CRISPR-Based Genetic Switches and Other Complex Circuits: Research and Application
Abstract
:1. Introduction
2. CRISPR-Based Genetic Switches in Transcription Level
3. CRISPR-Based Genetic Switches in Translation Level
4. Application of CRISPR-Based Switches in Genetic Circuits
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gardner, T.S.; Cantor, C.R.; Collins, J.J. Construction of a genetic toggle switch in Escherichia coli. Nature 2000, 403, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Prindle, A.; Samayoa, P.; Razinkov, I.; Danino, T.; Tsimring, L.S.; Hasty, J. A sensing array of radically coupled genetic ‘biopixels’. Nature 2011, 481, 39–44. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Fu, X.; Liu, L.; Ren, X.; Chau, C.K.; Li, S.; Xiang, L.; Zeng, H.; Chen, G.; Tang, L.H.; et al. Sequential establishment of stripe patterns in an expanding cell population. Science 2011, 334, 238–241. [Google Scholar] [CrossRef] [Green Version]
- Schaerli, Y.; Munteanu, A.; Gili, M.; Cotterell, J.; Sharpe, J.; Isalan, M. A unified design space of synthetic stripe-forming networks. Nat. Commun. 2014, 5, 4905. [Google Scholar] [CrossRef] [Green Version]
- Tamsir, A.; Tabor, J.J.; Voigt, C.A. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature 2011, 469, 212–215. [Google Scholar] [CrossRef] [Green Version]
- Auslander, D.; Auslander, S.; Pierrat, X.; Hellmann, L.; Rachid, L.; Fussenegger, M. Programmable full-adder computations in communicating three-dimensional cell cultures. Nat. Methods 2018, 15, 57–60. [Google Scholar] [CrossRef]
- Stanton, B.C.; Nielsen, A.A.; Tamsir, A.; Clancy, K.; Peterson, T.; Voigt, C.A. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 2014, 10, 99–105. [Google Scholar] [CrossRef] [Green Version]
- Farzadfard, F.; Lu, T.K. Synthetic biology. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 2014, 346, 1256272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnet, J.; Subsoontorn, P.; Endy, D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl. Acad. Sci. USA 2012, 109, 8884–8889. [Google Scholar] [CrossRef] [Green Version]
- Du, P.; Zhao, H.; Zhang, H.; Wang, R.; Huang, J.; Tian, Y.; Luo, X.; Luo, X.; Wang, M.; Xiang, Y.; et al. De novo design of an intercellular signaling toolbox for multi-channel cell-cell communication and biological computation. Nat. Commun. 2020, 11, 4226. [Google Scholar] [CrossRef] [PubMed]
- Billerbeck, S.; Brisbois, J.; Agmon, N.; Jimenez, M.; Temple, J.; Shen, M.; Boeke, J.D.; Cornish, V.W. A scalable peptide-GPCR language for engineering multicellular communication. Nat. Commun. 2018, 9, 5057. [Google Scholar] [CrossRef] [Green Version]
- Bacchus, W.; Lang, M.; El-Baba, M.D.; Weber, W.; Stelling, J.; Fussenegger, M. Synthetic two-way communication between mammalian cells. Nat. Biotechnol. 2012, 30, 991–996. [Google Scholar] [CrossRef]
- Chen, M.T.; Weiss, R. Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat. Biotechnol. 2005, 23, 1551–1555. [Google Scholar] [CrossRef]
