Enceladus as a Potential Niche for Methanogens and Estimation of Its Biomass
Abstract
:1. Introduction
2. Materials and Methods
2.1. Niche Model
2.2. Growth Model
3. Results
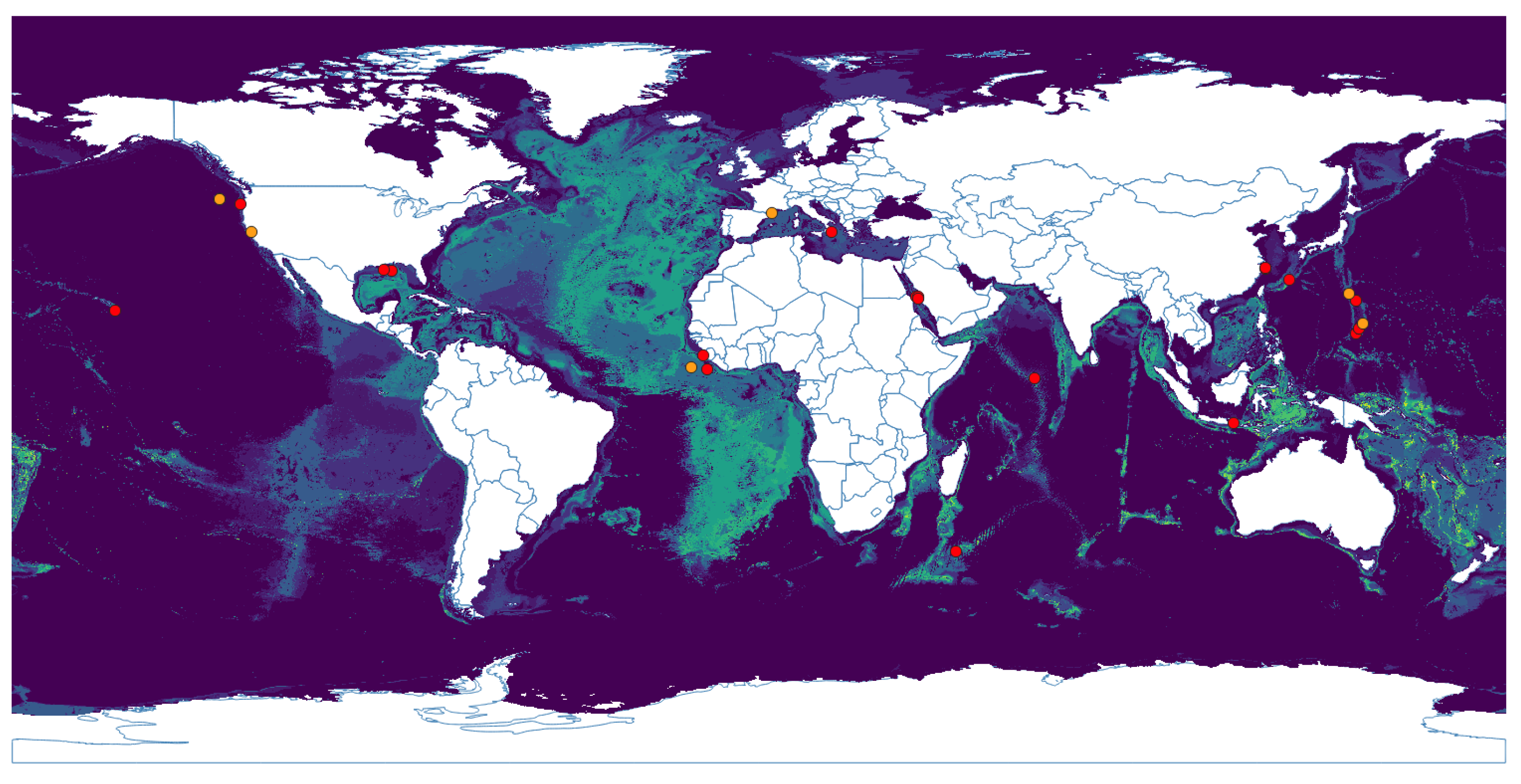
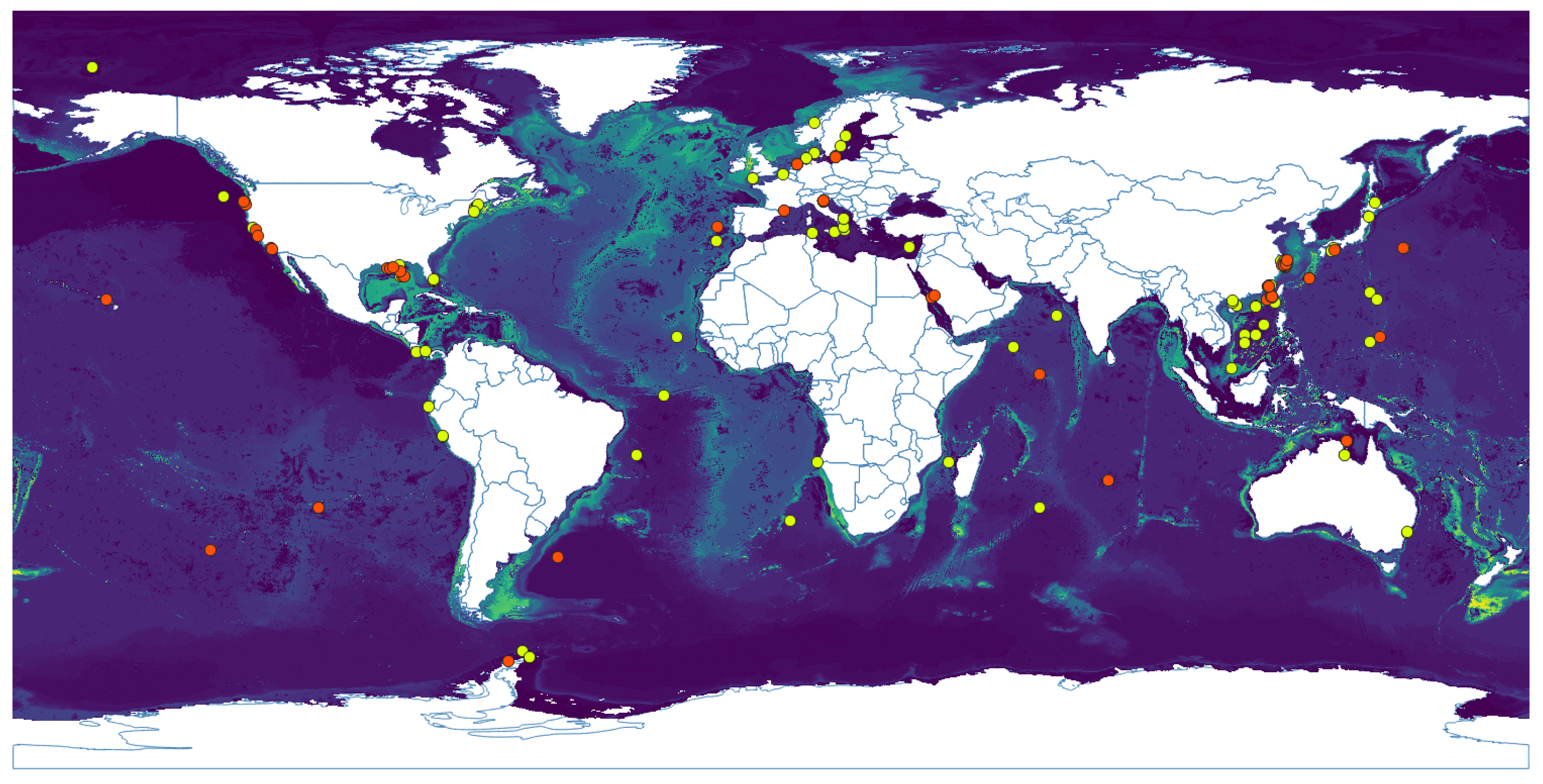
3.1. Niche Estimation
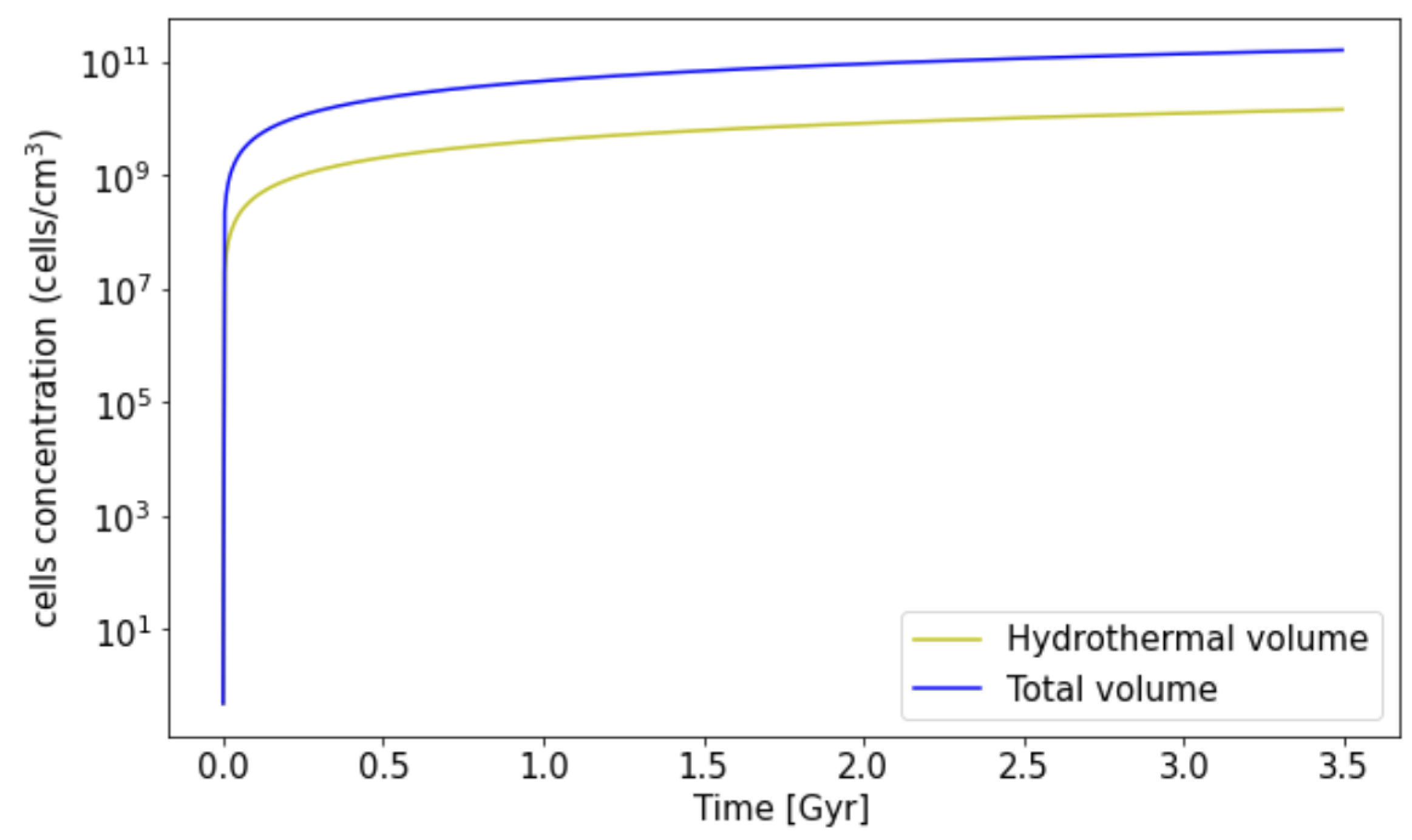
3.2. Biomass Estimation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Postberg, F.; Khawaja, N.; Abel, B.; Choblet, G.; Glein, C.R.; Gudipati, M.S.; Henderson, B.L.; Hsu, H.W.; Kempf, S.; Klenner, F.; et al. Macromolecular organic compounds from the depths of Enceladus. Nature 2018, 558, 564. [Google Scholar] [CrossRef] [PubMed]
- Waite, J.H.; Glein, C.R.; Perryman, R.S.; Teolis, B.D.; Magee, B.A.; Miller, G.; Grimes, J.; Perry, M.E.; Miller, K.E.; Bouquet, A.; et al. Cassini finds molecular hydrogen in the Enceladus plume: Evidence for hydrothermal processes. Science 2017, 356, 155–159. [Google Scholar] [CrossRef] [Green Version]
- Bouquet, A.; Mousis, O.; Waite, J.H.; Picaud, S. Possible evidence for a methane source in Enceladus’ ocean. Geophys. Res. Lett. 2015, 42, 1334–1339. [Google Scholar] [CrossRef]
- Waite, J.H.; Combi, M.R.; Ip, W.H.; Cravens, T.E.; McNutt, R.L.; Kasprzak, W.; Yelle, R.; Luhmann, J.; Niemann, H.; Gell, D.; et al. Cassini ion and neutral mass spectrometer: Enceladus plume composition and structure. Science 2006, 311, 1419–1422. [Google Scholar] [CrossRef] [Green Version]
