An Episomal CRISPR/Cas12a System for Mediating Efficient Gene Editing
Abstract
:1. Introduction
2. Materials and Methods
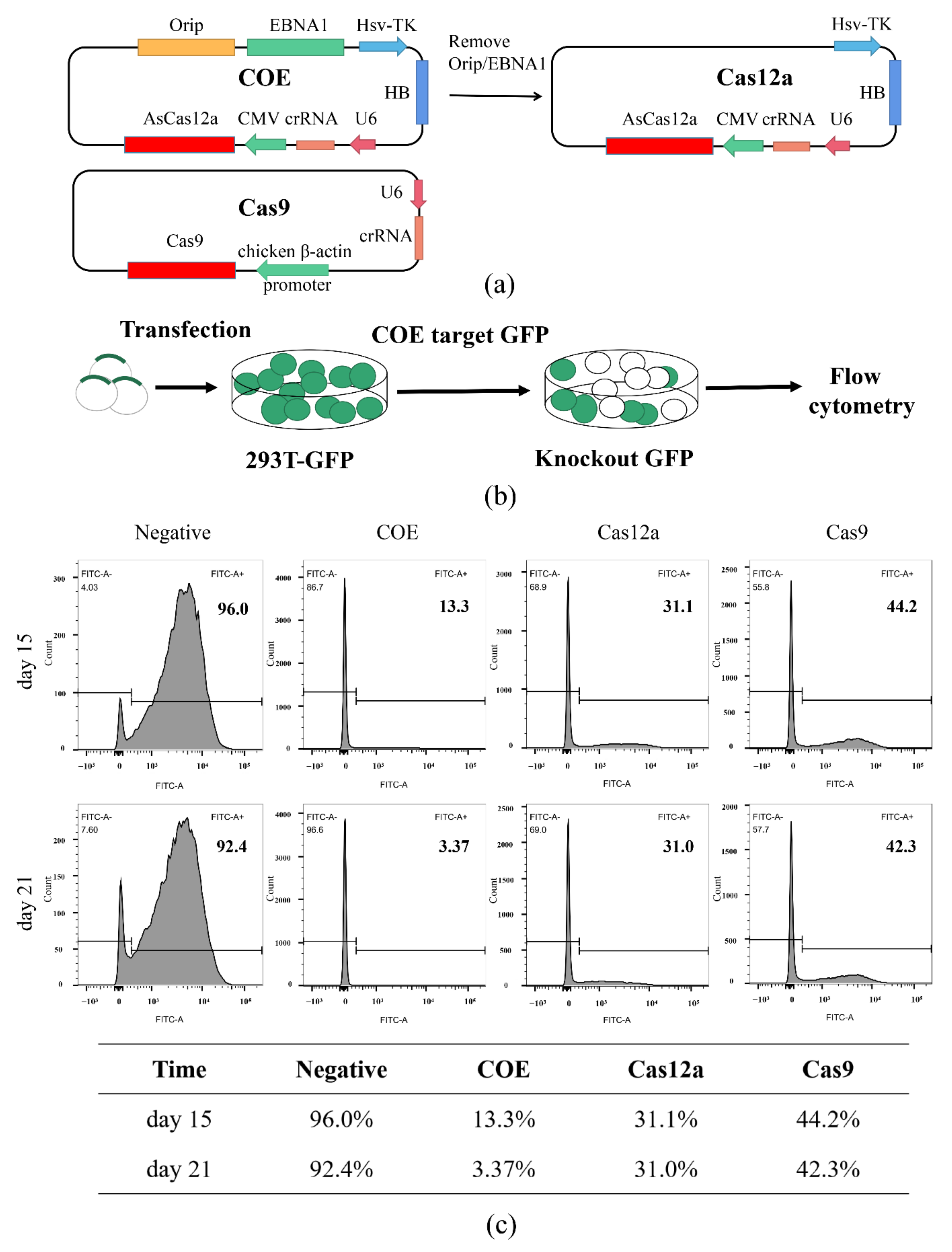
2.1. Design and Construction of Episomal CRISPR/Cas12a
2.2. Cell Culture and Maintenance
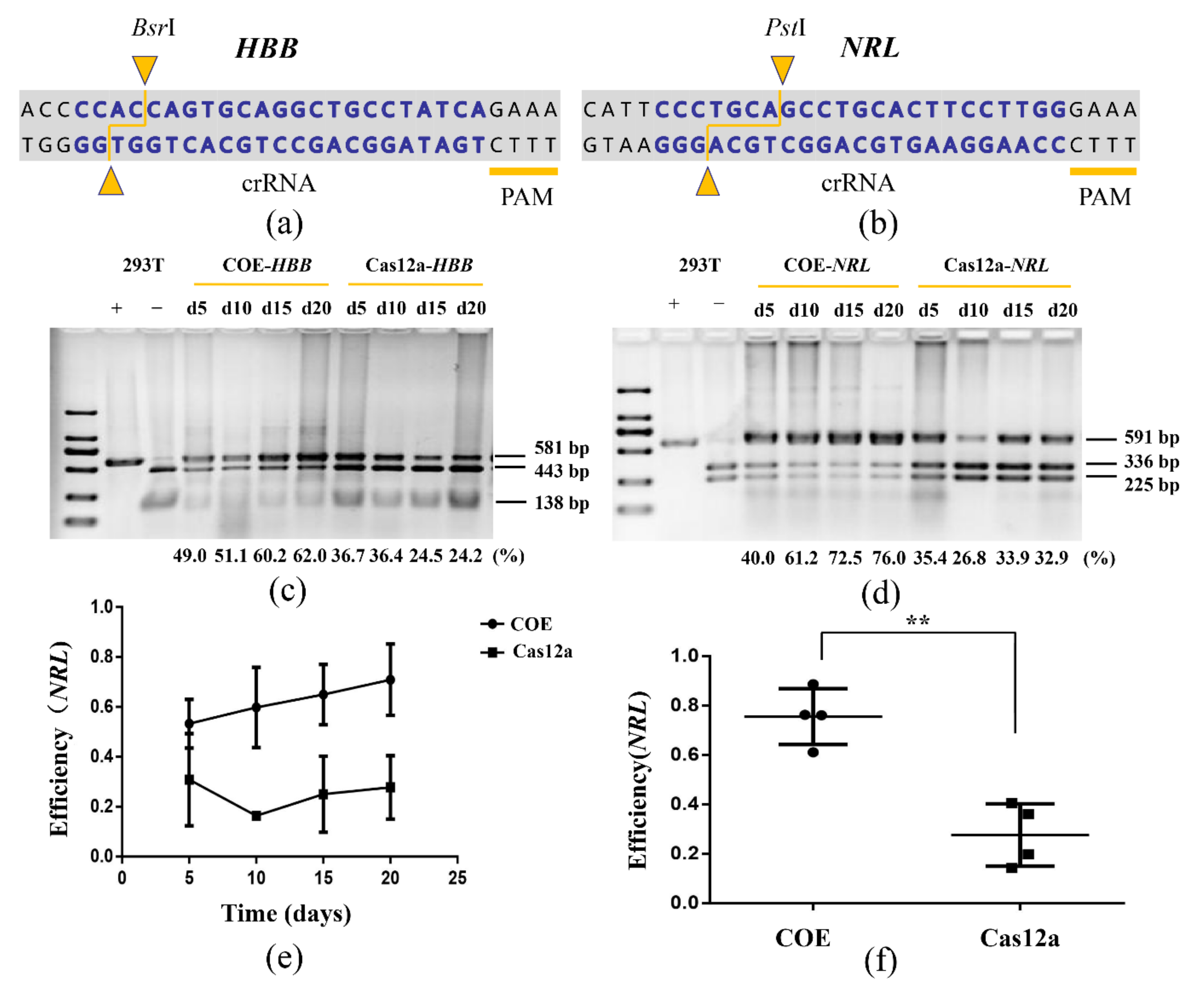
2.3. Genome Editing of HEK-293T with COE System
2.4. Restriction Fragment Length Polymorphism (RFLP) Analysis
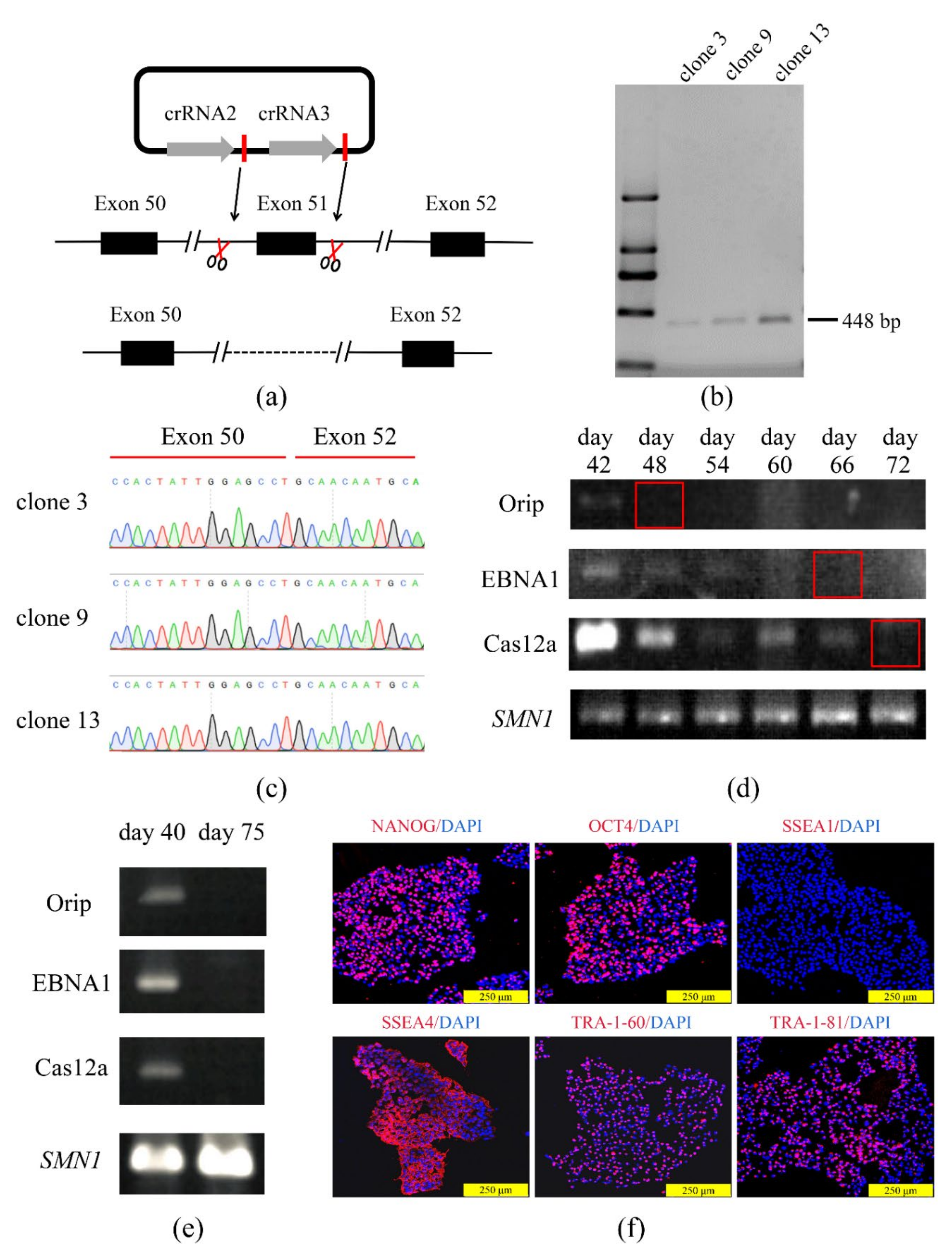
2.5. Genome Editing of iPSCs with COE System
2.6. RNA Isolation and Quantitative Polymerase Chain Reaction (qPCR)
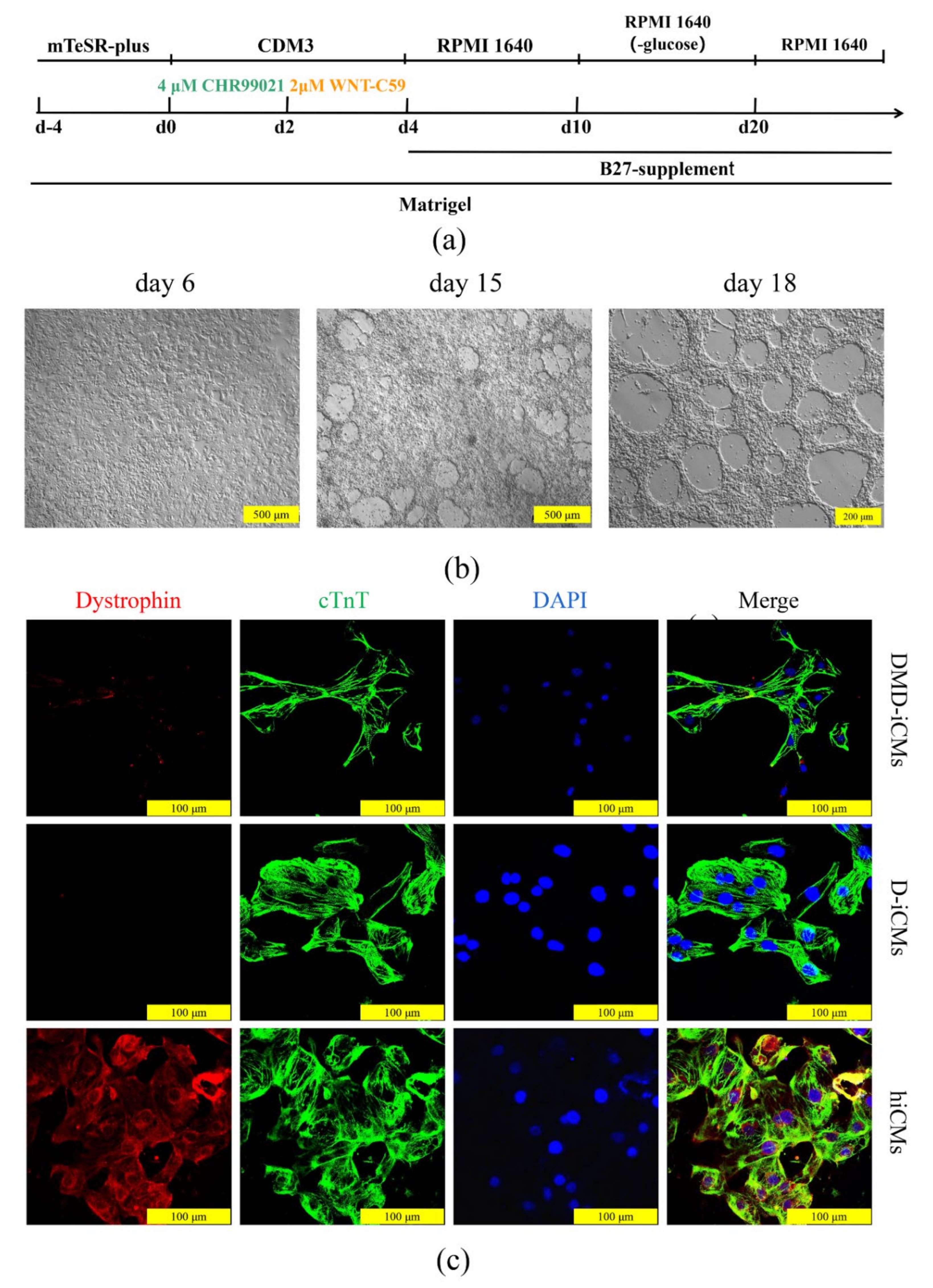
2.7. Cardiac Differentiation
2.8. Immunofluorescence Staining
2.9. Residue of Plasmid Components and Off-Target Analysis
2.10. Statistical Analysis
3. Results
3.1. Establishment of the Episomal CRISPR/Cas12a System
3.2. The COE System Significantly Promotes Gene Editing in HEK-293T
3.3. The COE System Efficient for Gene Deletion in Human iPSCs
3.4. The Edited iPSCs Were Directed to Differentiate into Cardiomyocytes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laoharawee, K.; DeKelver, R.C.; Podetz-Pedersen, K.M.; Rohde, M.; Sproul, S.; Nguyen, H.O.; Nguyen, T.; St Martin, S.J.; Ou, L.; Tom, S.; et al. Dose-Dependent Prevention of Metabolic and Neurologic Disease in Murine MPS II by ZFN-Mediated In Vivo Genome Editing. Mol. Ther. 2018, 26, 1127–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Yu, J.; Niu, Y.; Qin, D.; Liu, H.; Li, G.; Hu, Y.; Wang, J.; Lu, Y.; Kang, Y.; et al. Modeling Rett Syndrome Using TALEN-Edited MECP2 Mutant Cynomolgus Monkeys. Cell 2017, 169, 945–955.e910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qasim, W.; Zhan, H.; Samarasinghe, S.; Adams, S.; Amrolia, P.; Stafford, S.; Butler, K.; Rivat, C.; Wright, G.; Somana, K.; et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.X.; Zhang, Y.; Yin, H. Genome Editing with mRNA Encoding ZFN, TALEN, and Cas9. Mol. Ther. 2019, 27, 735–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidenreich, M.; Zhang, F. Applications of CRISPR-Cas systems in neuroscience. Nat. Rev. Neurosci. 