- Gupta, A.; Reizman, I.M.; Reisch, C.R.; Prather, K.L. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 2017, 35, 273–279. [Google Scholar] [CrossRef] [Green Version]
- Soma, Y.; Hanai, T. Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab. Eng. 2015, 30, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Tsao, C.Y.; Hooshangi, S.; Wu, H.C.; Valdes, J.J.; Bentley, W.E. Autonomous induction of recombinant proteins by minimally rewiring native quorum sensing regulon of E. coli. Metab. Eng. 2010, 12, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Stephens, K.; Pozo, M.; Tsao, C.Y.; Hauk, P.; Bentley, W.E. Bacterial co-culture with cell signaling translator and growth controller modules for autonomously regulated culture composition. Nat. Commun. 2019, 10, 4129. [Google Scholar] [CrossRef] [PubMed]
- Kitada, T.; DiAndreth, B.; Teague, B.; Weiss, R. Programming gene and engineered-cell therapies with synthetic biology. Science 2018, 359. [Google Scholar] [CrossRef] [Green Version]
- Sedlmayer, F.; Jaeger, T.; Jenal, U.; Fussenegger, M. Quorum-Quenching Human Designer Cells for Closed-Loop Control of Pseudomonas aeruginosa Biofilms. Nano Lett. 2017, 17, 5043–5050. [Google Scholar] [CrossRef]
- Saeidi, N.; Wong, C.K.; Lo, T.M.; Nguyen, H.X.; Ling, H.; Leong, S.S.; Poh, C.L.; Chang, M.W. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol. Syst. Biol. 2011, 7, 521. [Google Scholar] [CrossRef]
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev. Biochem. 2013, 82, 237–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef] [Green Version]
- Farzadfard, F.; Perli, S.D.; Lu, T.K. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth. Biol. 2013, 2, 604–613. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef] [Green Version]
- Thakore, P.I.; D’Ippolito, A.M.; Song, L.; Safi, A.; Shivakumar, N.K.; Kabadi, A.M.; Reddy, T.E.; Crawford, G.E.; Gersbach, C.A. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 2015, 12, 1143–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandegar, M.A.; Huebsch, N.; Frolov, E.B.; Shin, E.; Truong, A.; Olvera, M.P.; Chan, A.H.; Miyaoka, Y.; Holmes, K.; Spencer, C.I.; et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell 2016, 18, 541–553. [Google Scholar] [CrossRef] [Green Version]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W., III; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant. Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef] [Green Version]
- Miao, C.; Zhao, H.; Qian, L.; Lou, C. Systematically investigating the key features of the DNase deactivated Cpf1 for tunable transcription regulation in prokaryotic cells. Synth. Syst. Biotechnol. 2019, 4, 1–9. [Google Scholar] [CrossRef]
- Kim, S.K.; Kim, S.H.; Subhadra, B.; Woo, S.G.; Rha, E.; Kim, S.W.; Kim, H.; Lee, D.H.; Lee, S.G. A Genetically Encoded Biosensor for Monitoring Isoprene Production in Engineered Escherichia coli. ACS Synth. Biol. 2018, 7, 2379–2390. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Han, J.; Chen, Z.; Wu, H.; Dong, H.; Nie, G. Engineering cell signaling using tunable CRISPR-Cpf1-based transcription factors. Nat. Commun. 2017, 8, 2095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Chen, J.; Wang, Y.; Liu, J.; Huang, J.; Chen, N.; Zheng, P.; Sun, J. Efficient Multiplex Gene Repression by CRISPR-dCpf1 in Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 2020, 8, 357. [Google Scholar] [CrossRef]
- Campa, C.C.; Weisbach, N.R.; Santinha, A.J.; Incarnato, D.; Platt, R.J. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat. Methods 2019, 16, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.L.; Peng, Y.Z.; Liu, D.; Liu, H.; Cao, Y.X.; Li, B.Z.; Li, C.; Yuan, Y.J. Gene repression via multiplex gRNA strategy in Y. lipolytica. Microb. Cell Fact. 2018, 17, 62. [Google Scholar] [CrossRef]
- Ciurkot, K.; Gorochowski, T.E.; Roubos, J.A.; Verwaal, R. Efficient multiplexed gene regulation in Saccharomyces cerevisiae using dCas12a. Nucleic Acids Res. 2021, 49, 7775–7790. [Google Scholar] [CrossRef]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q.; et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 17103. [Google Scholar] [CrossRef]
- Borchardt, E.K.; Vandoros, L.A.; Huang, M.; Lackey, P.E.; Marzluff, W.F.; Asokan, A. Controlling mRNA stability and translation with the CRISPR endoribonuclease Csy4. RNA 2015, 21, 1921–1930. [Google Scholar] [CrossRef] [Green Version]
- Nissim, L.; Perli, S.D.; Fridkin, A.; Perez-Pinera, P.; Lu, T.K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell 2014, 54, 698–710. [Google Scholar] [CrossRef] [Green Version]
- Zalatan, J.G.; Lee, M.E.; Almeida, R.; Gilbert, L.A.; Whitehead, E.H.; La Russa, M.; Tsai, J.C.; Weissman, J.S.; Dueber, J.E.; Qi, L.S.; et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 2015, 160, 339–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarty, N.S.; Shaw, W.M.; Ellis, T.; Ledesma-Amaro, R. Rapid Assembly of gRNA Arrays via Modular Cloning in Yeast. ACS Synth. Biol. 2019, 8, 906–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, J.; HamediRad, M.; Hu, S.; Zhao, H. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system. Nat. Commun. 2017, 8, 1688. [Google Scholar] [CrossRef]
- Ho, H.I.; Fang, J.R.; Cheung, J.; Wang, H.H. Programmable CRISPR-Cas transcriptional activation in bacteria. Mol. Syst. Biol. 2020, 16, e9427. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Fontana, J.; Patel, A.; Carothers, J.M.; Zalatan, J.G. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat. Commun. 2018, 9, 2489. [Google Scholar] [CrossRef] [Green Version]
- Fontana, J.; Dong, C.; Kiattisewee, C.; Chavali, V.P.; Tickman, B.I.; Carothers, J.M.; Zalatan, J.G. Effective CRISPRa-mediated control of gene expression in bacteria must overcome strict target site requirements. Nat. Commun. 2020, 11, 1618. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wan, X.; Wang, B. Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria. Nat. Commun. 2019, 10, 3693. [Google Scholar] [CrossRef] [Green Version]