- Hsu, H.W.; Postberg, F.; Sekine, Y.; Kempf, S.; Juhasz, A.; Horanyi, M. Silica nanoparticles as an evidence of hydrothermal activities at Enceladus. In Proceedings of the AAS/Division for Planetary Sciences Meeting Abstracts #45, Denver, CO, USA, 6–11 October 2013; Volume 45, p. 416-04. [Google Scholar]
- Hsu, H.W.; Postberg, F.; Sekine, Y.; Shibuya, T.; Kempf, S.; Horányi, M.; Juhász, A.; Altobelli, N.; Suzuki, K.; Masaki, Y.; et al. Ongoing hydrothermal activities within Enceladus. Nature 2015, 519, 207–210. [Google Scholar] [CrossRef]
- Matson, D.L.; Castillo, J.C.; Lunine, J.; Johnson, T.V. Enceladus’ plume: Compositional evidence for a hot interior. Icarus 2007, 187, 569–573. [Google Scholar] [CrossRef]
- Meier, D.V.; Pjevac, P.; Bach, W.; Hourdez, S.; Girguis, P.R.; Vidoudez, C.; Amann, R.; Meyerdierks, A. Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J. 2017, 11, 1545. [Google Scholar] [CrossRef] [Green Version]
- Dodd, M.S.; Papineau, D.; Grenne, T.; Slack, J.F.; Rittner, M.; Pirajno, F.; O’Neil, J.; Little, C.T. Evidence for early life in Earth’s oldest hydrothermal vent precipitates. Nature 2017, 543, 60–64. [Google Scholar] [CrossRef]