2016, 17, 36–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendriks, D.; Clevers, H.; Artegiani, B. CRISPR-Cas Tools and Their Application in Genetic Engineering of Human Stem Cells and Organoids. Cell Stem Cell 2020, 27, 705–731. [Google Scholar] [CrossRef] [PubMed]
- Manghwar, H.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas System: Recent Advances and Future Prospects for Genome Editing. Trends Plant Sci. 2019, 24, 1102–1125. [Google Scholar] [CrossRef] [Green Version]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.; Li, Z.; Peterson, R.T.; Yeh, J.R.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [Green Version]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tóth, E.; Weinhardt, N.; Bencsura, P.; Huszár, K.; Kulcsár, P.I.; Tálas, A.; Fodor, E.; Welker, E. Cpf1 nucleases demonstrate robust activity to induce DNA modification by exploiting homology directed repair pathways in mammalian cells. Biol. Direct 2016, 11, 46. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Ren, Q.; Tang, X.; Liu, S.; Malzahn, A.A.; Zhou, J.; Wang, J.; Yin, D.; Pan, C.; Yuan, M.; et al. Expanding the scope of plant genome engineering with Cas12a orthologs and highly multiplexable editing systems. Nat. Commun. 2021, 12, 1944. [Google Scholar] [CrossRef]
- Bai, J.; Lin, H.; Li, H.; Zhou, Y.; Liu, J.; Zhong, G.; Wu, L.; Jiang, W.; Du, H.; Yang, J.; et al. Cas12a-Based On-Site and Rapid Nucleic Acid Detection of African Swine Fever. Front. Microbiol. 2019, 10, 2830. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Luk, K.; Shin, M.; Idrizi, F.; Kwok, S.; Roscoe, B.; Mintzer, E.; Suresh, S.; Morrison, K.; Frazão, J.B.; et al. Enhanced Cas12a editing in mammalian cells and zebrafish. Nucleic Acids Res. 2019, 47, 4169–4180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, B.; Montoya, G. CRISPR-Cas12a: Functional overview and applications. Biomed. J. 2020, 43, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Liu, D.; Jia, X.; Zheng, Y.; Liu, W.; Xiao, Y. CRISPR-Cas9/Cas12a biotechnology and application in bacteria. Synth. Syst. Biotechnol. 2018, 3, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Soldner, F.; Jaenisch, R. Stem Cells, Genome Editing, and the Path to Translational Medicine. Cell 2018, 175, 615–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timin, A.S.; Muslimov, A.R.; Lepik, K.V.; Epifanovskaya, O.S.; Shakirova, A.I.; Mock, U.; Riecken, K.; Okilova, M.V.; Sergeev, V.S.; Afanasyev, B.V.; et al. Efficient gene editing via non-viral delivery of CRISPR-Cas9 system using polymeric and hybrid microcarriers. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.B.; Zhang, S.; Kos, P.; Xiong, H.; Zhou, K.; Perelman, S.S.; Zhu, H.; Siegwart, D.J. Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew. Chem. 2017, 56, 1059–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Conboy, M.; Park, H.M.; Jiang, F.; Kim, H.J.; Dewitt, M.A.; Mackley, V.A.; Chang, K.; Rao, A.; Skinner, C.; et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017, 1, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Engle, S.J.; Blaha, L.; Kleiman, R.J. Best Practices for Translational Disease Modeling Using Human iPSC-Derived Neurons. Neuron 2018, 100, 783–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hockemeyer, D.; Jaenisch, R. Induced Pluripotent Stem Cells Meet Genome Editing. Cell Stem Cell 2016, 18, 573–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, J.H.; Fu, X.; Yang, P.C. Exosomes