- Perez-Pinera, P.; Kocak, D.D.; Vockley, C.M.; Adler, A.F.; Kabadi, A.M.; Polstein, L.R.; Thakore, P.I.; Glass, K.A.; Ousterout, D.G.; Leong, K.W.; et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 2013, 10, 973–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeder, M.L.; Linder, S.J.; Cascio, V.M.; Fu, Y.; Ho, Q.H.; Joung, J.K. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 2013, 10, 977–979. [Google Scholar] [CrossRef] [Green Version]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; Iyer, E.P.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 2015, 12, 326–328. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.; Peng, S.; Huang, W.; Cai, Z.; Xie, Z. Rational Design of Mini-Cas9 for Transcriptional Activation. ACS Synth. Biol. 2018, 7, 978–985. [Google Scholar] [CrossRef]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef] [Green Version]
- Baeumler, T.A.; Ahmed, A.A.; Fulga, T.A. Engineering Synthetic Signaling Pathways with Programmable dCas9-Based Chimeric Receptors. Cell Rep. 2017, 20, 2639–2653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tak, Y.E.; Kleinstiver, B.P.; Nunez, J.K.; Hsu, J.Y.; Horng, J.E.; Gong, J.; Weissman, J.S.; Joung, J.K. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat. Methods 2017, 14, 1163–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, P.; Miao, C.; Lou, Q.; Wang, Z.; Lou, C. Engineering Translational Activators with CRISPR-Cas System. ACS Synth. Biol. 2016, 5, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Fu, Y.; Sander, J.D.; Reyon, D.; Cascio, V.M.; Joung, J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014, 32, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.H.; Tee, L.Y.; Wang, X.G.; Huang, Q.S.; Yang, S.H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol. Ther. Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pattanayak, V.; Lin, S.; Guilinger, J.P.; Ma, E.; Doudna, J.A.; Liu, D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013, 31, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.A.; Voigt, C.A. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 2014, 10, 763. [Google Scholar] [CrossRef] [PubMed]
- Menn, D.J.; Pradhan, S.; Kiani, S.; Wang, X. Fluorescent Guide RNAs Facilitate Development of Layered Pol II-Driven CRISPR Circuits. ACS Synth. Biol. 2018, 7, 1929–1936. [Google Scholar] [CrossRef]
- Uddin, F.; Rudin, C.M.; Sen, T. CRISPR Gene Therapy: Applications, Limitations, and Implications for the Future. Front. Oncol. 2020, 10, 1387. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Cui, T.; Feng, G.; Guo, L.; Xu, K.; Gao, Q.; Li, T.; Li, J.; Zhou, Q.; Li, W. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov. 2018, 4, 63. [Google Scholar] [CrossRef]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. CRISPR-CasPhi from huge phages is a hypercompact genome editor. Science 2020, 369, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Orlova, N.; Oakes, B.L.; Ma, E.; Spinner, H.B.; Baney, K.L.M.; Chuck, J.; Tan, D.; Knott, G.J.; Harrington, L.B.; et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 2019, 566, 218–223. [Google Scholar] [CrossRef]