- Mullineaux, L.S. Deep-sea hydrothermal vent communities. In Marine Community Ecology and Conservation; Bertness, M., Bruno, M., Silliman, B., Stachowicz, J., Eds.; Sinauer Associates: Sunderland, MA, USA, 2014; pp. 383–400. [Google Scholar]
- Martin, W.; Baross, J.; Kelley, D.; Russell, M.J. Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 2008, 6, 805. [Google Scholar] [CrossRef]
- Russell, M.J.; Martin, W. The rocky roots of the acetyl-CoA pathway. Trends Biochem. Sci. 2004, 29, 358–363. [Google Scholar] [CrossRef] [Green Version]
- Baker, E.; Hey, R.; Lupton, J.; Resing, J.; Feely, R.; Gharib, J.; Massoth, G.; Sansone, F.; Kleinrock, M.; Martinez, F.; et al. Hydrothermal venting along Earth’s fastest spreading center: East Pacific Rise, 27.5°–32.3°. J. Geophys. Res. Solid Earth 2002, 107, EPM 2-1–EPM 2-14. [Google Scholar] [CrossRef]
- Joseph, A. Seafloor Hot Chimneys and Cold Seeps: Mysterious Life Around Them. In Investigating Seafloors and Oceans From Mud Volcanoes to Giant Squid; Joshep, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; Chapter 6; pp. 307–375. [Google Scholar]
- Bachraty, C.; Legendre, P.; Desbruyères, D. Biogeographic relationships among deep-sea hydrothermal vent faunas at global scale. Deep Sea Res. Part I Oceanogr. Res. Pap. 2009, 56, 1371–1378. [Google Scholar] [CrossRef] [Green Version]