Generated From iPSC-Derivatives: New Direction for Stem Cell Therapy in Human Heart Diseases. Circ. Res. 2017, 120, 407–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, Y.; Yamanaka, S. Induced Pluripotent Stem Cells 10 Years Later: For Cardiac Applications. Circ. Res. 2017, 120, 1958–1968. [Google Scholar] [CrossRef] [PubMed]
- Dheekollu, J.; Wiedmer, A.; Ayyanathan, K.; Deakyne, J.S.; Messick, T.E.; Lieberman, P.M. Cell-cycle-dependent EBNA1-DNA crosslinking promotes replication termination at oriP and viral episome maintenance. Cell 2021, 184, 643–654.e613. [Google Scholar] [CrossRef]
- Huertas, D.; Howe, S.; McGuigan, A.; Huxley, C. Expression of the human CFTR gene from episomal oriP-EBNA1-YACs in mouse cells. Hum. Mol. Genet. 2000, 9, 617–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanbo, A.; Sugden, A.; Sugden, B. The coupling of synthesis and partitioning of EBV’s plasmid replicon is revealed in live cells. EMBO J. 2007, 26, 4252–4262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Wang, D.; Lan, F.; Wei, G.; Ni, T.; Chai, R.; Liu, D.; Hu, S.; Li, M.; Li, D.; et al. An episomal vector-based CRISPR/Cas9 system for highly efficient gene knockout in human pluripotent stem cells. Sci. Rep. 2017, 7, 2320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Z.; Qi, W.H.; Zhao, G.; Liu, L.X.; Xue, H.; Hu, W.X.; Wang, Q.Q.; Li, C.S. Correlation between PTEN and P62 gene expression in rat colorectal cancer cell. Saudi J. Biol. Sci. 2019, 26, 1986–1990. [Google Scholar] [CrossRef] [PubMed]