- Ji, X.; Zhao, H.; Zhu, H.; Zhu, K.; Tang, S.Y.; Lou, C. CRISPRi/dCpf1-mediated dynamic metabolic switch to enhance butenoic acid production in Escherichia coli. Appl. Microbiol. Biotechnol. 2020, 104, 5385–5393. [Google Scholar] [CrossRef]
- Hou, J.; Zeng, W.; Zong, Y.; Chen, Z.; Miao, C.; Wang, B.; Lou, C. Engineering the Ultrasensitive Transcription Factors by Fusing a Modular Oligomerization Domain. ACS Synth. Biol. 2018, 7, 1188–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, W.; Du, P.; Lou, Q.; Wu, L.; Zhang, H.M.; Lou, C.; Wang, H.; Ouyang, Q. Rational Design of an Ultrasensitive Quorum-Sensing Switch. ACS Synth. Biol. 2017, 6, 1445–1452. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.V.; Nunez, J.K.; Doudna, J.A. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 2016, 164, 29–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, L.; Haurwitz, R.E.; Shao, W.; Doudna, J.A.; Arkin, A.P. RNA processing enables predictable programming of gene expression. Nat. Biotechnol. 2012, 30, 1002–1006. [Google Scholar] [CrossRef] [PubMed]
- Haurwitz, R.E.; Jinek, M.; Wiedenheft, B.; Zhou, K.; Doudna, J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 2010, 329, 1355–1358. [Google Scholar] [CrossRef] [Green Version]
- Haurwitz, R.E.; Sternberg, S.H.; Doudna, J.A. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 2012, 31, 2824–2832. [Google Scholar] [CrossRef] [Green Version]
- Ozcan, A.; Krajeski, R.; Ioannidi, E.; Lee, B.; Gardner, A.; Makarova, K.S.; Koonin, E.V.; Abudayyeh, O.O.; Gootenberg, J.S. Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature 2021, 597, 720–725. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [Green Version]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [Green Version]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef]
- Liao, C.; Ttofali, F.; Slotkowski, R.A.; Denny, S.R.; Cecil, T.D.; Leenay, R.T.; Keung, A.J.; Beisel, C.L. Modular one-pot assembly of CRISPR arrays enables library generation and reveals factors influencing crRNA biogenesis. Nat. Commun. 2019, 10, 2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Bao, Z.; Jain, S.; Jaroenpuntaruk, V.; Zhao, H. Orthogonal Genetic Regulation in Human Cells Using Chemically Induced CRISPR/Cas9 Activators. ACS Synth. Biol. 2017, 6, 686–693. [Google Scholar] [CrossRef]
- Zetsche, B.; Volz, S.E.; Zhang, F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015, 33, 139–142. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Xiong, X.; Wong, S.; Charles, E.J.; Lim, W.A.; Qi, L.S. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat. Methods 2016, 13, 1043–1049. [Google Scholar] [CrossRef]
- Levskaya, A.; Weiner, O.D.; Lim, W.A.; Voigt, C.A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 2009, 461, 997–1001. [Google Scholar] [CrossRef] [Green Version]
- Nihongaki, Y.; Furuhata, Y.; Otabe, T.; Hasegawa, S.; Yoshimoto, K.; Sato, M. CRISPR-Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat. Methods 2017, 14, 963–966. [Google Scholar] [CrossRef]