- Antranikian, G.; Suleiman, M.; Schäfers, C.; Adams, M.W.; Bartolucci, S.; Blamey, J.M.; Birkeland, N.K.; Bonch-Osmolovskaya, E.; da Costa, M.S.; Cowan, D.; et al. Diversity of bacteria and archaea from two shallow marine hydrothermal vents from Vulcano Island. Extremophiles 2017, 21, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Teske, A.; Hinrichs, K.U.; Edgcomb, V.; de Vera Gomez, A.; Kysela, D.; Sylva, S.P.; Sogin, M.L.; Jannasch, H.W. Microbial diversity of hydrothermal sediments in the Guaymas Basin: Evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 2002, 68, 1994–2007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baquero, F.; Coque, T.M.; Galán, J.C.; Martinez, J.L. The origin of niches and species in the bacterial world. Front. Microbiol. 2021, 12, 566. [Google Scholar] [CrossRef]
- Pearman, P.B.; Guisan, A.; Broennimann, O.; Randin, C.F. Niche dynamics in space and time. Trends Ecol. Evol. 2008, 23, 149–158. [Google Scholar] [CrossRef]
- Odling-Smee, F.J.; Laland, K.N.; Feldman, M.W. Niche construction. Am. Nat. 1996, 147, 641–648. [Google Scholar] [CrossRef]
- Whittaker, R.H.; Levin, S.A.; Root, R.B. Niche, habitat, and ecotope. Am. Nat. 1973, 107, 321–338. [Google Scholar] [CrossRef]
- Godsoe, W.; Jankowski, J.; Holt, R.D.; Gravel, D. Integrating biogeography with contemporary niche theory. Trends Ecol. Evol. 2017, 32, 488–499. [Google Scholar] [CrossRef]
- Soberón, J.; