, curculin. Chem. Senses 1995, 20, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Picone, D.; Temussi, P.A. Dissimilar sweet proteins from plants: Oddities or normal components? Plant Sci. 2012, 195, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Staszewski, L.; Xu, H.; Durick, K.; Zoller, M.; Adler, E. Human receptors for sweet and umami taste. Proc. Natl. Acad. Sci. USA 2002, 99, 4692–4696. [Google Scholar] [CrossRef]
- Nelson, G.; Hoon, M.A.; Chandrashekar, J.; Zhang, Y.; Ryba, N.J.P.; Zuker, C.S. Mammalian sweet taste receptors. Cell 2001, 106, 381–390. [Google Scholar] [CrossRef]
- Temussi, P.A. New insights into the characteristics of sweet and bitter taste receptors. Int. Rev. Cell Mol. Biol. 2011, 291, 191–226. [Google Scholar]
- Chandrashekar, J.; Hoon, M.A.; Ryba, N.J.P.; Zuker, C.S. The receptors and cells for mammalian taste. Nature 2006, 444, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Jiang, P.; Maillet, E.; Max, M.; Margolskee, R.F.; Osman, R. The heterodimeric sweet taste receptor has multiple potential ligand binding sites. Curr. Pharm. Des. 2006, 12, 4591–4600. [Google Scholar] [CrossRef]
- Morini, G.; Bassoli, A.; Temussi, P.A. From small sweeteners to sweet proteins: Anatomy of the binding sites of the human T1R2_T1R3 receptor. J. Med. Chem. 2005, 48, 5520–5529. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, R.; Ninomiya, Y. Taste information derived from T1R-expressing taste cells in mice. Biochem. J. 2016, 473, 525–536. [Google Scholar] [CrossRef]
- Masuda, K.; Koizumi, A.; Nakajima, K.; Tanaka, T.; Abe, K.; Misaka, T.; Ishiguro, M. Characterization of the modes of binding between human sweet taste receptor and low-molecular-weight sweet compounds. PLoS ONE 2012, 7, e0035380. [Google Scholar] [CrossRef]
- Tancredi, T.; Pastore, A.; Salvadori, S.; Esposito, V.; Temussi, P.A. Interaction of sweet proteins with their receptor: A conformational study of peptides corresponding to loops of brazzein, monellin and thaumatin. Eur. J. Biochem. 2004, 271, 2231–2240. [Google Scholar] [CrossRef]
- Temussi, P.A. Why are sweet proteins sweet? Interaction of brazzein, monellin and thaumatin with the T1R2-T1R3 receptor. FEBS Lett. 2002, 526, 1–4. [Google Scholar] [CrossRef]
- Leone, S.; Pica, A.; Merlino, A.; Sannino, F.; Temussi, P.A.; Picone, D. Sweeter and stronger: Enhancing sweetness and stability of the single chain monellin MNEI through molecular design. Sci. Rep. 2016, 6, 34045. [Google Scholar] [CrossRef] [PubMed]
- Ohta, K.; Masuda, T.; Ide, N.; Kitabatake, N. Critical molecular regions for elicitation of the sweetness of the sweet-tasting protein, thaumatin I. FEBS J. 2008, 275, 3644–3652. [Google Scholar] [CrossRef]
- Yoon, S.-Y.; Kong, J.-N.; Jo, D.-H.; Kong, K.-H. Residue mutations in the sweetness loops for the sweet-tasting protein brazzein. Food Chem. 2011, 129, 1327–1330. [Google Scholar] [CrossRef]
- Faus, I. Recent developments in the characterization and biotechnological production of sweet-tasting proteins. Appl. Microbiol. Biotechnol. 2000, 53, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Suez, J.; Korem, T.; Zeevi, D.; Zilberman-Schapira, G.; Thaiss, C.A.; Maza, O.; Israeli, D.; Zmora, N.; Gilad, S.; Weinberger, A. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 2014, 514, 181–186. [Google Scholar] [CrossRef]
- Esposito, V.; Gallucci, R.; Picone, D.; Saviano, G.; Tancredi, T.; Temussi, P.A. The importance of electrostatic potential in the interaction of sweet proteins with the sweet taste receptor. J. Mol. Biol. 2006, 360, 448–456. [Google Scholar] [CrossRef]
- Xue, W.-F.; Szczepankiewicz, O.; Thulin, E.; Linse, S.; Carey, J. Role of protein surface charge in monellin sweetness. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 410–420. [Google Scholar] [CrossRef]
- Liu, Q.; Li, L.; Yang, L.; Liu, T.; Cai, C.; Liu, B. Modification of the sweetness and stability of sweet-tasting protein monellin by gene mutation and protein engineering. BioMed Res. Int. 2016, 2016, 3647173. [Google Scholar] [CrossRef]