- Nihongaki, Y.; Kawano, F.; Nakajima, T.; Sato, M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015, 33, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Polstein, L.R.; Gersbach, C.A. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015, 11, 198–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nihongaki, Y.; Yamamoto, S.; Kawano, F.; Suzuki, H.; Sato, M. CRISPR-Cas9-based photoactivatable transcription system. Chem. Biol. 2015, 22, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef]
- Ooi, K.H.; Liu, M.M.; Tay, J.W.D.; Teo, S.Y.; Kaewsapsak, P.; Jin, S.; Lee, C.K.; Hou, J.; Maurer-Stroh, S.; Lin, W.; et al. An engineered CRISPR-Cas12a variant and DNA-RNA hybrid guides enable robust and rapid COVID-19 testing. Nat. Commun. 2021, 12, 1739. [Google Scholar] [CrossRef]
- Xiong, D.; Dai, W.; Gong, J.; Li, G.; Liu, N.; Wu, W.; Pan, J.; Chen, C.; Jiao, Y.; Deng, H.; et al. Rapid detection of SARS-CoV-2 with CRISPR-Cas12a. PLoS Biol. 2020, 18, e3000978. [Google Scholar] [CrossRef] [PubMed]
- Fozouni, P.; Son, S.; Diaz de Leon Derby, M.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333.e9. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qian, L.; Wei, W.; Wang, Y.; Wang, B.; Lin, P.; Liu, W.; Xu, L.; Li, X.; Liu, D.; et al. Paired Design of dCas9 as a Systematic Platform for the Detection of Featured Nucleic Acid Sequences in Pathogenic Strains. ACS Synth. Biol. 2017, 6, 211–216. [Google Scholar] [CrossRef]
- Santos-Moreno, J.; Tasiudi, E.; Stelling, J.; Schaerli, Y. Multistable and dynamic CRISPRi-based synthetic circuits. Nat. Commun. 2020, 11, 2746. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J.; Yuan, R.; Sanchez, C.; Paulsson, J.; Silver, P.A. Toward a translationally independent RNA-based synthetic oscillator using deactivated CRISPR-Cas. Nucleic Acids Res. 2020, 48, 8165–8177. [Google Scholar] [CrossRef]
- Henningsen, J.; Schwarz-Schilling, M.; Leibl, A.; Gutierrez, J.N.; Sagredo, S.; Simmel, F.C. Single Cell Characterization of a Synthetic Bacterial Clock with a Hybrid Feedback Loop Containing dCas9-sgRNA. ACS Synth. Biol. 2020, 9, 3377–3387. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.D.; Qian, Y.; Siciliano, V.; DiAndreth, B.; Huh, J.; Weiss, R.; Del Vecchio, D. An endoribonuclease-based feedforward controller for decoupling resource-limited genetic modules in mammalian cells. Nat. Commun. 2020, 11, 5690. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Srinivasan, P.; Chavez, M.; Carter, M.A.; Dominguez, A.A.; La Russa, M.; Lau, M.B.; Abbott, T.R.; Xu, X.; Zhao, D.; et al. Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nat. Commun. 2019, 10, 194. [Google Scholar] [CrossRef] [Green Version]
- Gander, M.W.; Vrana, J.D.; Voje, W.E.; Carothers, J.M.; Klavins, E. Digital logic circuits in yeast with CRISPR-dCas9 NOR gates. Nat. Commun. 2017, 8, 15459. [Google Scholar] [CrossRef]
