Harnessing CRISPR-Cas to Combat COVID-19: From Diagnostics to Therapeutics
Abstract
:1. Introduction
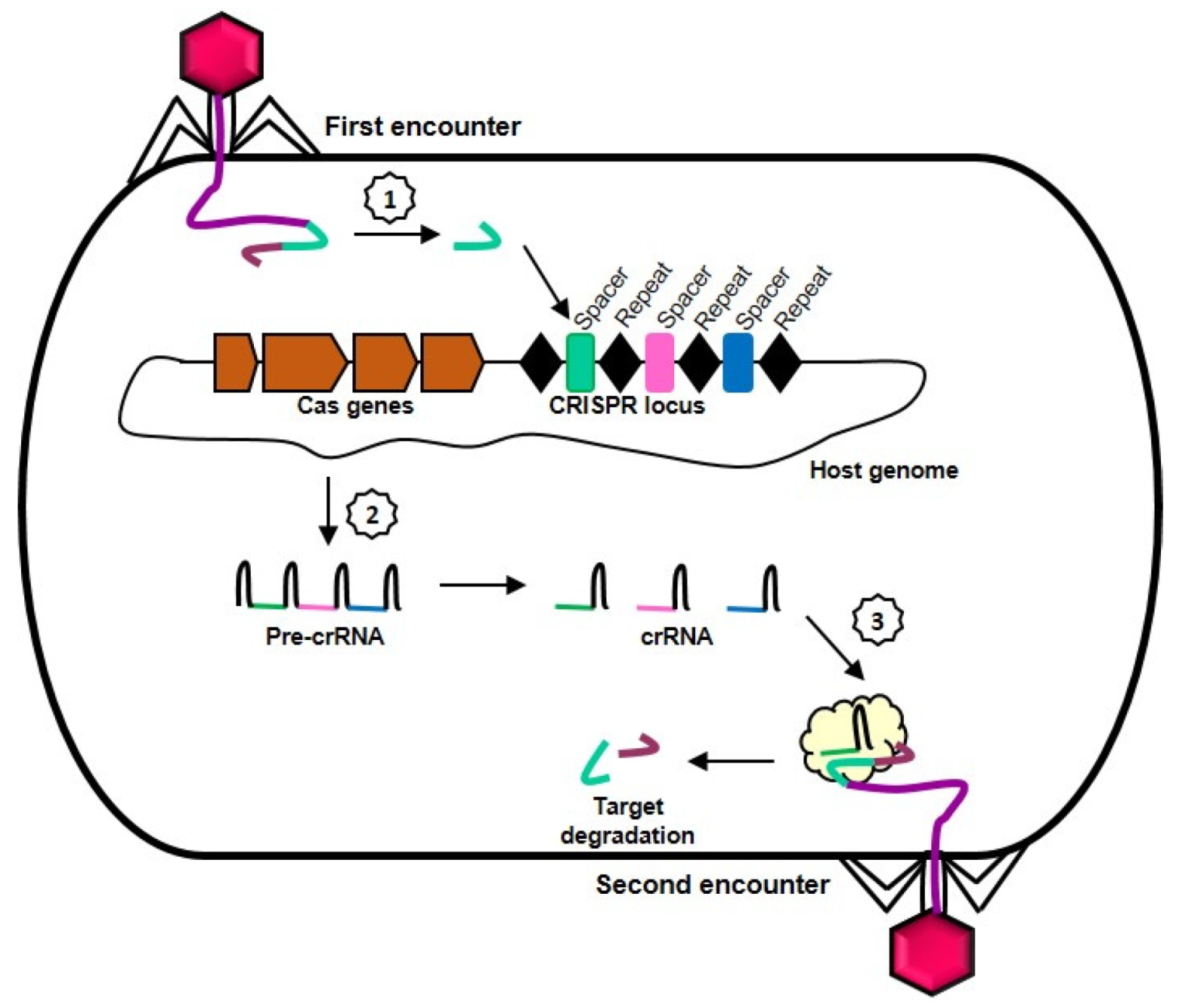
2. Molecular Mechanism of CRISPR-Cas
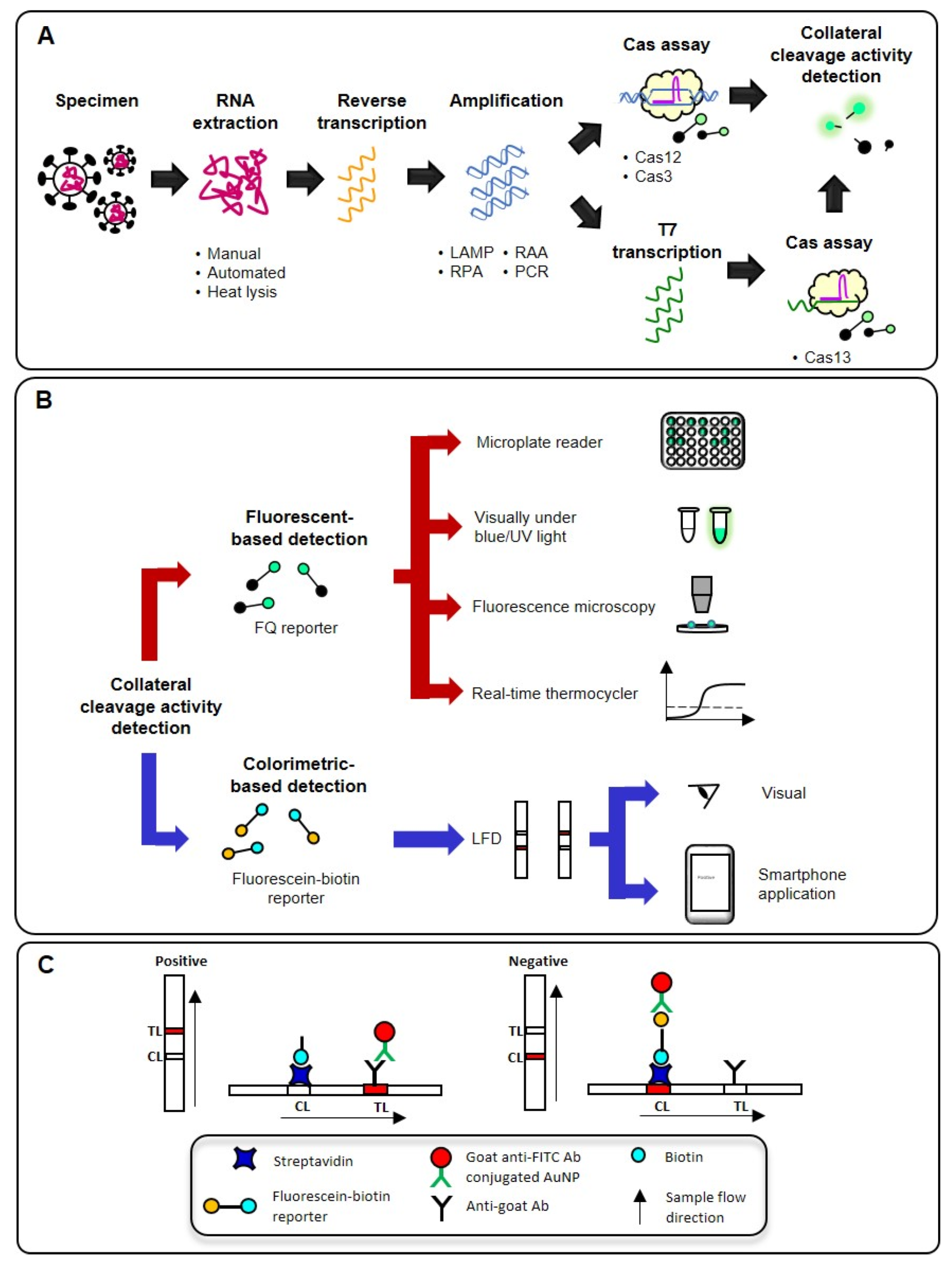
3. An Overview of CRISPR-Dx Workflow
4. Cas12-Based CRISPR-Dx
4.1. Two-Pot Assays
4.2. One-Pot Assays
4.3. Other Assay Formats
4.4. RNA Extraction-Free Protocols
4.5. Sensitivity and Specificity Enhancement Strategies
5. Cas13-Based CRISPR-Dx
5.1. Two-Pot Assays
5.2. Label-Free Assay
5.3. Amplification-Free Assay
5.4. Strategies for High-Throughput Analysis
6. Cas13d-Based Assay
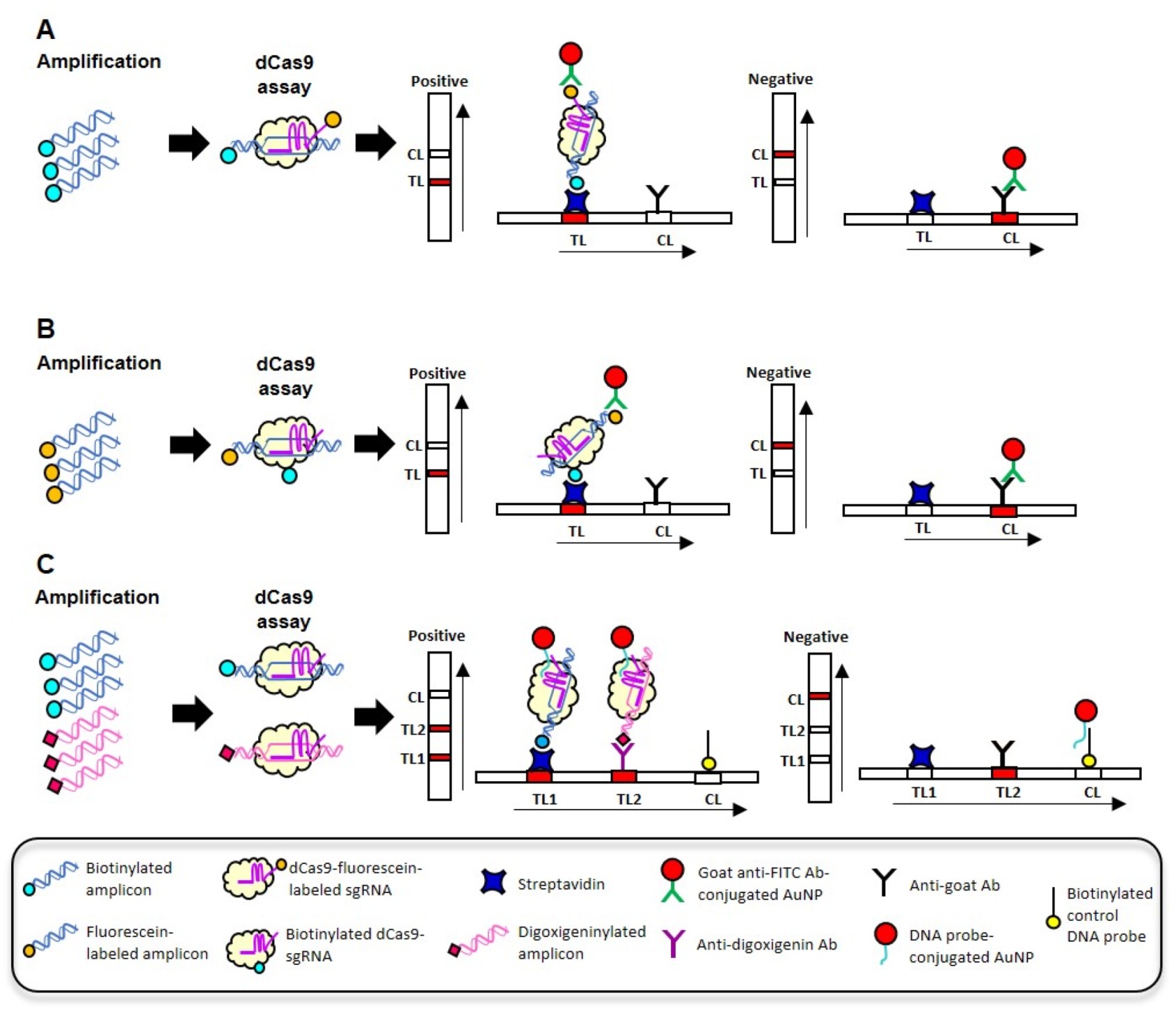
7. Cas9-Based CRISPR-Dx
8. Cas3-Based CRISPR-Dx
9. CRISPR-Cas as an Antiviral Agent
10. Summary and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 27 September 2021).
- Flaxman, S.; Mishra, S.; Gandy, A.; Unwin, H.J.T.; Mellan, T.; Coupland, H.; Whittaker, C.; Zhu, H.; Berah, T.; Eaton, J.W.; et al. Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature 2020, 584, 257–261. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; McGoogan, J.M. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China. JAMA 2020, 323, 1239. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir–Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, M.; Yin, L.; Wang, K.; Zhou, Y.; Zhou, M.; Lu, Y. COVID-19 treatment: Close to a cure? A rapid review of pharmacotherapies for the novel coronavirus (SARS-CoV-2). Int. J. Antimicrob. Agents 2020, 56, 106080. [Google Scholar] [CrossRef] [PubMed]
- Kimball, A.; Hatfield, K.M.; Arons, M.; James, A.; Taylor, J.; Spicer, K.; Bardossy, A.C.; Oakley, L.P.; Tanwar, S.; Chisty, Z.; et al. Asymptomatic and Presymptomatic SARS-CoV-2 Infections in Residents of a Long-Term Care Skilled Nursing Facility—King County, Washington, March 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 377–381. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.-Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323, 1406. [Google Scholar] [CrossRef] [Green Version]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Tsatsakis, A.; Calina, D.; Falzone, L.; Petrakis, D.; Mitrut, R.; Siokas, V.; Pennisi, M.; Lanza, G.; Libra, M.; Doukas, S.G.; et al. SARS-CoV-2 pathophysiology and its clinical implications: An integrative overview of the pharmacotherapeutic management of COVID-19. Food Chem. Toxicol. 2020, 146, 111769. [Google Scholar] [CrossRef]
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115. [Google Scholar] [CrossRef]
- Vallamkondu, J.; John, A.; Wani, W.Y.; Ramadevi, S.P.; Jella, K.K.; Reddy, P.H.; Kandimalla, R. SARS-CoV-2 pathophysiology and assessment of coronaviruses in CNS diseases with a focus on therapeutic targets. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165889. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Chan, K.G.; Yean, C.Y.; Ang, G.Y. Nucleic Acid-Based Diagnostic Tests for the Detection SARS-CoV-2: An Update. Diagnostics 2021, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-López, B.; Mir, M. Commercialized diagnostic technologies to combat SARS-CoV2: Advantages and disadvantages. Talanta 2020, 225, 121898. [Google Scholar] [CrossRef]
- Hardinge, P.; Murray, J.A.H. Reduced False Positives and Improved Reporting of Loop-Mediated Isothermal Amplification using Quenched Fluorescent Primers. Sci. Rep. 2019, 9, 7400. [Google Scholar] [CrossRef]
