Stochastic Effects in Retrotransposon Dynamics Revealed by Modeling under Competition for Cellular Resources
Abstract
:1. Introduction
2. Methods
2.1. Basic Predator–Prey Model of Transposon Dynamics
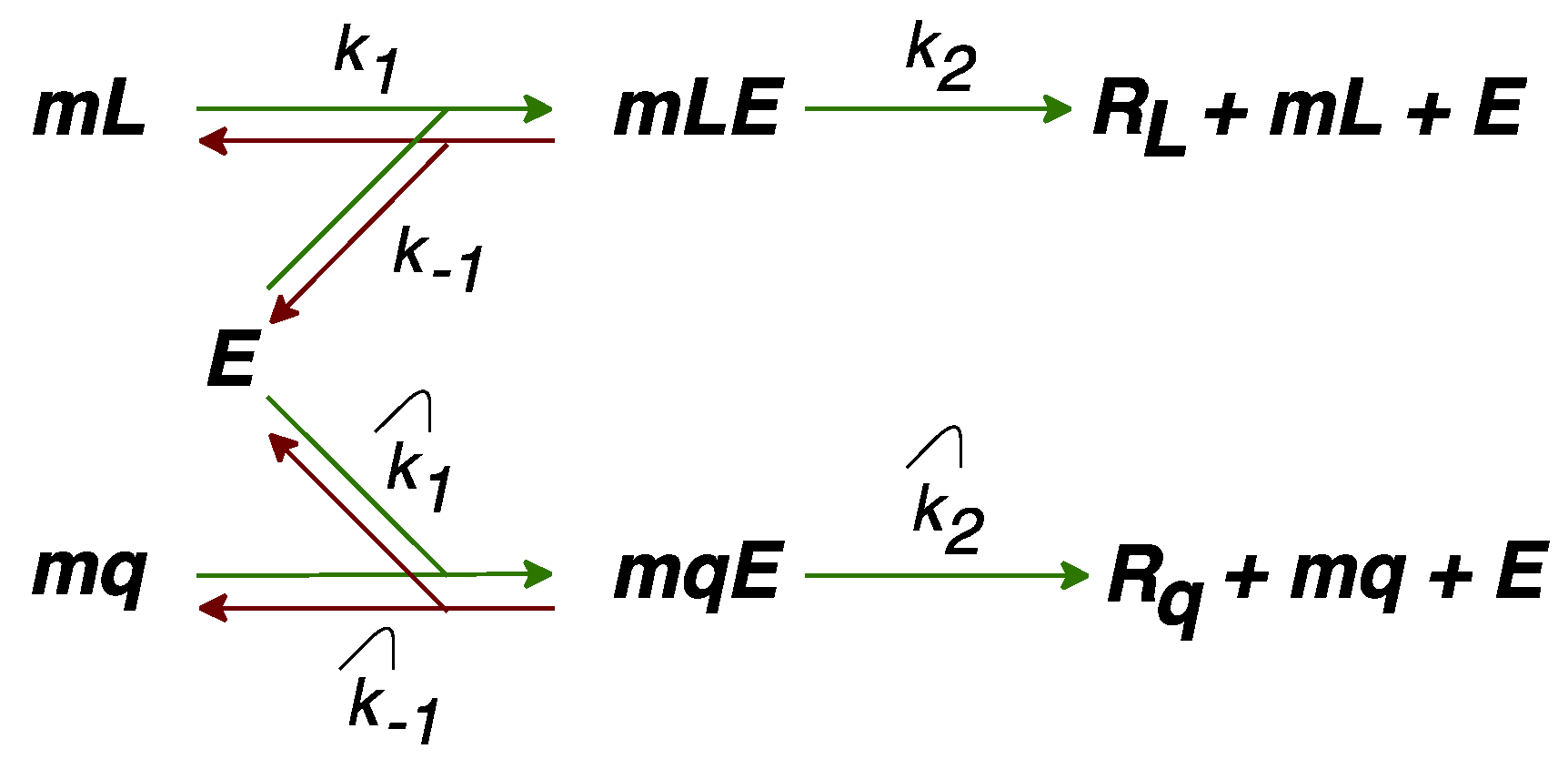
2.2. Modification of the Model to Include Competition for Ribosomes
2.3. Modification of the Model to Include Competition for Energy
2.4. Parameter Values
2.5. Stability Analysis
2.6. Stochastic Simulation in Cell Population
3. Results
3.1. Model of Transposon Dynamics Under Competition for Free Ribosomes
3.2. Stability Analysis Predicts Two Different Attractors for L1
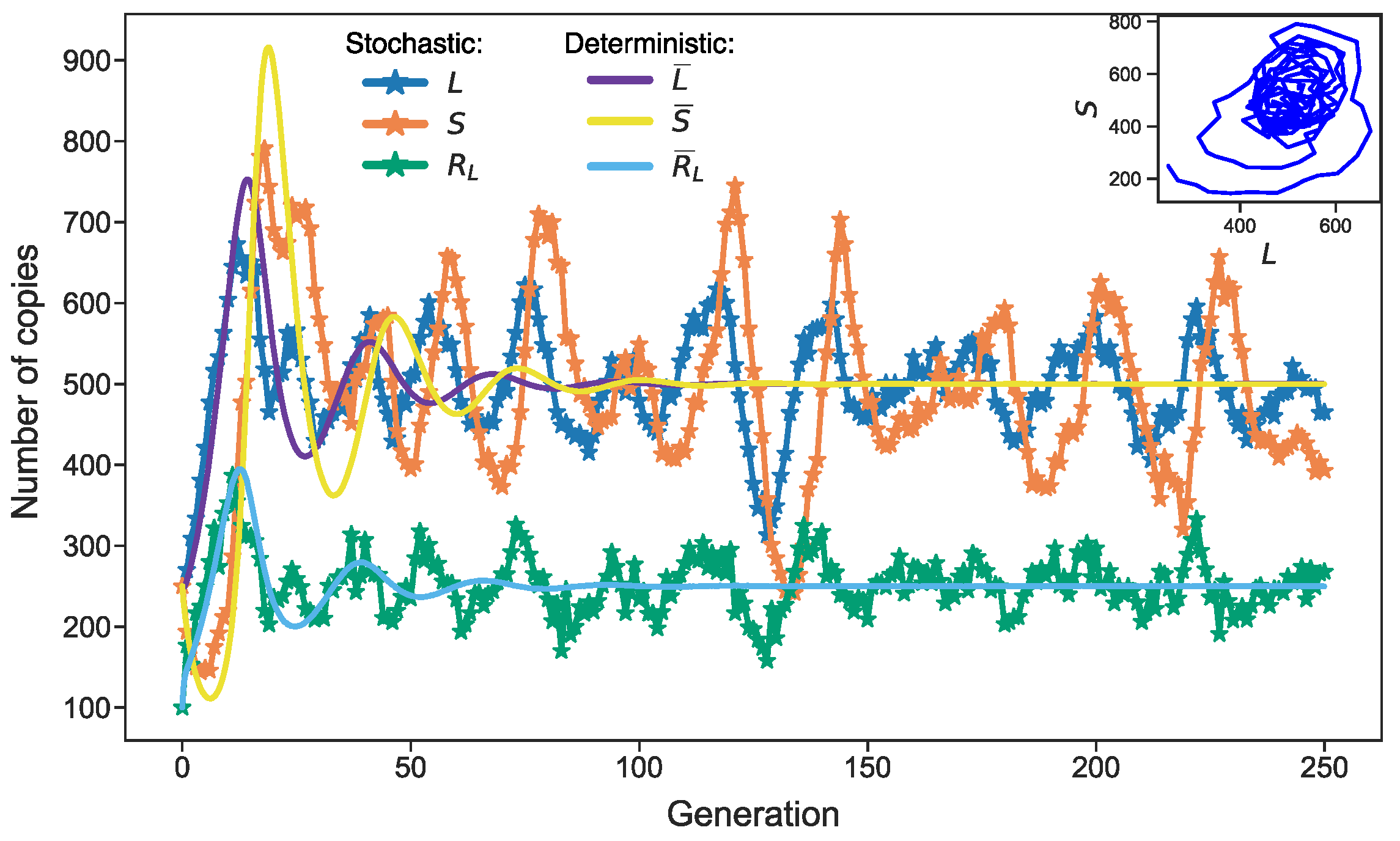
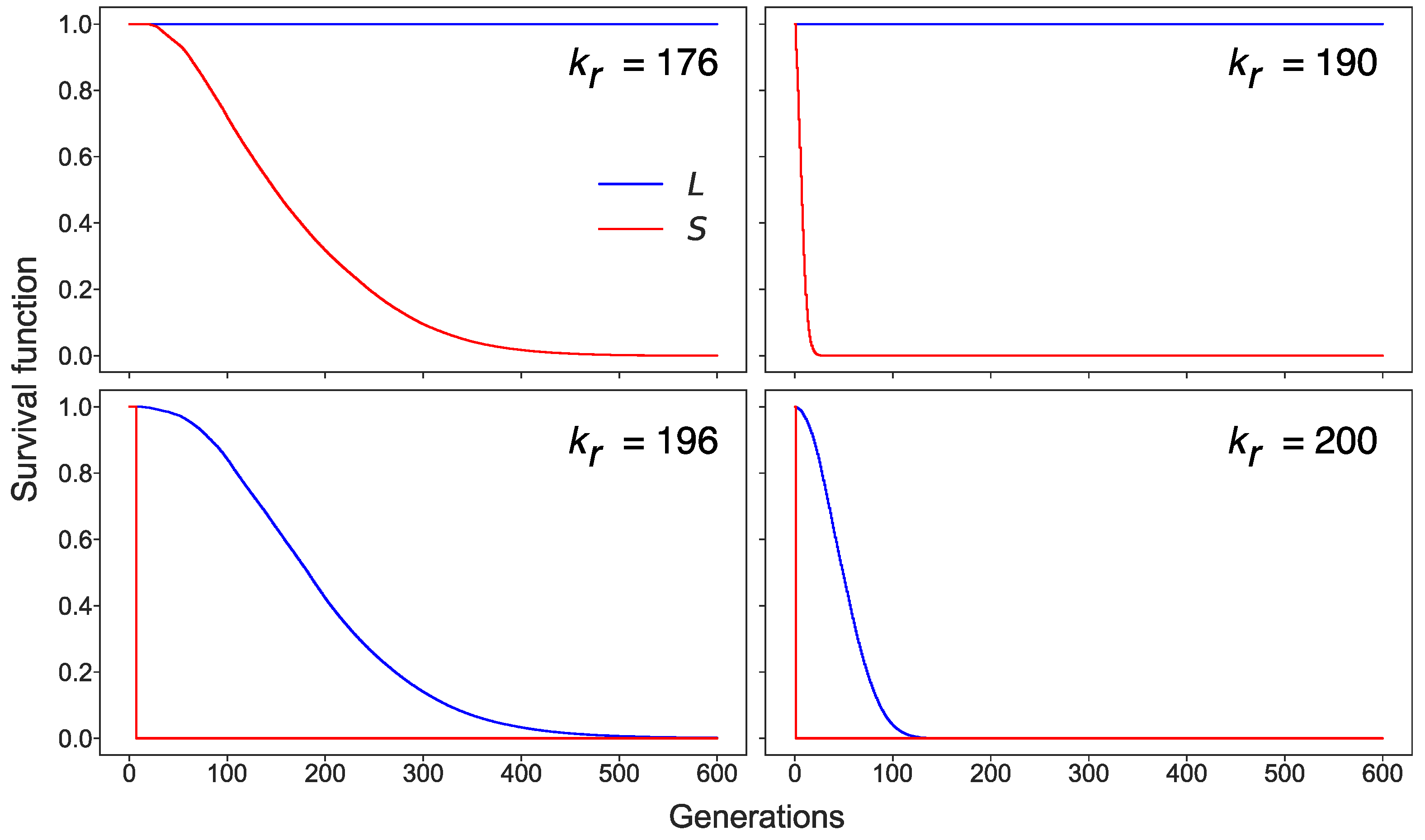
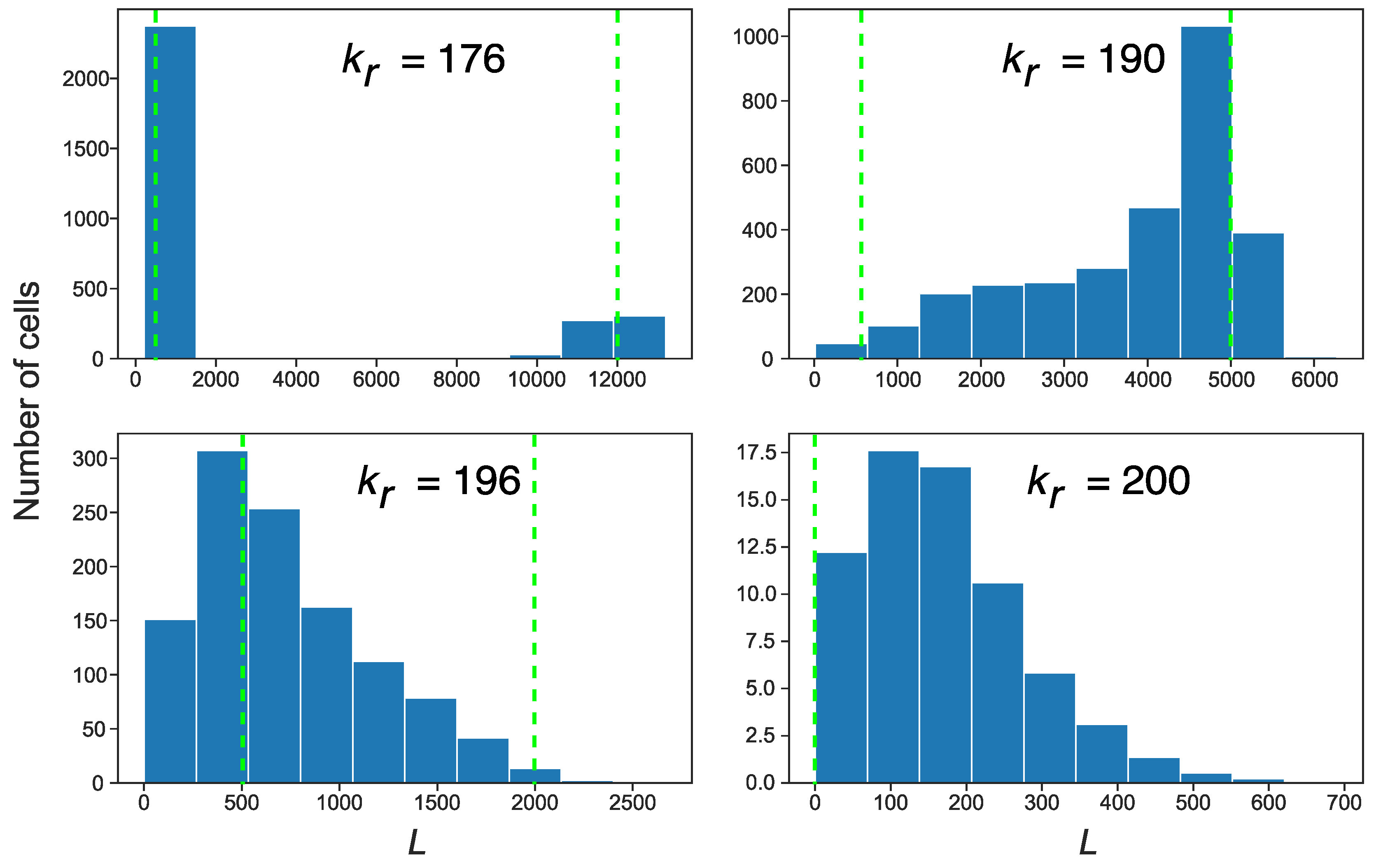
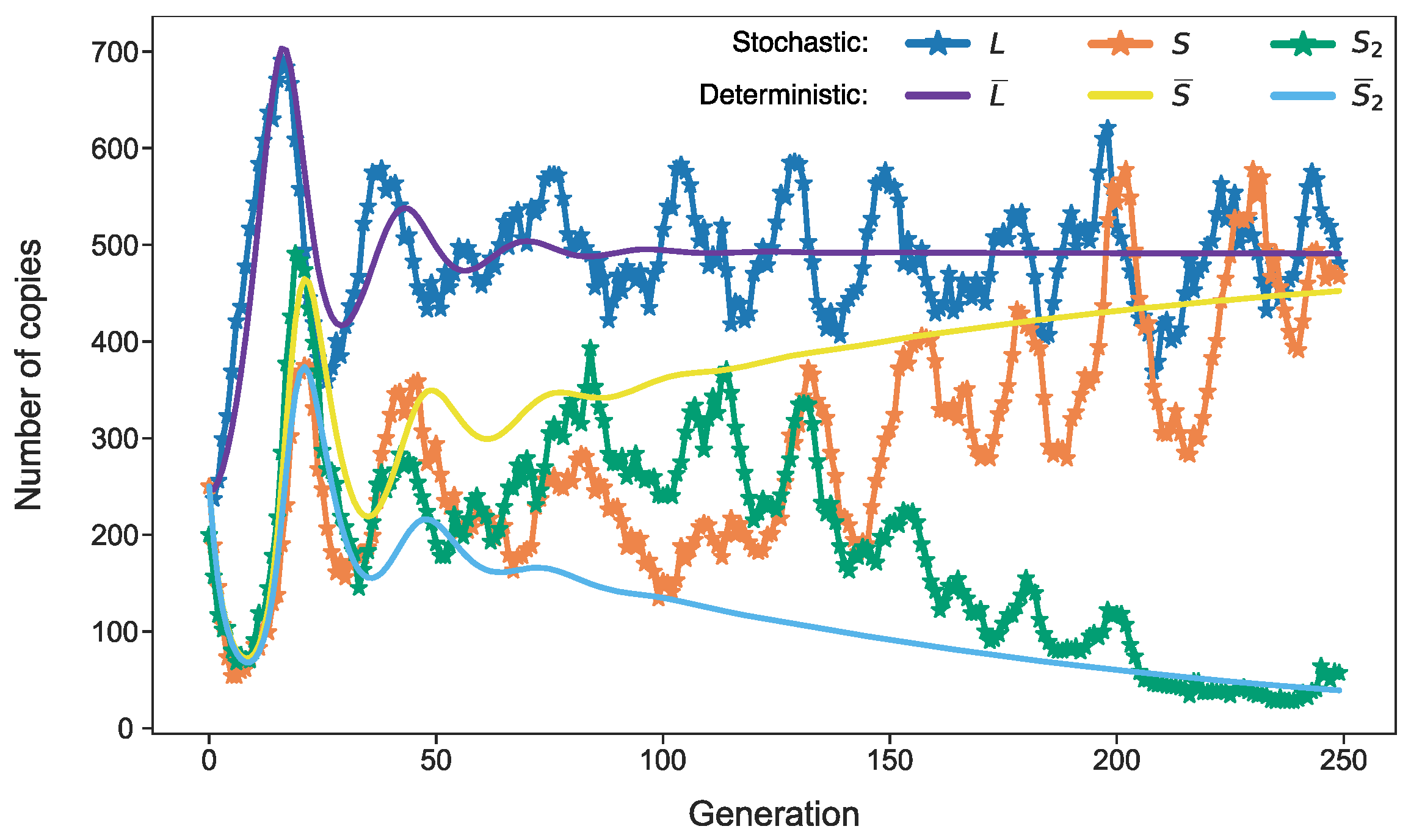
3.3. Stochastic Effects in Transposon Dynamics Divide Cell Population into Pools of Cells with Essentially Different Numbers of L1
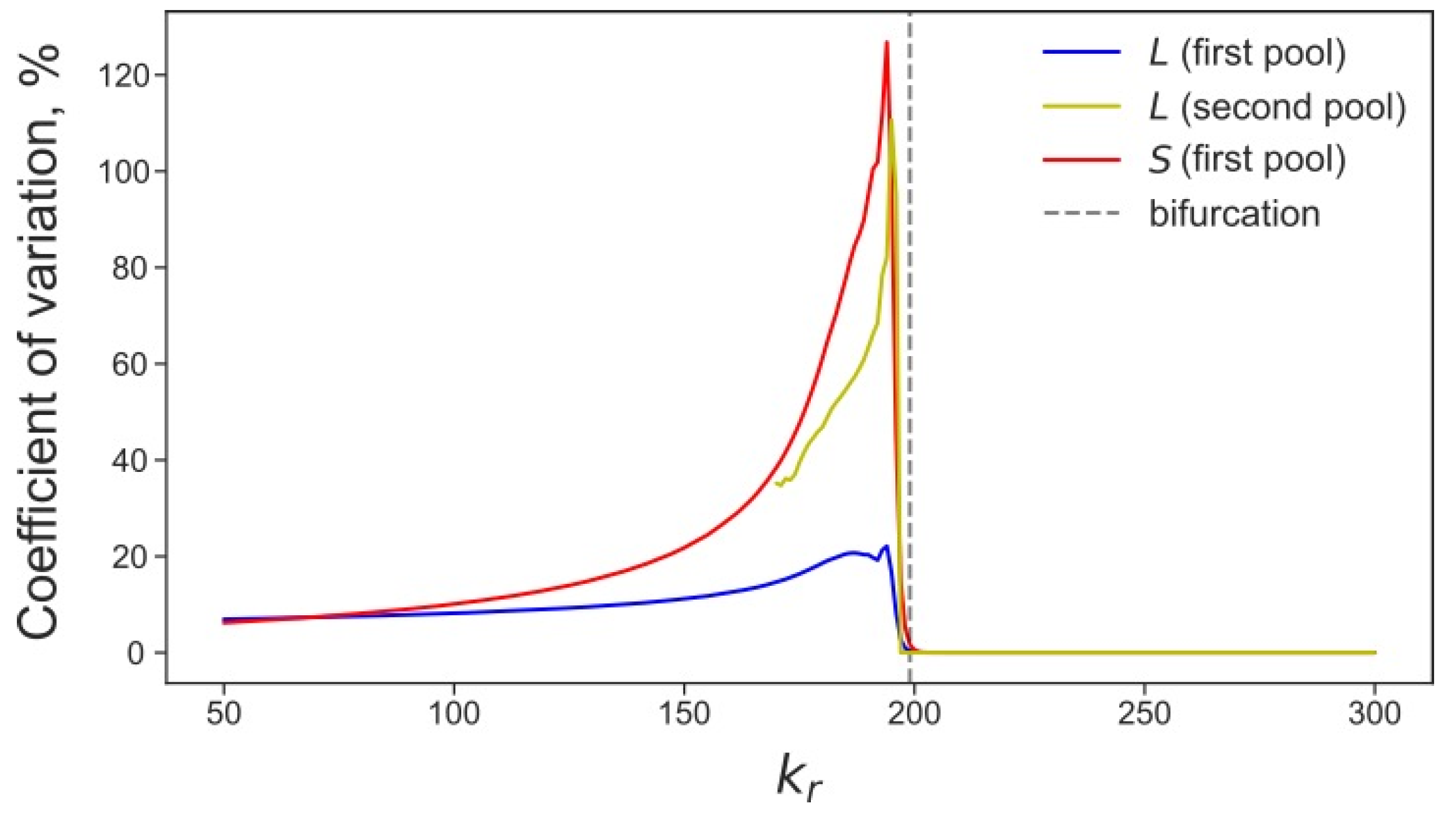
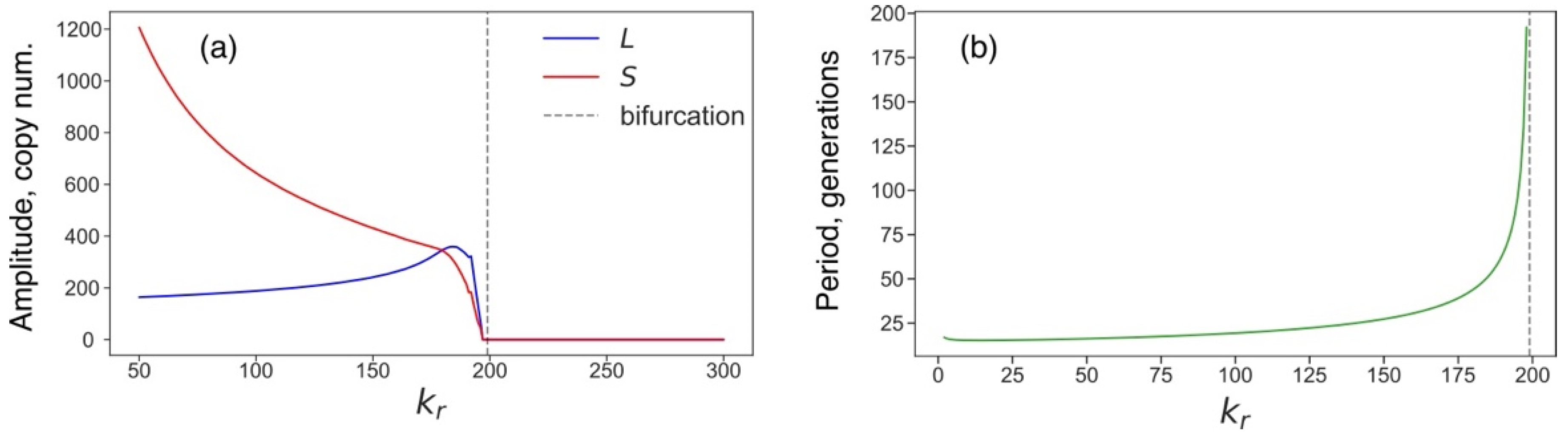
3.4. Rising Competition for Ribosomes Affects Noise Levels and Oscillation Parameters of L and S
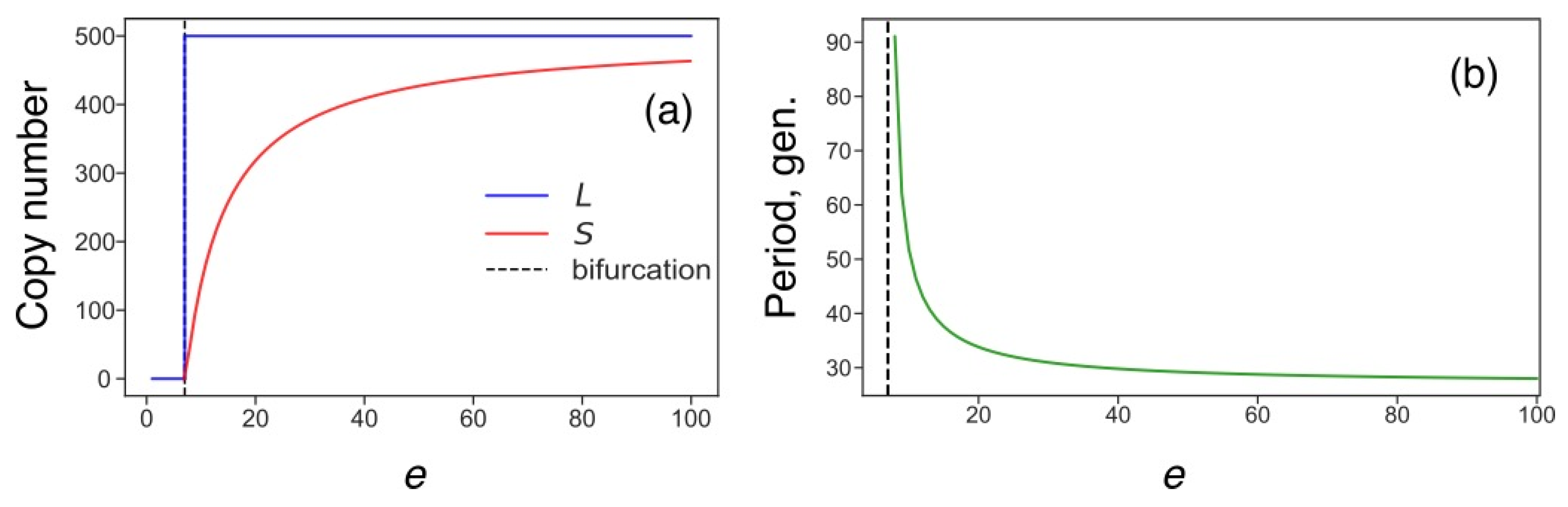
3.5. Transposon Dynamics under Competition for Energy