- Somoza, J.R.; Cho, J.M.; Kim, S.-H. The taste-active regions of monellin, a potently sweet protein. Chem. Senses 1995, 20, 61–68. [Google Scholar] [CrossRef]
- Masuda, T.; Ohta, K.; Ojiro, N.; Murata, K.; Mikami, B.; Tani, F.; Temussi, P.A.; Kitabatake, N. A hypersweet protein: Removal of the specific negative charge at Asp21 enhances thaumatin sweetness. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef]
- Kaneko, R.; Kitabatake, N. Structure-sweetness relationship in thaumatin: Importance of lysine residues. Chem. Senses 2001, 26, 167–177. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ohta, K.; Masuda, T.; Tani, F.; Kitabatake, N. Introduction of a negative charge at Arg82 in thaumatin abolished responses to human T1R2–T1R3 sweet receptors. Biochem. Biophys. Res. Commun. 2011, 413, 41–45. [Google Scholar] [CrossRef]
- Assadi-Porter, F.M.; Tonelli, M.; Maillet, E.L.; Markley, J.L.; Max, M. Interactions between the human sweet-sensing T1R2–T1R3 receptor and sweeteners detected by saturation transfer difference NMR spectroscopy. Biochim. Biophys. Acta Biomembr. 2010, 1798, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Walters, D.E.; Cragin, T.; Jin, Z.; Rumbley, J.N.; Hellekant, G. Design and evaluation of new analogs of the sweet protein brazzein. Chem. Senses 2009, 34, 679–683. [Google Scholar] [CrossRef]
- Inglett, G.E.; MAY, J.F. Serendipity berries—Source of a new intense sweetener. J. Food Sci. 1969, 34, 408–411. [Google Scholar] [CrossRef]
- Ogata, C.; Hatada, M.; Tomlinson, G.; Shin, W.-C.; Kim, S.-H. Crystal structure of the intensely sweet protein monellin. Nature 1987, 328, 739–742. [Google Scholar] [CrossRef]
- Somoza, J.R.; Jiang, F.; Tong, L.; Kang, C.-H.; Cho, J.M.; Kim, S.-H. Two crystal structures of a potently sweet protein: Natural monellin at 2·75 Å resolution and single-chain monellin at 1 7 Å resolution. J. Mol. Biol. 1993, 234, 390–404. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.-F.; Szczepankiewicz, O.; Bauer, M.C.; Thulin, E.; Linse, S. Intra-versus intermolecular interactions in monellin: Contribution of surface charges to protein assembly. J. Mol. Biol. 2006, 358, 1244–1255. [Google Scholar] [CrossRef]
- Kim, S.-H.; Kang, C.-H.; Kim, R.; Cho, J.M.; Lee, Y.-B.; Lee, T.-K. Redesigning a sweet protein: Increased stability and renaturability. Protein Eng. Des. Sel. 1989, 2, 571–575. [Google Scholar] [CrossRef]
- Tancredi, T.; Iijima, H.; Saviano, G.; Amodeo, P.; Temussi, P.A. Structural determination of the active site of a sweet protein A 1H NMR investigation of pMNEI. FEBS Lett. 1992, 310, 27–30. [Google Scholar] [CrossRef]
- Kant, R. Sweet proteins—Potential replacement for artificial low calorie sweeteners. Nutr. J. 2005, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Weiffert, T.; Linse, S. Protein stabilization with retained function of monellin using a split GFP system. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Yang, L.; Cai, C.; Ni, J.; Liu, B. Expression, purification and characterization of a novel double-sites mutant of the single-chain sweet-tasting protein monellin (MNEI) with both improved sweetness and stability. Protein Expr. Purif. 2018, 143, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Leone, S.; Sannino, F.; Tutino, M.L.; Parrilli, E.; Picone, D. Acetate: Friend or foe? Efficient production of a sweet protein in Escherichia coli BL21 using acetate as a carbon source. Microb. Cell Fact. 2015, 14, 106. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 3095–3103. [Google Scholar] [CrossRef]
- Pagano, B.; Randazzo, A.; Fotticchia, I.; Novellino, E.; Petraccone, L.; Giancola, C. Differential scanning calorimetry to investigate G-quadruplexes structural stability. Methods 2013, 64, 43–51. [Google Scholar] [CrossRef]
- Aghera, N.; Dasgupta, I.; Udgaonkar, J.B. A buried ionizable residue destabilizes the native state and the transition state in the folding of monellin. Biochemistry 2012, 51, 9058–9066. [Google Scholar] [CrossRef]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef]
