A Framework for Stochastic Optimization of Parameters for Integrative Modeling of Macromolecular Assemblies
Abstract
:1. Introduction
2. Methods and Materials
2.1. Theory and the Algorithm
2.1.1. Example
2.1.2. Notation
2.1.3. Partitioning into Groups
2.1.4. Basic Optimization Strategies: Manual, Binary, m-ary
2.1.4.1. Manual Search
2.1.4.2. Binary Search
2.1.4.3. m-ary Search
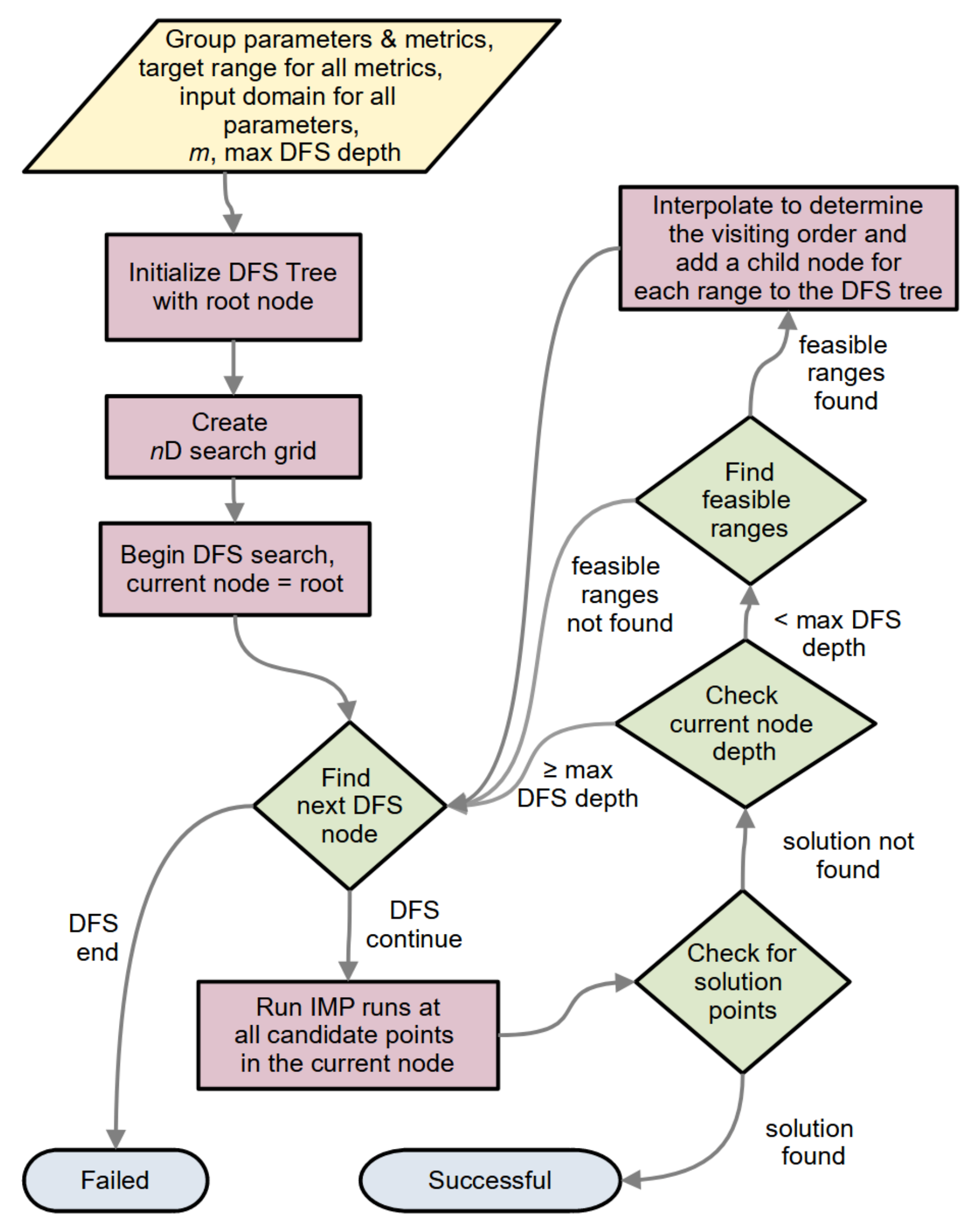
2.1.5. The General Algorithm
2.1.5.1. n-D Search and DFS
2.1.5.2. Maximum DFS Depth and DFS versus BFS
2.1.5.3. Multiple Metrics in a Group
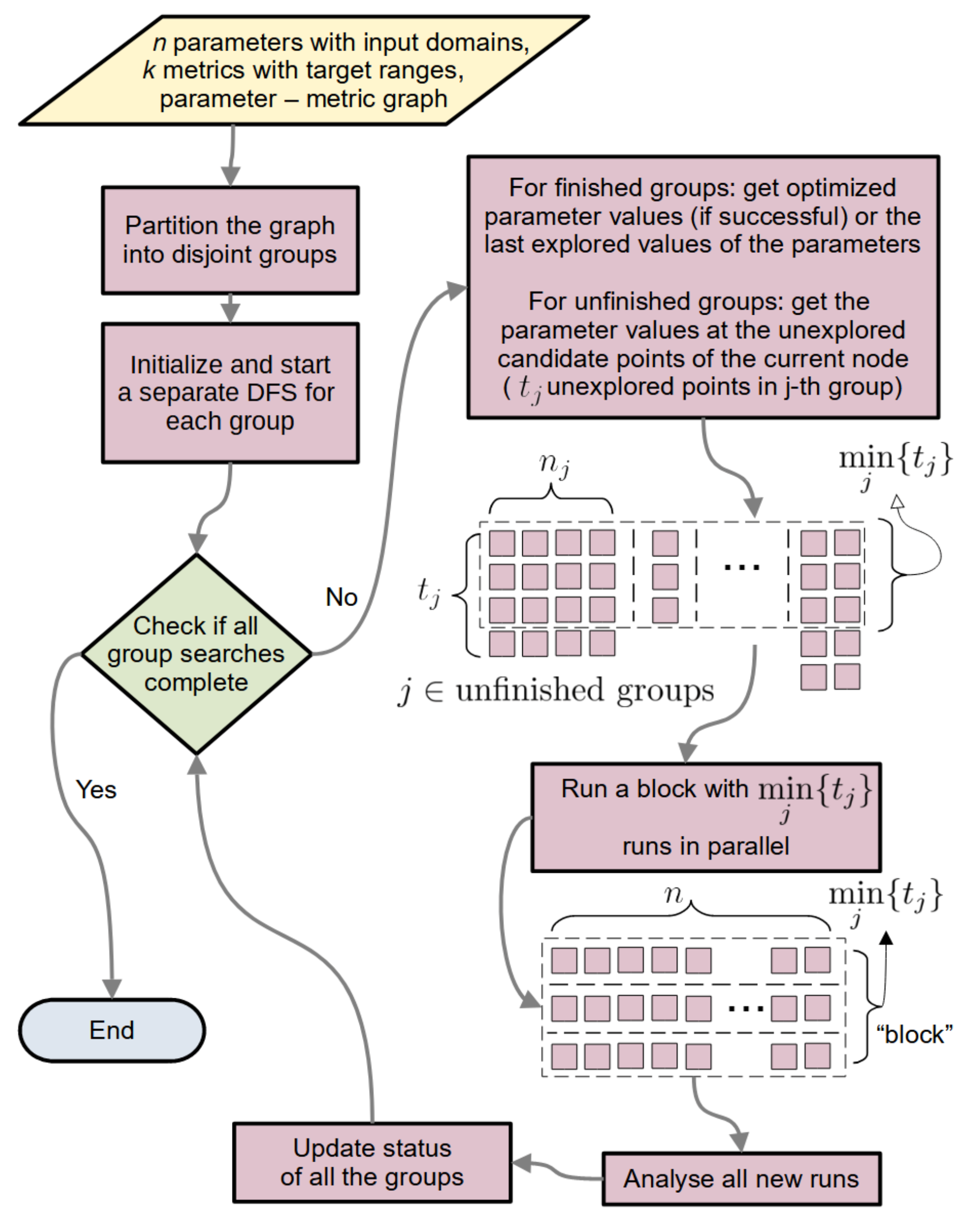
2.1.6. StOP: Algorithm and Practical Enhancements
2.1.6.1. General Flow
2.1.6.2. Updating the Group State
2.1.6.3. Setting the Visiting Order for the Nodes
2.1.6.4. CPU Economy and
2.2. Illustrative Examples
2.2.1. Example Functions
2.2.2. Comparison to Genetic Algorithm
2.2.3. Integrative Modeling Examples
3. Results
3.1. StOP Applied to Example Functions
3.2. Comparison of StOP to Binary Search
3.3. Comparison to Genetic Algorithm Search
3.4. IMP Systems
4. Discussion
4.1. Advantages
4.1.1. Parallelism
4.1.2. Local versus Global Search
4.1.3. Constraints on the Objective Function and Method Parameters
4.1.4. Number of Function Evaluations
4.2. Disadvantages
4.3. Uses
4.4. Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alber, F.; Dokudovskaya, S.; Veenhoff, L.M.; Zhang, W.; Kipper, J.; Devos, D.; Suprapto, A.; Karni-Schmidt, O.; Williams, R.; Chait, B.T.; et al. Determining the architectures of macromolecular assemblies. Nature 2007, 450, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.B.; Sali, A.; Wilson, I.A. Integrative Structural Biology. Science 2013, 339, 913–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rout, M.P.; Sali, A. Principles for Integrative Structural Biology Studies. Cell 2019, 177, 1384–1403. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Li, S.; Ser, Z.; Sanyal, T.; Choi, K.; Wan, B.; Kuang, H.; Sali, A.; Kentsis, A.; Patel, D.J.; et al. Integrative analysis reveals unique structural and functional features of the Smc5/6 complex. Proc. Natl. Acad. Sci. USA 2021, 118, e2026844118. [Google Scholar] [CrossRef]