- Kovač, A.; Miskey, C.; Menzel, M.; Grueso, E.; Gogol-Döring, A.; Ivics, Z. RNA-guided retargeting of Sleeping Beauty transposition in human cells. eLife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Hu, Z.; Qiu, L.; Zhou, T.; Feng, M.; Hu, Q.; Zeng, B.; Li, Z.; Sun, Q.; Wu, Y.; et al. Seamless Genetic Conversion of SMN2 to SMN1 via CRISPR/Cpf1 and Single-Stranded Oligodeoxynucleotides in Spinal Muscular Atrophy Patient-Specific Induced Pluripotent Stem Cells. Hum. Gene Ther. 2018, 29, 1252–1263. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Zhou, M.; Liu, B.; Shen, F.; Xiao, R.; Su, J.; Hu, Z.; Zhang, Y.; Gu, A.; Wu, L.; et al. Targeted addition of mini-dystrophin into rDNA locus of Duchenne muscular dystrophy patient-derived iPSCs. Biochem. Biophys. Res. Commun. 2021, 545, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liang, P.; Lan, F.; Wu, H.; Lisowski, L.; Gu, M.; Hu, S.; Kay, M.A.; Urnov, F.D.; Shinnawi, R.; et al. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J. Am. Coll. Cardiol. 2014, 64, 451–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egawa, N.; Kitaoka, S.; Tsukita, K.; Naitoh, M.; Takahashi, K.; Yamamoto, T.; Adachi, F.; Kondo, T.; Okita, K.; Asaka, I.; et al. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci. Transl. Med. 2012, 4, 145ra104. [Google Scholar] [CrossRef] [PubMed]
- Dewanjee, S.; Dua, T.K.; Khanra, R.; Das, S.; Barma, S.; Joardar, S.; Bhattacharjee, N.; Zia-Ul-Haq, M.; Jaafar, H.Z. Water Spinach, Ipomoea aquatic (Convolvulaceae), Ameliorates Lead Toxicity by Inhibiting Oxidative Stress and Apoptosis. PLoS ONE 2015, 10, e0139831. [Google Scholar] [CrossRef] [Green Version]
- Wakefield, A.E.; Pixley, F.J.; Banerji, S.; Sinclair, K.; Miller, R.F.; Moxon, E.R.; Hopkin, J.M. Detection of Pneumocystis carinii with DNA amplification. Lancet 1990, 336, 451–453. [Google Scholar] [CrossRef]
- Sharma, A.; McKeithan, W.L.; Serrano, R.; Kitani, T.; Burridge, P.W.; Del Álamo, J.C.; Mercola, M.; Wu, J.C. Use of human induced pluripotent stem cell-derived cardiomyocytes to assess drug cardiotoxicity. Nat. Protoc. 2018, 13, 3018–3041. [Google Scholar] [CrossRef]