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kazazian, H.H. Mobile Elements: Drivers of Genome Evolution. Science 2004, 303, 1626–1632. [Google Scholar] [CrossRef] [Green Version]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suh, A. Genome Size Evolution: Small Transposons with Large Consequences. Curr. Biol. 2019, 29, R241–R243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pace, J.K.; Feschotte, C. The evolutionary history of human DNA transposons: Evidence for intense activity in the primate lineage. Genome Res. 2007, 17, 422–432. [Google Scholar] [CrossRef] [Green Version]
- Hancks, D.C.; Kazazian, H.H. Roles for retrotransposon insertions in human disease. Mob. DNA 2016, 7, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, K.H. Transposable elements in cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef]
- Feschotte, C.; Pritham, E.J. DNA Transposons and the Evolution of Eukaryotic Genomes. Annu. Rev. Genet. 2007, 41, 331–368. [Google Scholar] [CrossRef] [Green Version]
- Goodier, J.L. Restricting retrotransposons: A review. Mob. DNA 2016, 7, 16. [Google Scholar] [CrossRef] [Green Version]
- Mills, R.E.; Bennett, E.A.; Iskow, R.C.; Devine, S.E. Which transposable elements are active in the human genome? Trends Genet. 2007, 23, 183–191. [Google Scholar] [CrossRef]