- Falzone, L.; Gattuso, G.; Tsatsakis, A.; Spandidos, D.A.; Libra, M. Current and innovative methods for the diagnosis of COVID-19 infection (Review). Int. J. Mol. Med. 2021, 47, 100. [Google Scholar] [CrossRef]
- Xu, M.; Wang, D.; Wang, H.; Zhang, X.; Liang, T.; Dai, J.; Li, M.; Zhang, J.; Zhang, K.; Xu, D.; et al. COVID-19 diagnostic testing: Technology perspective. Clin. Transl. Med. 2020, 10, e158. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, J. Immunologic Testing for SARS-CoV-2 Infection from the Antigen Perspective. J. Clin. Microbiol. 2021, 59, e02160-20. [Google Scholar] [CrossRef] [PubMed]
- Jameleddine Chtioui, M.; Grati, H.; Harzallah, N.; Odabachian Jebali, M.C.; Trabelsi, A.; Ben Ahmed, M.; Thabet, L.; Mhalla, S.; Ben Moussa, M.; Messaoud, T.; et al. Rapid diagnostic tests: Pros, cons and potential use in the COVID-19 management in Tunisia. Tunis Med. 2020, 98, 639–642. [Google Scholar]
- Li, Y.; Li, S.; Wang, J.; Liu, G. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 2019, 37, 730–743. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Mojica, F.J.M.; Soria, E. Intervening Sequences of Regularly Spaced Prokaryotic Repeats Derive from Foreign Genetic Elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef]
- Garneau, J.E.; Dupuis, M.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadan, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nat. Cell Biol. 2010, 468, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nat. Rev. Genet. 2019, 18, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Burmistrz, M.; Krakowski, K.; Krawczyk-Balska, A. RNA-Targeting CRISPR–Cas Systems and Their Applications. Int. J. Mol. Sci. 2020, 21, 1122. [Google Scholar] [CrossRef] [Green Version]
- Faure, G.; Shmakov, S.A.; Makarova, K.S.; Wolf, Y.I.; Crawley, A.B.; Barrangou, R.; Koonin, E.V. Comparative genomics and evolution of trans-activating RNAs in Class 2 CRISPR-Cas systems. RNA Biol. 2018, 16, 435–448. [Google Scholar] [CrossRef]
- Makarova, K.S.; Anantharaman, V.; Aravind, L.; Koonin, E.V. Live virus-free or die: Coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol. Direct 2012, 7, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellner, M.J.; Koob, J.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- Feng, W.; Newbigging, A.M.; Tao, J.; Cao, Y.; Peng, H.; Le, C.; Wu, J.; Pang, B.; Li, J.; Tyrrell, D.L.; et al. CRISPR technology incorporating amplification strategies: Molecular assays for nucleic acids, proteins, and small molecules. Chem. Sci. 2021, 12, 4683–4698. [Google Scholar] [CrossRef]
- Yoshimi, K.; Takeshita, K.; Yamayoshi, S.; Shibumura, S.; Yamauchi, Y.; Yamamoto, M.; Yotsuyanagi, H.; Kawaoka, Y.; Mashimo, T. Rapid and accurate detection of novel coronavirus SARS-CoV-2 using CRISPR-Cas3. medRxiv 2020. [Google Scholar] [CrossRef]
- Morisaka, H.; Yoshimi, K.; Okuzaki, Y.; Gee, P.; Kunihiro, Y.; Sonpho, E.; Xu, H.; Sasakawa, N.; Naito, Y.; Nakada, S.; et al. CRISPR-Cas3 induces broad and unidirectional genome editing in human cells. Nat. Commun. 2019, 10, 5302. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, R.; Li, J. CRISPR/cas systems redefine nucleic acid detection: Principles and methods. Biosens. Bioelectron. 2020, 165, 112430. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.; Oakes, B.L.; Sternberg, S.H.; Seletsky, A.; Kaplan, M.; Doudna, J.A. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014, 516, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Brandsma, E.; Verhagen, H.J.M.P.; van de Laar, T.J.W.; Claas, E.C.J.; Cornelissen, M.; Akker, E.V.D. Rapid, Sensitive, and Specific Severe Acute Respiratory Syndrome Coronavirus 2 Detection: A Multicenter Comparison Between Standard Quantitative Reverse-Transcriptase Polymerase Chain Reaction and CRISPR-Based DETECTR. J. Infect. Dis. 2020, 223, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Samacoits, A.; Nimsamer, P.; Mayuramart, O.; Chantaravisoot, N.; Sitthi-Amorn, P.; Nakhakes, C.; Luangkamchorn, L.; Tongcham, P.; Zahm, U.; Suphanpayak, S.; et al. Machine Learning-Driven and Smartphone-Based Fluorescence Detection for CRISPR Diagnostic of SARS-CoV-2. ACS Omega 2021, 6, 2727–2733. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Kim, N.-G.; Woolley, A.E.; Segel, M.; Barretto, R.P.; Ranu, A.; Macrae, R.K.; Faure, G.; et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020, 383, 1492–1494. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLOS Pathog. 2020, 16, e1008705. [Google Scholar] [CrossRef]
- Huang, Z.; Tian, D.; Liu, Y.; Lin, Z.; Lyon, C.J.; Lai, W.; Fusco, D.; Drouin, A.; Yin, X.; Hu, T.; et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020, 164, 112316. [Google Scholar] [CrossRef]
- Huang, W.; Yu, L.; Wen, D.; Wei, D.; Sun, Y.; Zhao, H.; Ye, Y.; Chen, W.; Zhu, Y.; Wang, L.; et al. A CRISPR-Cas12a-based specific enhancer for more sensitive detection of SARS-CoV-2 infection. EBioMedicine 2020, 61, 103036. [Google Scholar] [CrossRef]
- Ning, B.; Yu, T.; Zhang, S.; Huang, Z.; Tian, D.; Lin, Z.; Niu, A.; Golden, N.; Hensley, K.; Threeton, B.; et al. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci. Adv. 2021, 7, eabe3703. [Google Scholar] [CrossRef] [PubMed]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.-S.F.; Kemball, M.E.; et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020, 11, 5921. [Google Scholar] [CrossRef]
- Teng, F.; Guo, L.; Cui, T.; Wang, X.-G.; Xu, K.; Gao, Q.; Zhou, Q.; Li, W. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019, 20, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, 63. [Google Scholar] [CrossRef] [Green Version]
- Piepenburg, O.; Williams Ch Fau-Stemple, D.L.; Stemple Dl Fau-Armes, N.A.; Armes, N.A. DNA detection using recom-bination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef]
- Mammoth Biosciences. SARS-CoV-2 DETECTR Reagent Kit. Available online: https://www.fda.gov/media/141765/download (accessed on 29 July 2021).
- Lucia, C.; Federico, P.-B.; Alejandra, G.C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhong, M.; Liu, Y.; Ma, P.; Dang, L.; Meng, Q.; Wan, W.; Ma, X.; Liu, J.; Yang, G.; et al. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci. Bull. 2020, 65, 1436–1439. [Google Scholar] [CrossRef]