- Ganesan, S.J.; Feyder, M.J.; Chemmama, I.E.; Fang, F.; Rout, M.P.; Chait, B.T.; Shi, Y.; Munson, M.; Sali, A. Integrative structure and function of the yeast exocyst complex. Protein Sci. 2020, 29, 1486–1501. [Google Scholar] [CrossRef]
- Gutierrez, C.; Chemmama, I.E.; Mao, H.; Yu, C.; Echeverria, I.; Block, S.A.; Rychnovsky, S.D.; Zheng, N.; Sali, A.; Huang, L. Structural dynamics of the human COP9 signalosome revealed by cross-linking mass spectrometry and integrative modeling. Proc. Natl. Acad. Sci. USA 2020, 117, 4088–4098. [Google Scholar] [CrossRef]
- Kim, S.J.; Fernandez-Martinez, J.; Nudelman, I.; Shi, Y.; Zhang, W.; Raveh, B.; Herricks, T.; Slaughter, B.D.; Hogan, J.A.; Upla, P.; et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature 2018, 555, 475–482. [Google Scholar] [CrossRef] [Green Version]
- Viswanath, S.; Bonomi, M.; Kim, S.J.; Klenchin, V.A.; Taylor, K.C.; Yabut, K.C.; Umbreit, N.T.; Van Epps, H.A.; Meehl, J.; Jones, M.H.; et al. The molecular architecture of the yeast spindle pole body core determined by Bayesian integrative modeling. Mol. Biol. Cell 2017, 28, 3298–3314. [Google Scholar] [CrossRef] [Green Version]
- Viswanath, S.; Chemmama, I.E.; Cimermancic, P.; Sali, A. Assessing Exhaustiveness of Stochastic Sampling for Integrative Modeling of Macromolecular Structures. Biophys. J. 2017, 113, 2344–2353. [Google Scholar] [CrossRef] [Green Version]
- Webb, B.; Viswanath, S.; Bonomi, M.; Pellarin, R.; Greenberg, C.H.; Saltzberg, D.; Sali, A. Integrative structure modeling with the Integrative Modeling Platform: Integrative Structure Modeling with IMP. Protein Sci. 2018, 27, 245–258. [Google Scholar] [CrossRef] [Green Version]
- Saltzberg, D.; Greenberg, C.H.; Viswanath, S.; Chemmama, I.; Webb, B.; Pellarin, R.; Echeverria, I.; Sali, A. Modeling Biological Complexes Using Integrative Modeling Platform. In Biomolecular Simulations; Bonomi, M., Camilloni, C., Eds.; Springer: New York, NY, USA, 2019; Volume 2022, pp. 353–377. [Google Scholar]