- Young, C.S.; Hicks, M.R.; Ermolova, N.V.; Nakano, H.; Jan, M.; Younesi, S.; Karumbayaram, S.; Kumagai-Cresse, C.; Wang, D.; Zack, J.A.; et al. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell Stem Cell 2016, 18, 533–540. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Gao, F.; Wu, S. An episomal CRISPR/Cas9 system to derive vector-free gene modified mammalian cells. Protein Cell 2016, 7, 689–691. [Google Scholar] [CrossRef] [Green Version]
- Min, Y.L.; Li, H.; Rodriguez-Caycedo, C.; Mireault, A.A.; Huang, J.; Shelton, J.M.; McAnally, J.R.; Amoasii, L.; Mammen, P.P.A.; Bassel-Duby, R.; et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci. Adv. 2019, 5, eaav4324. [Google Scholar] [CrossRef] [Green Version]
- Yates, J.L.; Warren, N.; Sugden, B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 1985, 313, 812–815. [Google Scholar] [CrossRef]
- Ren, X.; Yang, Z.; Xu, J.; Sun, J.; Mao, D.; Hu, Y.; Yang, S.J.; Qiao, H.H.; Wang, X.; Hu, Q.; et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014, 9, 1151–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.; Gore, A.; Yan, W.; Abalde-Atristain, L.; Li, Z.; He, C.; Wang, Y.; Brodsky, R.A.; Zhang, K.; Cheng, L.; et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 2014, 15, 12–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veres, A.; Gosis, B.S.; Ding, Q.; Collins, R.; Ragavendran, A.; Brand, H.; Erdin, S.; Cowan, C.A.; Talkowski, M.E.; Musunuru, K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 2014, 15, 27–30. [Google Scholar] [CrossRef] [Green Version]