- Hofmann, A.; Falk, J.; Prangemeier, T.; Happel, D.; Kober, A.; Christmann, A.; Koeppl, H.; Kolmar, H. A tightly regulated and adjustable CRISPR-dCas9 based AND gate in yeast. Nucleic Acids Res. 2019, 47, 509–520. [Google Scholar] [CrossRef] [Green Version]
- Santos-Moreno, J.; Schaerli, Y. A Framework for the Modular and Combinatorial Assembly of Synthetic Gene Circuits. ACS Synth. Biol. 2019, 8, 1691–1697. [Google Scholar] [CrossRef]
- Kiani, S.; Beal, J.; Ebrahimkhani, M.R.; Huh, J.; Hall, R.N.; Xie, Z.; Li, Y.; Weiss, R. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat. Methods 2014, 11, 723–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Didovyk, A.; Borek, B.; Hasty, J.; Tsimring, L. Orthogonal Modular Gene Repression in Escherichia coli Using Engineered CRISPR/Cas9. ACS Synth. Biol. 2016, 5, 81–88. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Bojar, D.; Fussenegger, M. A CRISPR/Cas9-based central processing unit to program complex logic computation in human cells. Proc. Natl. Acad. Sci. USA 2019, 116, 7214–7219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
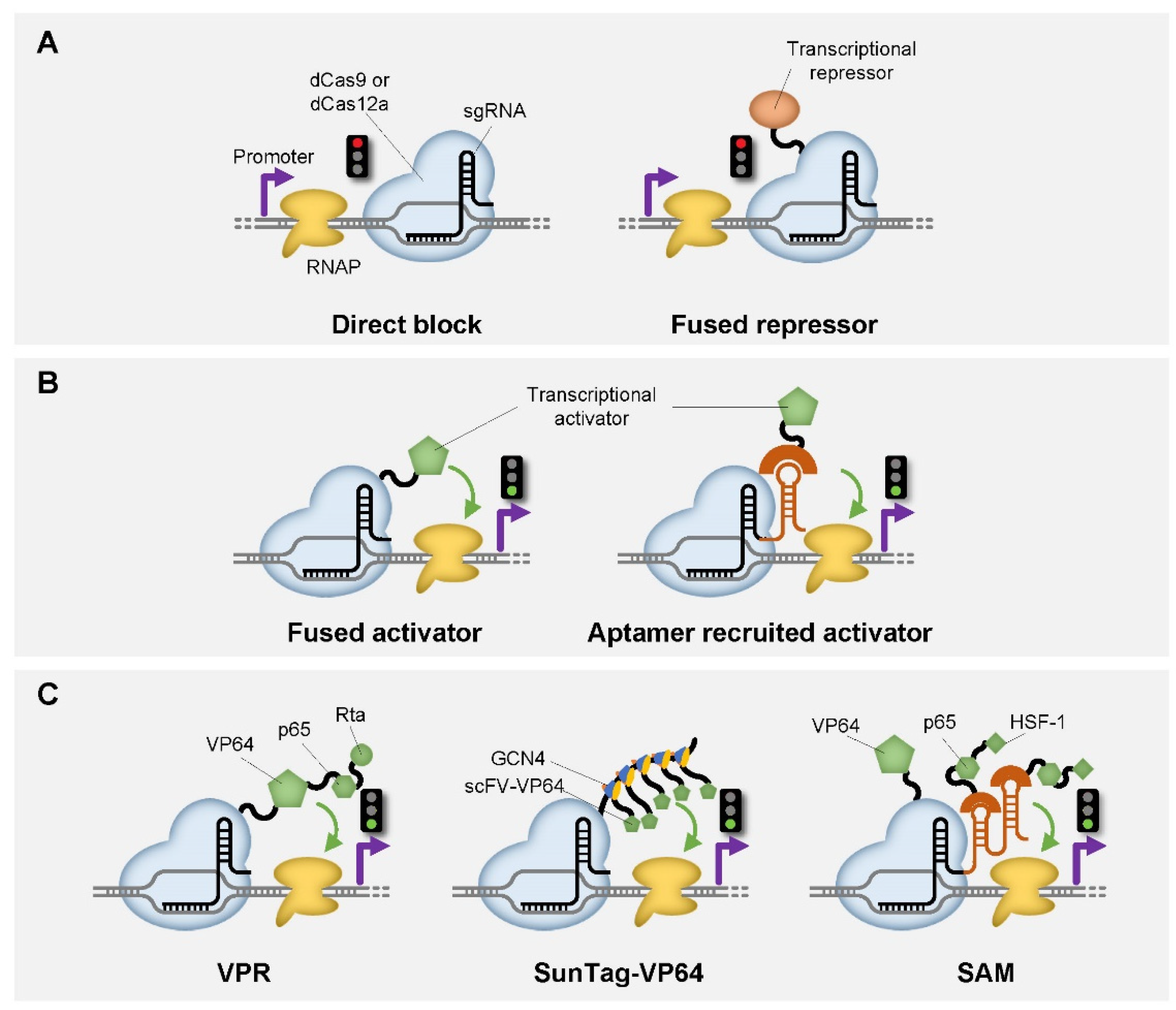

| CRISPR Enzyme | Additional Module | Function | Host | Reference |
|---|---|---|---|---|
| dCas9 | / | Transcriptional repression | bacteria, yeast | [23,24,25,26] |
| dCas9 | KRAB (F) | Transcriptional repression | yeast, mammalian cell | [25,26,27,28,29,30] |
| dCas9 | Mxi1 (F) | Transcriptional repression | yeast | [26] |
| dCas9 | SRDX (F) | Transcriptional repression | Arabidopsis | [30] |
| dCas12a | / | Transcriptional repression | bacteria, mammalian cell | [31,32,33,34,35] |
| dCas12a | KRAB (F) | Transcriptional repression | mammalian cell | [33,36] |
| dCas12a | Mxi1 (F) | Transcriptional repression | yeast | [37] |
| dCas12a | SRDX (F) | Transcriptional repression | Arabidopsis | [38] |
| Csy4 | / | Translational repression | E. coli, mammalian cell | [39] |
| Csy4 | dCas9 | Translational repression | mammalian cell | [40] |
| Csy4 | dCas9 | Translational repression | yeast, mammalian cell | [41] |
| Csy4 | dCas9 | Translational repression | yeast | [42] |
| Csy4 | dCas9/dCas12a | Translational repression | yeast | [43] |
| dCas9 | RNAP ω subunit (F) | Transcriptional activation | E. coli | [24] |
| dCas9 | AsiA (F) | Transcriptional activation | E. coli | [44] |
| dCas9 | MCP-SoxS (R) | Transcriptional activation | E. coli | [45,46] |
| dCas9 | λN22plus-PspFΔHTH (R) | Transcriptional activation | E. coli | [47] |
| dCas9 | VP64 (F) | Transcriptional activation | yeast, mammalian cell, Arabidopsis | [25,26,30,48,49] |
| dCas9 | p65 (F) | Transcriptional activation | mammalian cell | [25] |
| dCas9 | VPR (F) | Transcriptional activation | yeast, mammalian cell | [50] |
| dCas9 | VTR3 (F) | Transcriptional activation | mammalian cell | [51] |
| dCas9 | SunTag (F) | Transcriptional activation | mammalian cell | [27,52] |
| dCas9 | SAM (F) | Transcriptional activation | E. coli, mammalian cell | [53,54] |
| dCas12a | VP64 (F) | Transcriptional activation | mammalian cell | [33] |
| dCas12a | p65 (F) | Transcriptional activation | mammalian cell | [55] |