- Saltzberg, D.J.; Viswanath, S.; Echeverria, I.; Chemmama, I.E.; Webb, B.; Sali, A. Using Integrative Modeling Platform to compute, validate, and archive a model of a protein complex structure. Protein Sci. 2021, 30, 250–261. [Google Scholar] [CrossRef]
- Russel, D.; Lasker, K.; Webb, B.; Velázquez-Muriel, J.; Tjioe, E.; Schneidman-Duhovny, D.; Peterson, B.; Sali, A. Putting the Pieces Together: Integrative Modeling Platform Software for Structure Determination of Macromolecular Assemblies. PLoS Biol. 2012, 10, e1001244. [Google Scholar] [CrossRef] [Green Version]
- Rieping, W. Inferential Structure Determination. Science 2005, 309, 303–306. [Google Scholar] [CrossRef] [Green Version]
- Roberts, G.O.; Rosenthal, J.S. Optimal scaling for various Metropolis-Hastings algorithms. Stat. Sci. 2001, 16, 351–367. [Google Scholar] [CrossRef]
- Rosenthal, S. Optimal Proposal Distributions and Adaptive MCMC. In Handbook of Markov Chain Monte Carlo; Chapman and Hall/CRC: Boca Raton, FL, USA, 2009. [Google Scholar]
- Roberts, G.O.; Rosenthal, J.S. Coupling and Ergodicity of Adaptive Markov Chain Monte Carlo Algorithms. J. Appl. Probab. 2007, 44, 458–475. [Google Scholar] [CrossRef] [Green Version]
- Di Pierro, M.; Elber, R. Automated Optimization of Potential Parameters. J. Chem. Theory Comput. 2013, 9, 3311–3320. [Google Scholar] [CrossRef]
- Larson, J.; Menickelly, M.; Wild, S.M. Derivative-free optimization methods. Acta Numer. 2019, 28, 287–404. [Google Scholar] [CrossRef] [Green Version]
- Nelder, J.A.; Mead, R. A Simplex Method for Function Minimization. Comput. J. 1965, 7, 308–313. [Google Scholar] [CrossRef]
- Fermi, E.; Metropolis, N. Numerical Solution of a Minimum Problem; Los Alamos Scientific Laboratory of the University of California: Los Alamos, NM, USA, 1952. [Google Scholar]
- Hooke, R.; Jeeves, T.A. “ Direct Search” Solution of Numerical and Statistical Problems. J. ACM 1961, 8, 212–229. [Google Scholar] [CrossRef]