- Ah-Fong, A.M.V.; Boyd, A.M.; Matson, M.E.H.; Judelson, H.S. A Cas12a-based gene editing system for Phytophthora infestans reveals monoallelic expression of an elicitor. Mol. Plant Pathol. 2021, 22, 737–752. [Google Scholar] [CrossRef]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.X.; Li, M.; Lee, C.M.; Chakraborty, S.; Kim, H.W.; Bao, G.; Leong, K.W. CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem. Rev. 2017, 117, 9874–9906. [Google Scholar] [CrossRef]
- Chew, W.L. Immunity to CRISPR Cas9 and Cas12a therapeutics. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018, 10. [Google Scholar] [CrossRef]
- Yin, H.; Kauffman, K.J.; Anderson, D.G. Delivery technologies for genome editing. Nat. Rev. Drug Discov. 2017, 16, 387–399. [Google Scholar] [CrossRef]
- Leonhardt, C.; Schwake, G.; Stögbauer, T.R.; Rappl, S.; Kuhr, J.T.; Ligon, T.S.; Rädler, J.O. Single-cell mRNA transfection studies: Delivery, kinetics and statistics by numbers. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 679–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahin, U.; Karikó, K.; Türeci, Ö. mRNA-based therapeutics—Developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, N.; Tang, S.; Zeng, B.; Hu, Z.; Hu, Q.; Wu, L.; Zhou, M.; Liang, D. An Episomal CRISPR/Cas12a System for Mediating Efficient Gene Editing. Life 2021, 11, 1262. https://doi.org/10.3390/life11111262
Duan N, Tang S, Zeng B, Hu Z, Hu Q, Wu L, Zhou M, Liang D. An Episomal CRISPR/Cas12a System for Mediating Efficient Gene Editing. Life. 2021; 11(11):1262. https://doi.org/10.3390/life11111262
Chicago/Turabian StyleDuan, Nannan, Shuqing Tang, Baitao Zeng, Zhiqing Hu, Qian Hu, Lingqian Wu, Miaojin Zhou, and Desheng Liang. 2021. "An Episomal CRISPR/Cas12a System for Mediating Efficient Gene Editing" Life 11, no. 11: 1262. https://doi.org/10.3390/life11111262
APA StyleDuan, N., Tang, S., Zeng, B., Hu, Z., Hu, Q., Wu, L., Zhou, M., & Liang, D. (2021). An Episomal CRISPR/Cas12a System for Mediating Efficient Gene Editing. Life, 11(11), 1262. https://doi.org/10.3390/life11111262