| dCas12a | VPR (F) | Transcriptional activation | mammalian cell | [33] |
| Csy4 | / | Translational activation | E. coli | [56] |
| Csy4 | Cas9 | Translational activation | yeast, mammalian cell | [41] |
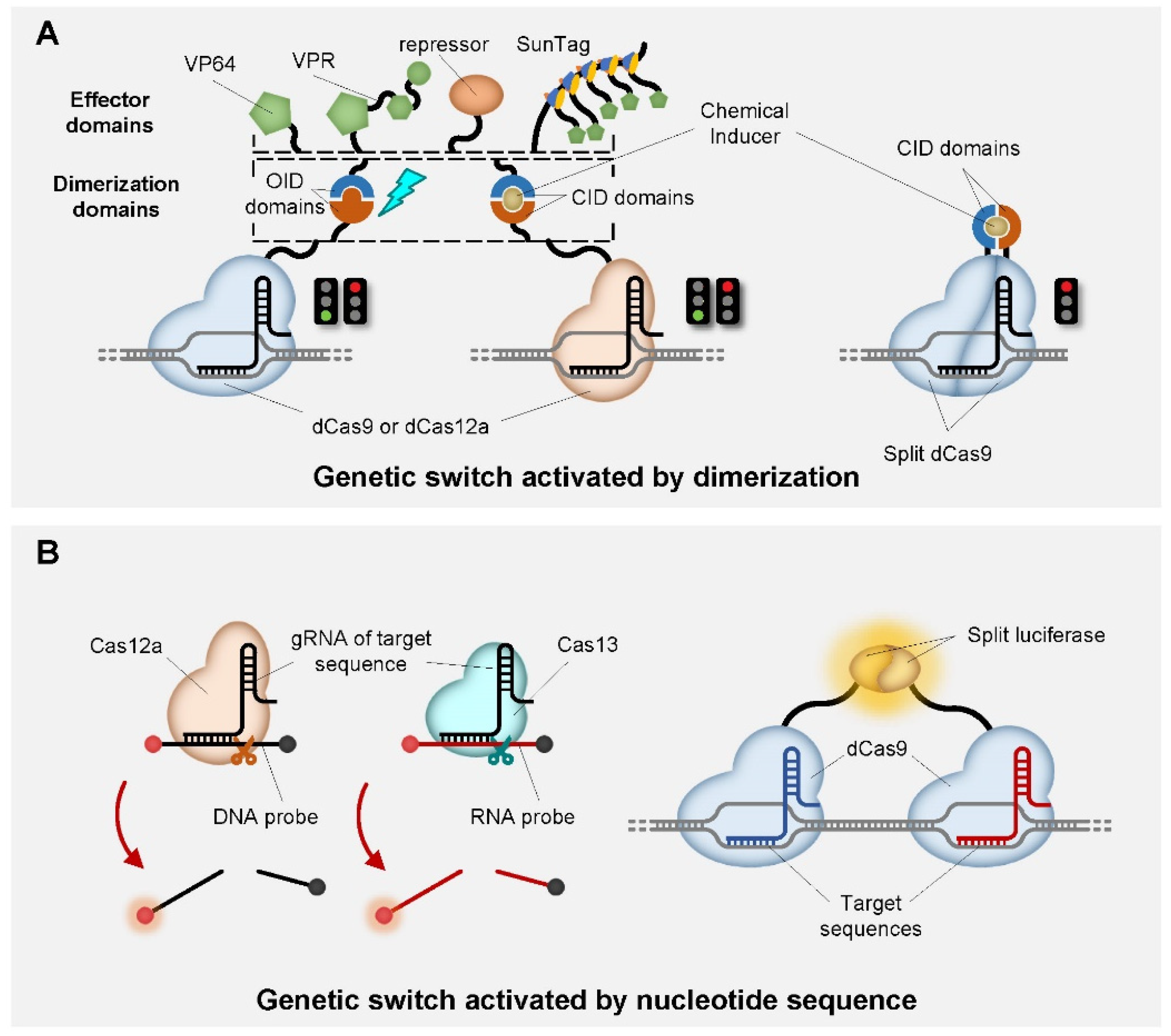
| CRISPR Enzyme | Dimerization Domain | Effector Domain | Input Signal | Circuit Type | Reference |
|---|---|---|---|---|---|
| dCas9 | FKBP-FRB | VPR | rapamycin | ligand-inducible genetic switch | [84] |
| Split dCas9 | FKBP-FRB | VP64 | rapamycin | ligand-inducible genetic switch | [85] |
| dCas12a | DmrA-DmrC | p65, VPR | rapamycin | ligand-inducible genetic switch | [55] |
| dCas9 | ABI-PYL1 | VPR | ABA | ligand-inducible genetic switch | [84,86] |
| dCas9 | GID1-GAI24 | VPR | GA | ligand-inducible genetic switch | [86] |
| dCas9 | PhyB-PIF | / | Red light | Light-inducible genetic switch | [87] |
| dCas9 | pMag-nMag | p65, VP64 | Blue light | Light-inducible genetic switch | [88] |
| Split Cas9 | pMag-nMag | / | Blue light | Light-inducible genome editor | [89] |
| dCas9 | CRY2-CIB1 | p65, VP64 | Blue light | Light-inducible genetic switch | [88,89,90,91] |
| Cas12a | / | fluorescent DNA probe | nucleotide sequence | in vitro diagnostic toolbox | [92,93,94,95] |
| Cas13 | / | fluorescent RNA probe | nucleotide sequence | in vitro diagnostic toolbox | [96,97] |
| dCas9 | split luciferease | luciferase | nucleotide sequence | in vitro diagnostic toolbox | [98] |
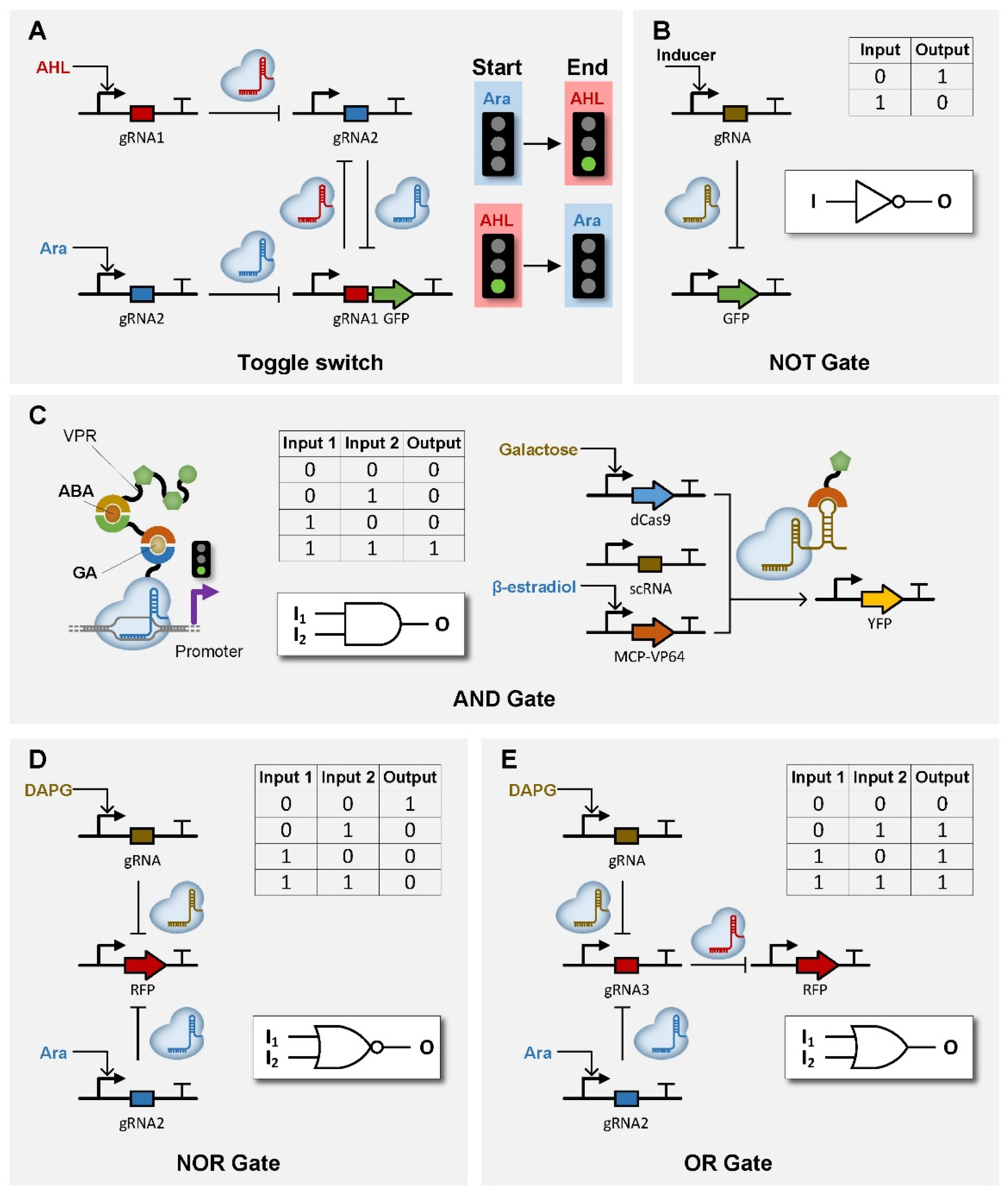
| dCas9 | / | / | AHL & Ara | toggle switch | [99] |
| dCas9 | / | / | / | oscillator | [99,100,101] |
| dCas9 | / | / | Ara | IFFL | [99] |
| CasE | / | VPR | DNA copy number | IFFL | [102] |
| dCas9 | / | KRAB, VPR | Anti-CRISPR protein | IFFL | [103] |
| dCas9 | / | / | DAPG and Ara | NOR/AND/OR gate | [65] |
| dCas9 | / | Mxi1 | gRNA | NOR gateNOT gate | [104] |
| dCas9 | GID1-GAI24 | VPR | GA and ABA | AND gate | [86] |
| dCas9 | VP64 | Galactose and β-estradiol | AND gate | [105] | |
| dCas9 | / | / | Ara | NOT gate | [99,106] |
| dCas9 | / | VP16 | gRNA | NOT gate | [107] |
| dCas9 | / | / | aTc | NOT gate | [108] |
| dCas9 | / | KRAB | gRNA | NOT gate | [109] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, P.; Lou, C.; Zhao, X.; Wang, Q.; Ji, X.; Wei, W. CRISPR-Based Genetic Switches and Other Complex Circuits: Research and Application. Life 2021, 11, 1255. https://doi.org/10.3390/life11111255
Du P, Lou C, Zhao X, Wang Q, Ji X, Wei W. CRISPR-Based Genetic Switches and Other Complex Circuits: Research and Application. Life. 2021; 11(11):1255. https://doi.org/10.3390/life11111255
Chicago/Turabian StyleDu, Pei, Chunbo Lou, Xuejin Zhao, Qihui Wang, Xiangyu Ji, and Weijia Wei. 2021. "CRISPR-Based Genetic Switches and Other Complex Circuits: Research and Application" Life 11, no. 11: 1255. https://doi.org/10.3390/life11111255
APA StyleDu, P., Lou, C., Zhao, X., Wang, Q., Ji, X., & Wei, W. (2021). CRISPR-Based Genetic Switches and Other Complex Circuits: Research and Application. Life, 11(11), 1255. https://doi.org/10.3390/life11111255