- Mayuramart, O.; Nimsamer, P.; Rattanaburi, S.; Chantaravisoot, N.; Khongnomnan, K.; Chansaenroj, J.; Puenpa, J.; Suntronwong, N.; Vichaiwattana, P.; Poovorawan, Y.; et al. Detection of severe acute respiratory syndrome coronavirus 2 and influenza viruses based on CRISPR-Cas12a. Exp. Biol. Med. 2020, 246, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020, 288, 198129. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, Y.; Chen, Y.; Yang, Z.; Wu, H.; Zhou, Z.; Li, J.; Ping, J.; He, L.; Shen, H.; et al. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection. Biosens. Bioelectron. 2020, 169, 112642. [Google Scholar] [CrossRef]
- Wang, R.; Qian, C.; Pang, Y.; Li, M.; Yang, Y.; Ma, H.; Zhao, M.; Qian, F.; Yu, H.; Liu, Z.; et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2020, 172, 112766. [Google Scholar] [CrossRef] [PubMed]
- Pang, B.; Xu, J.; Liu, Y.; Peng, H.; Feng, W.; Cao, Y.; Wu, J.; Xiao, H.; Pabbaraju, K.; Tipples, G.; et al. Isothermal Amplification and Ambient Visualization in a Single Tube for the Detection of SARS-CoV-2 Using Loop-Mediated Amplification and CRISPR Technology. Anal. Chem. 2020, 92, 16204–16212. [Google Scholar] [CrossRef]
- Sun, Y.; Yu, L.; Liu, C.; Ye, S.; Chen, W.; Li, D.; Huang, W. One-tube SARS-CoV-2 detection platform based on RT-RPA and CRISPR/Cas12a. J. Transl. Med. 2021, 19, 74. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Sun, X.; Wang, X.; Liang, C.; Jiang, H.; Gao, Q.; Dai, M.; Qu, B.; Fang, S.; Mao, Y.; et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020, 6, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, A.; Huyke, D.A.; Sharma, E.; Sahoo, M.K.; Huang, C.; Banaei, N.; Pinsky, B.A.; Santiago, J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 29518–29525. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Hsieh, K.; Chen, L.; Kaushik, A.; Trick, A.Y.; Wang, T. Digital CRISPR/Cas-Assisted Assay for Rapid and Sensitive Detection of SARS-CoV-2. Adv. Sci. 2020, 8, 2003564. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, Y.; Shi, Y.; Wang, L.; Zhang, M.; Wu, J.; Chen, H. Carrying out pseudo dual nucleic acid detection from sample to visual result in a polypropylene bag with CRISPR/Cas12a. Biosens. Bioelectron. 2021, 178, 113001. [Google Scholar] [CrossRef]
- Xiong, D.; Dai, W.; Gong, J.; Li, G.; Liu, N.; Wu, W.; Pan, J.; Chen, C.; Jiao, Y.; Deng, H.; et al. Rapid detection of SARS-CoV-2 with CRISPR-Cas12a. PLoS Biol. 2020, 18, e3000978. [Google Scholar] [CrossRef]
- Garcia-Venzor, A.; Rueda-Zarazua, B.; Marquez-Garcia, E.; Maldonado, V.; Moncada-Morales, A.; Olivera, H.; Lopez, I.; Zuñiga, J.; Melendez-Zajgla, J. SARS-CoV-2 direct detection without RNA isolation with loop-mediated isothermal amplification (LAMP) and CRISPR-Cas12. Front. Med. 2021, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Ooi, K.H.; Liu, M.M.; Tay, J.W.D.; Teo, S.Y.; Kaewsapsak, P.; Jin, S.; Lee, C.K.; Hou, J.; Maurer-Stroh, S.; Lin, W.; et al. An engineered CRISPR-Cas12a variant and DNA-RNA hybrid guides enable robust and rapid COVID-19 testing. Nat. Commun. 2021, 12, 1739. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Meng, Q.; Sun, B.; Zhao, B.; Dang, L.; Zhong, M.; Liu, S.; Xu, H.; Mei, H.; Liu, J.; et al. MeCas12a, a Highly Sensitive and Specific System for COVID-19 Detection. Adv. Sci. 2020, 7, 2001300. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Smith, B.M.; Jain, P.K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 2020, 11, 4906. [Google Scholar] [CrossRef] [PubMed]
- Crone, M.A.; Priestman, M.; Ciechonska, M.; Jensen, K.; Sharp, D.J.; Anand, A.; Randell, P.; Storch, M.; Freemont, P.S. A role for Biofoundries in rapid development and validation of automated SARS-CoV-2 clinical diagnostics. Nat. Commun. 2020, 11, 4464. [Google Scholar] [CrossRef]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Rauch, J.N.; Valois, E.; Solley, S.C.; Braig, F.; Lach, R.S.; Audouard, M.; Ponce-Rojas, J.C.; Costello, M.S.; Baxter, N.J.; Kosik, K.S.; et al. A scalable, easy-to-deploy, protocol for Cas13-based detection of SARS-CoV-2 genetic material. J. Clin. Microbiol. 2021, 59, e02402-20. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Chen, J.; Wang, M.; Zhang, T.; Luo, W.; Li, Y.; Wu, Y.; Zeng, B.; Zhang, K.; et al. Detection of SARS-CoV-2 and Its Mutated Variants via CRISPR-Cas13-Based Transcription Amplification. Anal. Chem. 2021, 93, 3393–3402. [Google Scholar] [CrossRef]
- Fozouni, P.; Son, S.; Derby, M.D.D.L.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2020, 184, 323–333.e9. [Google Scholar] [CrossRef]
- Brogan, D.J.; Chaverra-Rodriguez, D.; Lin, C.P.; Smidler, A.L.; Yang, T.; Alcantara, L.M.; Antoshechkin, I.; Liu, J.; Raban, R.R.; Belda-Ferre, P.; et al. A sensitive, rapid, and portable CasRx-based diagnostic assay for SARS-CoV-2. medRxiv 2020. [Google Scholar] [CrossRef]
- Li, H.; Dong, X.; Wang, Y.; Yang, L.; Cai, K.; Zhang, X.; Kou, Z.; He, L.; Sun, S.; Li, T.; et al. Sensitive and Easy-Read CRISPR Strip for COVID-19 Rapid Point-of-Care Testing. CRISPR J. 2021, 4, 392–399. [Google Scholar] [CrossRef]
- SHERLOCK Biosciences. Sherlock CRISPR SARS-CoV-2 kit. Available online: https://www.fda.gov/media/137746/download (accessed on 29 July 2021).
- Azhar, M.; Phutela, R.; Ansari, A.H.; Sinha, D.; Sharma, N.; Kumar, M.; Aich, M.; Sharma, S.; Rauthan, R.; Singhal, K.; et al. Rapid, field-deployable nucleobase detection and identification using FnCas9. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Osborn, M.; Bhardwaj, A.; Bingea, S.; Knipping, F.; Feser, C.; Lees, C.; Collins, D.; Steer, C.; Blazar, B.; Tolar, J. CRISPR/Cas9-Based Lateral Flow and Fluorescence Diagnostics. Bioengineering 2021, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Xiong, E.; Jiang, L.; Tian, T.; Hu, M.; Yue, H.; Huang, M.; Lin, W.; Jiang, Y.; Zhu, D.; Zhou, X. Simultaneous Dual-Gene Diagnosis of SARS-CoV-2 Based on CRISPR/Cas9-Mediated Lateral Flow Assay. Angew. Chem. Int. Ed. 2020, 60, 5307–5315. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Kwon, H.-J.; Yong, D.; Lee, I.-C.; Kim, H.; Kang, H.; Lim, E.-K.; Lee, K.-S.; Jung, J.; Park, H.G.; et al. Colorimetric Detection of SARS-CoV-2 and Drug-Resistant pH1N1 Using CRISPR/dCas9. ACS Sens. 2020, 5, 4017–4026. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration. In Vitro Diagnostics EUAs—Molecular Diagnostic Tests for SARS-CoV-2. Available online: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-sars-cov-2 (accessed on 28 July 2021).
- UCSF Health Clinical Laboratories. SARS-CoV-2 RNA DETECTR Assay. Available online: https://www.fda.gov/media/139937/download (accessed on 2 November 2020).
- Centers for Disease Control and Prevention. CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel (CDC). Available online: https://www.fda.gov/media/134922/download (accessed on 24 August 2020).
- Bin Moon, S.; Lee, J.M.; Kang, J.G.; Lee, N.-E.; Ha, D.-I.; Kim, D.Y.; Kim, S.H.; Yoo, K.; Kim, D.; Ko, J.-H.; et al. Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3′-overhang. Nat. Commun. 2018, 9, 3651. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Zhang, Y.; Pandolfi, P.P. Virus against virus: A potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020, 30, 189–190. [Google Scholar] [CrossRef] [Green Version]
- Abbott, T.R.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.S.; Debs, R.; et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell 2020, 181, 865–876.e12. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, J.; Wang, Q.; Wang, Y.; Kang, C. Rapid design and development of CRISPR-Cas13a targeting SARS-CoV-2 spike protein. Theranostics 2021, 11, 649–664. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Adriano, A.; Berhane, S.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; Beese, S.; et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020. [Google Scholar] [CrossRef]
- Falzone, L.; Musso, N.; Gattuso, G.; Bongiorno, D.; Palermo, C.I.; Scalia, G.; Libra, M.; Stefani, S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020, 46, 957–964. [Google Scholar] [CrossRef]
- Jakobsen, K.K.; Jensen, J.S.; Todsen, T.; Tolsgaard, M.G.; Kirkby, N.; Lippert, F.; Vangsted, A.M.; Martel, C.J.M.; Klokker, M.; von Buchwald, C. Accuracy and cost description of rapid antigen test compared with reverse transcriptase-polymerase chain reaction for SARS-CoV-2 detection. Dan Med. J. 2021, 68, A03210217. [Google Scholar] [PubMed]
- Uwamino, Y.; Wakui, M.; Aoki, W.; Kurafuji, T.; Yanagita, E.; Morita, M.; Nagata, M.; Inose, R.; Noguchi, M.; Yokota, H.; et al. Evaluation of the usability of various rapid antibody tests in the diagnostic application for COVID-19. Ann. Clin. Biochem. Int. J. Lab. Med. 2021, 58, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Javalkote, V.S.; Kancharla, N.; Bhadra, B.; Shukla, M.; Soni, B.; Sapre, A.; Goodin, M.; Bandyopadhyay, A.; Dasgupta, S. CRISPR-based assays for rapid detection of SARS-CoV-2. Methods 2020. [Google Scholar] [CrossRef] [PubMed]
- Rahman, R.; Hossain, A.; Mozibullah, M.; Al Mujib, F.; Afrose, A.; Mahmud, S.A.; Apu, A.I. CRISPR is a useful biological tool for detecting nucleic acid of SARS-CoV-2 in human clinical samples. Biomed. Pharmacother. 2021, 140, 111772. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ang, G.Y.; Yu, C.Y.; Yean, C.Y. Ambient temperature detection of PCR amplicons with a novel sequence-specific nucleic acid lateral flow biosensor. Biosens. Bioelectron. 2012, 38, 151–156. [Google Scholar] [CrossRef]
- PlexBio. IntelliPlex SARS-CoV-2 Detection Kit. Available online: https://www.fda.gov/media/139527/download (accessed on 2 November 2020).
- Applied BioCode. BioCode SARS-CoV-2 Assay. Available online: https://www.fda.gov/media/139049/download (accessed on 2 November 2020).
- Koujah, L.; Shukla, D.; Naqvi, A.R. CRISPR-Cas based targeting of host and viral genes as an antiviral strategy. Semin. Cell Dev. Biol. 2019, 96, 53–64. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Wu, Y.-H.; Yeh, I.-J.; Phan, N.N.; Yen, M.-C.; Hung, J.-H.; Chiao, C.-C.; Chen, C.-F.; Sun, Z.; Hsu, H.-P.; Wang, C.-Y.; et al. Gene signatures and potential therapeutic targets of Middle East respiratory syndrome coronavirus (MERS-CoV)-infected human lung adenocarcinoma epithelial cells. J. Microbiol. Immunol. Infect. 2021, 54, 845–857. [Google Scholar] [CrossRef]
- Liu, H.-L.; Yeh, I.-J.; Phan, N.N.; Wu, Y.-H.; Yen, M.-C.; Hung, J.-H.; Chiao, C.-C.; Chen, C.-F.; Sun, Z.; Jiang, J.-Z.; et al. Gene signatures of SARS-CoV/SARS-CoV-2-infected ferret lungs in short- and long-term models. Infect. Genet. Evol. 2020, 85, 104438. [Google Scholar] [CrossRef]
| CRISPR-Cas12a | CRISPR-Cas13a | CRISPR-Cas3 | CRISPR-Cas9 | |
|---|---|---|---|---|
| Class | 2 | 2 | 1 | 2 |
| Type | V | VI | I | II |
| Effector Cas protein complex | Single unit | Single unit | Multi-subunit | Single unit |
| Size (amino acid) | ~1200 (LbCas12a) | ~1200 (LwaCas13a) | ~900 (EcoCas3) | ~1400 (SpCas9) |
| Nuclease domain | RuvC | 2 HEPN domains | HD | RuvC, HNH |
| PAM/PFS | 5′ T-rich PAM | 3′ non-G PFS | Variable PAM (recognition by Cascade) | 3′ G-rich PAM |
| Pre-crRNA processing | Yes | Yes | Yes | No |
| tracrRNA | No | No | No | Yes |
| On target substrate (activator) | ssDNA, dsDNA | ssRNA | dsDNA | dsDNA (ssDNA and ssRNA with PAMmer) |
| Collateral cleavage activity | Yes | Yes | Yes | No |
| Off target substrate | ssDNA | ssRNA | ssDNA | NA |
| Cas Protein | Assay Name | RNA Extraction | Assay Component | Time Required a | Target Gene(s) | Result Interpretation | LoD | PPA (n) b | NPA (n) b | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Cas12-based CRISPR-Dx | ||||||||||