- Pica, A.; Leone, S.; Di Girolamo, R.; Donnarumma, F.; Emendato, A.; Rega, M.F.; Merlino, A.; Picone, D. pH driven fibrillar aggregation of the super-sweet protein Y65R-MNEI: A step-by-step structural analysis. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 808–815. [Google Scholar] [CrossRef]
- Leone, S.; Picone, D. Molecular dynamics driven design of pH-stabilized mutants of MNEI, a sweet protein. PLoS ONE 2016, 11, e0158372. [Google Scholar] [CrossRef]
- Rega, M.F.; Di Monaco, R.; Leone, S.; Donnarumma, F.; Spadaccini, R.; Cavella, S.; Picone, D. Design of sweet protein based sweeteners: Hints from structure-function relationships. Food Chem. 2015, 173, 1179–1186. [Google Scholar] [CrossRef]
- Donnarumma, F.; Leone, S.; Delfi, M.; Emendato, A.; Ami, D.; Laurents, D.V.; Natalello, A.; Spadaccini, R.; Picone, D. Probing structural changes during amyloid aggregation of the sweet protein MNEI. FEBS J. 2020, 287, 2808–2822. [Google Scholar] [CrossRef] [PubMed]
- Rega, M.F.; Siciliano, A.; Gesuele, R.; Lofrano, G.; Carpentieri, A.; Picone, D.; Guida, M. Ecotoxicological survey of MNEI and Y65R-MNEI proteins as new potential high-intensity sweeteners. Environ. Sci. Pollut. Res. 2017, 24, 9734–9740. [Google Scholar] [CrossRef]
- Cancelliere, R.; Leone, S.; Gatto, C.; Mazzoli, A.; Ercole, C.; Iossa, S.; Liverini, G.; Picone, D.; Crescenzo, R. Metabolic effects of the sweet protein MNEI as a sweetener in drinking water. A pilot study of a high fat dietary regimen in a rodent model. Nutrients 2019, 11, 2643. [Google Scholar] [CrossRef] [PubMed]
- Donnarumma, F.; Emendato, A.; Leone, S.; Ercole, C.; D’Errico, G.; Picone, D. Salt modulated fibrillar aggregation of the sweet protein MNEI in aqueous solution. J. Solut. Chem. 2018, 47, 939–949. [Google Scholar] [CrossRef]
- Garcia-Seisdedos, H.; Ibarra-Molero, B.; Sanchez-Ruiz, J.M. How many ionizable groups can sit on a protein hydrophobic core? Proteins 2012, 80, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Krap, D.A.; Gittis, A.G.; Stahley, M.R.; Fitch, C.G.; Stites, W.E.; Garcia-Moreno, E.B. High apparent dielectric constant inside a protein reflects structural reorganization coupled to the ionization of an internal asp. Biophys. J. 2007, 92, 2041–2053. [Google Scholar] [CrossRef] [PubMed]

| pH 2.5 | pH 5.1 | pH 6.8 | |
|---|---|---|---|
| α-helix | 19.8 | 13.7 | 17.3 |
| β-sheet (antiparallel) | 42.2 | 38.3 | 38.8 |
| β-sheet (parallel) | 1.8 | 0 | 0 |
| Turn | 4.0 | 8.3 | 4.7 |
| Random coil | 32.2 | 39.7 | 39.2 |
| Properties | Mut9 pH 2.5 | MNEI pH 2.5 | Mut9 pH 5.1 | MNEI pH 5.1 | Mut9 pH 6.8 | MNEI pH 6.8 |
|---|---|---|---|---|---|---|
| Tm (°C) | 78.0 | 71.4 | 96.2 | 84.4 | 95.8 | 74.2 |
| ΔHcal (kJ/mol) | 327 | 241 | 384 | 253 | 336 | 247 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delfi, M.; Emendato, A.; Leone, S.; Lampitella, E.A.; Porcaro, P.; Cardinale, G.; Petraccone, L.; Picone, D. A Super Stable Mutant of the Plant Protein Monellin Endowed with Enhanced Sweetness. Life 2021, 11, 236. https://doi.org/10.3390/life11030236
Delfi M, Emendato A, Leone S, Lampitella EA, Porcaro P, Cardinale G, Petraccone L, Picone D. A Super Stable Mutant of the Plant Protein Monellin Endowed with Enhanced Sweetness. Life. 2021; 11(3):236. https://doi.org/10.3390/life11030236
Chicago/Turabian StyleDelfi, Masoud, Alessandro Emendato, Serena Leone, Eros Antonio Lampitella, Piero Porcaro, Gaetano Cardinale, Luigi Petraccone, and Delia Picone. 2021. "A Super Stable Mutant of the Plant Protein Monellin Endowed with Enhanced Sweetness" Life 11, no. 3: 236. https://doi.org/10.3390/life11030236
APA StyleDelfi, M., Emendato, A., Leone, S., Lampitella, E. A., Porcaro, P., Cardinale, G., Petraccone, L., & Picone, D. (2021). A Super Stable Mutant of the Plant Protein Monellin Endowed with Enhanced Sweetness. Life, 11(3), 236. https://doi.org/10.3390/life11030236