- Rosenbrock, H.H. An Automatic Method for Finding the Greatest or Least Value of a Function. Comput. J. 1960, 3, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Torczon, V. On the Convergence of the Multidirectional Search Algorithm. SIAM J. Optim. 1991, 1, 123–145. [Google Scholar] [CrossRef]
- Booker, A.J.; Dennis, J.E.; Frank, P.D.; Serafini, D.B.; Torczon, V.; Trosset, M.W. A rigorous framework for optimization of expensive functions by surrogates. Struct. Optim. 1999, 17, 1–13. [Google Scholar] [CrossRef]
- Audet, C.; Dennis, J.E. Mesh Adaptive Direct Search Algorithms for Constrained Optimization. SIAM J. Optim. 2006, 17, 188–217. [Google Scholar] [CrossRef] [Green Version]
- Robbins, H.; Monro, S. A Stochastic Approximation Method. Ann. Math. Stat. 1951, 22, 400–407. [Google Scholar] [CrossRef]
- Kiefer, J.; Wolfowitz, J. Stochastic Estimation of the Maximum of a Regression Function. Ann. Math. Stat. 1952, 23, 462–466. [Google Scholar] [CrossRef]
- Rastrigrin, L. The Convergence of the Random Search Method in the External Control of Many-Parameter System. Autom. Remote Control. 1963, 24, 1337–1342. [Google Scholar]
- Cormen, T.H.; Leiserson, C.E.; Rivest, R.L.; Stein, C. Introduction to Algorithms, 3rd ed.; Cormen, T.H., Ed.; MIT Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Peterson, W.W. Addressing for Random-Access Storage. IBM J. Res. Dev. 1957, 1, 130–146. [Google Scholar] [CrossRef]
- Kiefer, J. Sequential minimax search for a maximum. Proc. Am. Math. Soc. 1953, 4, 502. [Google Scholar] [CrossRef]
- Shubert, B.O. A Sequential Method Seeking the Global Maximum of a Function. SIAM J. Numer. Anal. 1972, 9, 379–388. [Google Scholar] [CrossRef]
- Jones, D.R.; Perttunen, C.D.; Stuckman, B.E. Lipschitzian optimization without the Lipschitz constant. J. Optim. Theory Appl. 1993, 79, 157–181. [Google Scholar] [CrossRef]
- Huyer, W.; Neumaier, A. Global Optimization by Multilevel Coordinate Search. J. Glob. Optim. 1999, 14, 331–355. [Google Scholar] [CrossRef]
- Holland, J.H. Outline for a Logical Theory of Adaptive Systems. J. ACM 1962, 9, 297–314. [Google Scholar] [CrossRef]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. Proc. Int. Conf. Neural Netw. 1995, 4, 1942–1948. [Google Scholar] [CrossRef]
- Mockus, J. Application of Bayesian approach to numerical methods of global and stochastic optimization. J. Glob. Optim. 1994, 4, 347–365. [Google Scholar] [CrossRef]
- Brilot, A.F.; Lyon, A.S.; Zelter, A.; Viswanath, S.; Maxwell, A.; MacCoss, M.J.; Muller, E.G.; Sali, A.; Davis, T.N.; Agard, D.A. CM1-driven assembly and activation of yeast γ-tubulin small complex underlies microtubule nucleation. eLife 2021, 10, e65168. [Google Scholar] [CrossRef]
- Tange, O. GNU Parallel 20200622 (’Privacy Shield’); Zenodo. Available online: https://zenodo.org/record/3956817#.YYEFRJpByUk (accessed on 20 October 2021).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Viswanath, S.; Sali, A. Optimizing model representation for integrative structure determination of macromolecular assemblies. Proc. Natl. Acad. Sci. USA 2019, 116, 540–545. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pasani, S.; Viswanath, S. A Framework for Stochastic Optimization of Parameters for Integrative Modeling of Macromolecular Assemblies. Life 2021, 11, 1183. https://doi.org/10.3390/life11111183
Pasani S, Viswanath S. A Framework for Stochastic Optimization of Parameters for Integrative Modeling of Macromolecular Assemblies. Life. 2021; 11(11):1183. https://doi.org/10.3390/life11111183
Chicago/Turabian StylePasani, Satwik, and Shruthi Viswanath. 2021. "A Framework for Stochastic Optimization of Parameters for Integrative Modeling of Macromolecular Assemblies" Life 11, no. 11: 1183. https://doi.org/10.3390/life11111183
APA StylePasani, S., & Viswanath, S. (2021). A Framework for Stochastic Optimization of Parameters for Integrative Modeling of Macromolecular Assemblies. Life, 11(11), 1183. https://doi.org/10.3390/life11111183