- Beck, C.R.; Garcia-Perez, J.L.; Badge, R.M.; Moran, J.V. LINE-1 Elements in Structural Variation and Disease. Annu. Rev. Genom. Hum. Genet. 2011, 12, 187–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shedlock, A.M.; Okada, N. SINE insertions: Powerful tools for molecular systematics. Bioessays 2000, 22, 148–160. [Google Scholar] [CrossRef]
- Kramerov, D.A.; Vassetzky, N.S. SINEs. Wiley Interdiscip. Rev. Rna 2011, 2, 772–786. [Google Scholar] [CrossRef]
- Kumar, A. Jump around: Transposons in and out of the laboratory. F1000research 2020, 9, 135. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B.; Charlesworth, D. The population dynamics of transposable elements. Genet. Res. 1983, 42, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Rouzic, A.L.; Deceliere, G. Models of the population genetics of transposable elements. Genet. Res. 2005, 85, 171–181. [Google Scholar] [CrossRef]
- Iranzo, J.; Gómez, M.J.; de Saro, F.J.L.; Manrubia, S. Large-Scale Genomic Analysis Suggests a Neutral Punctuated Dynamics of Transposable Elements in Bacterial Genomes. PLoS Comput. Biol. 2014, 10, e1003680. [Google Scholar] [CrossRef] [Green Version]
- Szitenberg, A.; Cha, S.; Opperman, C.H.; Bird, D.M.; Blaxter, M.L.; Lunt, D.H. Genetic Drift, Not Life History or RNAi, Determine Long-Term Evolution of Transposable Elements. Genome Biol. Evol. 2016, 8, 2964–2978. [Google Scholar] [CrossRef] [Green Version]
- Rouzic, A.L.; Capy, P. Population Genetics Models of Competition Between Transposable Element Subfamilies. Genetics 2006, 174, 785–793. [Google Scholar] [CrossRef] [Green Version]
- Banuelos, M.; Sindi, S. Modeling transposable element dynamics with fragmentation equations. Math. Biosci. 2018, 302, 46–66. [Google Scholar] [CrossRef]
- Rouzic, A.L.; Dupas, S.; Capy, P. Genome ecosystem and transposable elements species. Gene 2007, 390, 214–220. [Google Scholar] [CrossRef]
- Rouzic, A.L.; Boutin, T.S.; Capy, P. Long-term evolution of transposable elements. Proc. Natl. Acad. Sci. USA 2007, 104, 19375–19380. [Google Scholar] [CrossRef] [Green Version]
- Xue, C.; Goldenfeld, N. Stochastic Predator-Prey Dynamics of Transposons in the Human Genome. Phys. Rev. Lett. 2016, 117, 208101. [Google Scholar] [CrossRef] [Green Version]
- Lane, N.; Martin, W. The energetics of genome complexity. Nature 2010, 467, 929–934. [Google Scholar] [CrossRef]
- Lynch, M.; Marinov, G.K. The bioenergetic costs of a gene. Proc. Natl. Acad. Sci. USA 2015, 112, 15690–15695. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, D.T. Stochastic Simulation of Chemical Kinetics. Annu. Rev. Phys. Chem. 2007, 58, 35–55. [Google Scholar] [CrossRef] [PubMed]
- Milo, R.; Jorgensen, P.; Moran, U.; Weber, G.; Springer, M. BioNumbers—the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010, 38, D750–D753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, A.F.; Schmeckpeper, B.J.; Abdelrazik, M.; Comey, C.T.; O’Hara, B.; Rossiter, J.P.; Cooley, T.; Heath, P.; Smith, K.D.; Margolet, L. Origin of the human L1 elements: Proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1987, 1, 113–125. [Google Scholar] [CrossRef]
- Weiße, A.Y.; Oyarzún, D.A.; Danos, V.; Swain, P.S. Mechanistic links between cellular trade-offs, gene expression, and growth. Proc. Natl. Acad. Sci. USA 2015, 112, E1038–E1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reardon, J.E. Human immunodeficiency virus reverse transcriptase: Steady-state and pre-steady-state kinetics of nucleotide incorporation. Biochemistry 1992, 31, 4473–4479. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, E.L.; Meier, P. Nonparametric Estimation from Incomplete Observations. J. Am. Stat. Assoc. 2012, 53, 457–481. [Google Scholar] [CrossRef]
- Wagner, A. Energy Constraints on the Evolution of Gene Expression. Mol. Biol. Evol. 2005, 22, 1365–1374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasperski, A.; Kasperska, R. Bioenergetics of life, disease and death phenomena. Theory Biosci. 2018, 137, 155–168. [Google Scholar] [CrossRef] [Green Version]
- Lieberthal, W.; Menza, S.A.; Levine, J.S. Graded ATP depletion can cause necrosis or apoptosis of cultured mouse proximal tubular cells. Am. J. Physiol. 1998, 274, F315–F327. [Google Scholar] [CrossRef]
- Eguchi, Y.; Shimizu, S.; Tsujimoto, Y. Intracellular ATP levels determine cell death fate by apoptosis or necrosis. Cancer Res. 1997, 57, 1835–1840. [Google Scholar]
- Skulachev, V.P. Bioenergetic aspects of apoptosis, necrosis and mitoptosis. Apoptosis 2006, 11, 473–485. [Google Scholar] [CrossRef]
- Raveh, A.; Margaliot, M.; Sontag, E.D.; Tuller, T. A model for competition for ribosomes in the cell. J. R. Soc. Interface 2016, 13, 20151062. [Google Scholar] [CrossRef]
- Gyorgy, A.; Jiménez, J.I.; Yazbek, J.; Huang, H.-H.; Chung, H.; Weiss, R.; Del Vecchio, D. Isocost Lines Describe the Cellular Economy of Genetic Circuits. Biophys. J. 2015, 109, 639–646. [Google Scholar] [CrossRef] [Green Version]