| LbCas12a | SENA | Yes | rRT-PCR (~64 min); Cas12 (48 °C, 10 min) | ~74 min | N, Orf1ab | FR (real-time thermocycler) | 1.6 copies/reaction | 92% (24 d) | 98% (240 e) | [41] |
| LbCas12a | COVID-19 CRISPR-FDS | Yes | RT-PCR (~38 min)/RT-RPA (42 °C, 20 min); Cas12 (37 °C, 20 min) | 40–60 min | N, Orf1ab | FR (plate reader) | 2 copies/reaction | 100% (15) | 71.4% (14) | [40] |
| LbCas12a | SARS-CoV-2 RNA DETECTR | Yes | RT-LAMP (62 °C, 30 min); Cas12 (37 °C, 15 min) | 45 min | N | FR (real-time thermocycler) | 20 copies/µL | 95% (40) | 100% (62) | [47] |
| LbCas12a | DETECTR | Yes | RT-LAMP (62 °C, 20–30 min); Cas12 (37 °C, 10 min) | 30–40 min | E, N2 | FR (plate reader), LFD | 10 copies/µL | 95% (36) | 100% (42) | [14] |
| LbCas12a | DETECTR | Yes | RT-LAMP (62 °C, 20 min); Cas12 (37 °C, 10 min) | 30 min | N | FR (plate reader), LFD | 50 copies (plasmid) | 93% (155) | 96% (223) | [35] |
| LbCas12a | - | Yes | RT-RPA (42 °C, 30 min); Cas12 (37 °C, up to 90 min) | 60–120 min | Orf1ab | FR (plate reader), LFD | 10 copies/µL | - | - | [48] † |
| LbCas12a | CRISPR-Cas12a-NER | Yes | RT-RAA (39 °C, 30 min); Cas12 (37 °C, 15 min) | 45 min | E | Visual (under blue light) | 10 copies | 100% (16) | 100% (15) | [49] |
| LbCas12a | - | Yes | RT-RPA (39 °C, 30 min; 75 °C, 5 min); Cas12 (39 °C, 15 min) | 50 min | S | Visual (under blue light) | 10 copies/reaction | 96% (53) | 100% (111) | [50] |
| LbCas12a | - | Yes | RT-RPA (39 °C, 30 min; 75 °C, 5 min); Cas12 (39 °C, 30 min) | 65 min | N | Cloud-based analysis (smartphone-based FR device) | 6.25 copies/µL | 87% (52) | 92% (63) | [36] |
| LbCas12a | Two-pot iSCAN | Yes | RT-LAMP (62 °C, 30 min); Cas12 (37 °C, 10 min) | 40 min | E, N | Visual (under UV light), LFD | 10 copies/reaction | 86% (N, 21); 38% (E, 21) | 100% (N, 3); 100% (E, 3) | [51] |
| LbCas12a | AIOD-CRISPR | Yes | RT-RPA and Cas12 (37 °C, 40 min) | 40 min | N | Visual (under blue/UV light) | ̴ 5 copies | 100% (8) | 100% (20) | [52] |
| LbCas12a | - | Yes | RT-LAMP (65 °C, 40 min); Cas12 (37 °C, 5 min) | 45 min | Orf | Visual (smart phone-based FR device) | 20 copies/reaction | 100% (7) | 100% (3) | [53] |
| LbCas12a | opvCRISPR | Yes | RT-LAMP (65 °C, 40 min); Cas12 (37 °C, 5 min) | 45 min | S | Visual (under blue light) | 5 copies | 100% (26) | 100% (24) | [54] |
| LbCas12a | - | Yes | RT-LAMP (62 °C, 30 min); Cas12a (r.t., 10 min) | 40 min | E, N | Visual (under UV light) | 30 copies/µL (N); 45 copies/µL (E) | 94% (50) | 100% (50) | [55] |
| LbCas12a | OR-DETECTR | Yes | RT-RPA (42 °C, 30 min); Cas12 (42 °C, 20 min) | 50 min | N, RdRp | FR (plate reader), LFD | Synthetic RNA: 10 copies/µL (RdRp), 2.5 copies/µL (N); pseudo-virus: 20 copies/µL (RdRp), 1 copy/µL (N) | 100% (6) | 100% (8) | [56] |
| AapCas12b | CASdetec | Optional (lysis: 50 °C, 5 min; 64 °C, 5 min) | RT-RAA (42 °C, 30 min); Cas12 (42 °C, 30 min) | 60 min | RdRp | FR (real-time thermocycler), visual (under blue LED) | FR: 1 × 104 copies/mL; visual: 5 × 104 copies/mL (pseudo-virus) | - | - | [57] |
| AapCas12b | STOPCovid. v2 | Yes (lysis and magnetic bead-based purification: r.t., 10 min) | RT-LAMP and Cas12 (60 °C, 45 min for FR/80 min for LFD) | 45–80 min | N | FR (real-time thermocycler), LFD | FR: 33 copies/mL; LFD: 83 copies/ml | 93% (202) | 99% (200) | [37] |
| AapCas12b | One-pot iSCAN | Yes | RT-LAMP (62 °C, 30 min); Cas12 (62 °C, 15 min) | 60 min | N | Visual (under UV light), LFD | 10 copies/reaction | 86% (N, 21) | 100% (N, 3) | [51] |
| LbCas12a | ITP-CRISPR | Yes (Off-CHIP, 95 °C, 2 min; on-chip, 3 min) | RT-LAMP (off-chip, 62 °C, 30 min); Cas12a (on-chip, 5 min) | 35 min | E, N | Fluorescent microscopy | 10 copies/µL | 94% (32) | 100% (32) | [58] |
| LbCas12a | deCOViD | Optional (heat inactivation: 65 °C, 30 min) | RT-RPA and Cas12a (42 °C, 30–60 min) | 30–60 min | N | Fluorescent microscopy | Synthetic RNA: 1 GE/µL; heat-inactivated virus: 20 GE/µL | 100% (2) | 100% (2) | [59] |
| LbCas12a | CRISPR-FDS | No (lysis: 37 °C, 5 min) | RT-RPA & Cas12 (r.t., 10 min) | 15 min | Orf1ab | Smartphone-based fluorescent microscopy | 0.38 copies/µL | - | - | [42] |
| LbCas12a | - | No (lysis: 65 °C, 10 min) | RT-LAMP (65 °C, 30 min); Cas12a (37 °C, 10–20 min) | 40–50 min | Orf | Visual (under UV light) | 20 copies/reaction (pseudo-virus) | 100% (4) | 100% (4) | [60] |
| LbCas12a | - | No (lysis: 80 °C, 5 min) | RT-RPA (42 °C, 15–20 min); Cas12 (37 °C, 15-20 min) | 30–40 min | N, Orf1ab | Visual (under UV light), LFD | Visual: 1 copy/reaction LFD: 1 copy/µL | 100% (11) | 100% (11) | [61] |
| LbCas12a | - | No (lysis: 42 °C, 20 min; 64 °C, 5 min) | RT-LAMP (65 °C, 30 min); Cas12 (37 °C, 10 min) | 40 min | N | FR (real-time thermocycler), Visual (under blue light) | 16 copies/µL | 100% (6) | 100% (6) | [62] |