- Rogalla, P.S.; Rudge, T.J.; Ciandrini, L. An equilibrium model for ribosome competition. Phys. Biol. 2019, 17, 015002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gale, M.; Tan, S.-L.; Katze, M.G. Translational Control of Viral Gene Expression in Eukaryotes. Microbiol. Mol. Biol. R. 2000, 64, 239–280. [Google Scholar] [CrossRef] [Green Version]
- Stern-Ginossar, N.; Thompson, S.R.; Mathews, M.B.; Mohr, I. Translational Control in Virus-Infected Cells. Cold Spring Harbor Perspect. Biol. 2019, 11, a033001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapointe, C.P.; Grosely, R.; Johnson, A.G.; Wang, J.; Fernández, I.S.; Puglisi, J.D. Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proc. Natl. Acad. Sci. USA 2021, 118, e2017715118. [Google Scholar] [CrossRef] [PubMed]
- Maupetit-Mehouas, S.; Vaury, C. Transposon Reactivation in the Germline May Be Useful for Both Transposons and Their Host Genomes. Cells 2020, 9, 1172. [Google Scholar] [CrossRef]
- Raiz, J.; Damert, A.; Chira, S.; Held, U.; Klawitter, S.; Hamdorf, M.; Löwer, J.; Strätling, W.H.; Löwer, R.; Schumann, G.G. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2012, 40, 1666–1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esnault, C.; Maestre, J.; Heidmann, T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000, 24, 363–367. [Google Scholar] [CrossRef]