| enAsCas12a | VaNGuard (quasi-one-pot) | Optional (proteinase K and heat treatment: 95 °C, 5 min) | RT-LAMP (60 °C/63 °C, 22 min); Cas12 (60 °C, 5 min) | 27 min | S | LFD | RNA extract: 2 copies/µL; proteinase K and heat treatment: 40 copies/µL | RNA extract: 84% (51); proteinase K and heat treatment: 76% (21) | RNA extract: 100% (36); proteinase K and heat treatment: 100% (21) | [63] |
| LbCas12a | MeCas12a | Yes | RT-RAA (39 °C, 30 min); desalting (~3 min); Cas12 (37 °C, 15 min) | 45 min | E | Visual (under blue light) | 5 copies | 100% (13) | 100% (11) | [64] |
| LbCas12a | CRISPR-ENHANCE | Yes | RT-LAMP (63 °C, 20–30 min); Cas12 (37°, 20 min) | 40–50 min | N | LFD | 3–300 copies | - | - | [65] |
| Cas13-based CRISPR-Dx | ||||||||||
| LwaCas13a | SHERLOCK | Yes | RT-RPA (42 °C, 25 min); T7 transcription and Cas13 (37 °C, 30–60 min for FR/30 min for LFD) | 55–85 min | S | FR (plate reader/real-time thermocycler), LFD | 42 copies/reaction | FR: 96% LFD: 8% (81) | FR: 100% LFD: 88% (73) | [38] |
| Cas13a | CRISPR-COVID | Yes | RT-RPA (42 °C, 30 min); T7 transcription and Cas13 (42 °C, 10 min) | 40 min | Orf1ab | FR | 7.5 copies/reaction (plasmid) | 100% ** (52) | 100% ** (62) | [39] |
| LwaCas13a | - | Yes | RT-PCR (~40 min)/RT-RPA (42 °C, 30 min); T7 transcription and Cas13 (37 °C, 120 min) | 150–160 min | N (RT-PCR); Orf1ab (RT-RPA) | FR (plate reader) | N: ~2.5 copies/reaction Orf1ab: ~200 copies/reaction (Virus-like particle) | - | - | [66] |
| LwaCas13a | CARMEN | Yes | Complex workflow | (SAMPLE-to-result: ~6.5 h) | - | Fluorescent microscopy | - | - | - | [67] |
| LwaCas13a | CREST | Optional (PEARL: 25 min) | RT (42 °C, 30 min); PCR (22 min); Cas13 (37 °C, 5–30 min) | 57–82 min | N1, N2, N3 | Visual (under blue light) | 10 copies/µL | 97% (65) | 98% (153) | [68] |
| LwaCas13a | - | Yes | Ligation (37 °C, 30 min); Transcription amplification (37 °C, duration not specified); Cas13 (37 °C, 20 min) | - | E, N | FR | 82 copies (pseudo-virus) | 100% (5) | 100% (1) | [69] |
| LbuCas13a | - | Yes | Cas13 (37 °C, 30 min) | 30 min | E, N | Smartphone-based fluorescent microscopy | ~100 copies/µL | 100% (5) | - | [70] |
| RfxCas13d (CasRx) | SENSR | Yes | RT-RPA (42 °C, 45 min); T7 transcription and Cas13 (37 °C, 90 min for FR/60 min for LFD) | 105–135 min | E, N | FR (real-time thermocycler), LFD | ~100 copies/µL | 57% (21) | 100% (21) | [71] † |
| LwaCas13a | SHINE | No (HUDSON: 40 °C, 5 min; 70 °C (95 °C for saliva), 5 min) | RT-RPA, T7 transcription and Cas13 (37 °C, 40 min) | 40 min | Orf1a | FR (under blue light with a smartphone application), LFD | FR: 10 copies/µL LFD: 100 copies/µL | 90% (30) | 100% (20) | [43] |
| LwaCas13a | ERASE | Yes | RT-RAA (42 °C, 30 min); T7 transcription and Cas13 (37 °C, 30 min) | 60 min | N | LFD | 1 copy/µL | 91% (286) | 99% (379) | [72] |
| LwaCas13a | Sherlock CRISPR SARS-CoV-2 kit | Yes | RT-LAMP (61 °C, 40 min); T7 transcription and Cas13 (37 °C, 10 min) | 50 min | N, Orf1ab | FR (plate reader) | 6.75 copies/µL (Orf1ab); 1.35 copies/µL (N) | 100% (30) | 100% (30) | [73] |
| Cas9-based CRISPR-Dx | ||||||||||
| dFnCas9 | FELUDA | Yes | RT-PCR (42 min); dCas9 (37 °C, 10 min) | 52 min | N, S | LFD | 10 copies | 100% (21) | 97% (60) | [74] † |
| dSpCas9 | - | - | RPA (r.t., 20 min); dCas9 (37 °C, 60 min) | 80 min | Orf8a | LFD | 50 copies (synthetic DNA) | - | - | [75] |
| SpCas9 | - | - | Hybridization (95 °C, 5 min; cooling to r.t. at -0.1 °C/s); Cas9 (37 °C, 60 min) | ~77 min | Orf8a | FR (real-time thermocycler) | 5 copies (synthetic DNA) | - | - | [75] |
| dSpCas9 | - | Yes | Multiplex RT-RPA (37 °C, 30 min); dCas9 (37 °C, 5 min) | 35 min | E, Orf1ab | LFD | 100 copies/reaction | 97% (35) | 100% (29) | [76] |
| dSpCas9 | - | No (Lysis: 50 °C, 5 min; 64 °C, 5 min) | Complex workflow | (Sample-to-result: 90 min) | N1, N2, N3 | Colorimetry (plate reader) | 10 PFU/ml | 100% (5) | 100% (3) | [77] |
| Cas3-based CRISPR-Dx | ||||||||||
| EcoCas3 | CONAN | Yes | RT-LAMP (62 °C, 30 min); CONAN (37 °C, 10 min) | 40 min | N1, N2 | LFD | 100 copies | - | - | [31] † |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan, K.G.; Ang, G.Y.; Yu, C.Y.; Yean, C.Y. Harnessing CRISPR-Cas to Combat COVID-19: From Diagnostics to Therapeutics. Life 2021, 11, 1210. https://doi.org/10.3390/life11111210
Chan KG, Ang GY, Yu CY, Yean CY. Harnessing CRISPR-Cas to Combat COVID-19: From Diagnostics to Therapeutics. Life. 2021; 11(11):1210. https://doi.org/10.3390/life11111210
Chicago/Turabian StyleChan, Kok Gan, Geik Yong Ang, Choo Yee Yu, and Chan Yean Yean. 2021. "Harnessing CRISPR-Cas to Combat COVID-19: From Diagnostics to Therapeutics" Life 11, no. 11: 1210. https://doi.org/10.3390/life11111210
APA StyleChan, K. G., Ang, G. Y., Yu, C. Y., & Yean, C. Y. (2021). Harnessing CRISPR-Cas to Combat COVID-19: From Diagnostics to Therapeutics. Life, 11(11), 1210. https://doi.org/10.3390/life11111210