- Kazazian, H.H. Processed pseudogene insertions in somatic cells. Mob. DNA 2014, 5, 20. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlov, S.; Gursky, V.V.; Samsonova, M.; Kanapin, A.; Samsonova, A. Stochastic Effects in Retrotransposon Dynamics Revealed by Modeling under Competition for Cellular Resources. Life 2021, 11, 1209. https://doi.org/10.3390/life11111209
Pavlov S, Gursky VV, Samsonova M, Kanapin A, Samsonova A. Stochastic Effects in Retrotransposon Dynamics Revealed by Modeling under Competition for Cellular Resources. Life. 2021; 11(11):1209. https://doi.org/10.3390/life11111209
Chicago/Turabian StylePavlov, Sergey, Vitaly V. Gursky, Maria Samsonova, Alexander Kanapin, and Anastasia Samsonova. 2021. "Stochastic Effects in Retrotransposon Dynamics Revealed by Modeling under Competition for Cellular Resources" Life 11, no. 11: 1209. https://doi.org/10.3390/life11111209
APA StylePavlov, S., Gursky, V. V., Samsonova, M., Kanapin, A., & Samsonova, A. (2021). Stochastic Effects in Retrotransposon Dynamics Revealed by Modeling under Competition for Cellular Resources. Life, 11(11), 1209. https://doi.org/10.3390/life11111209






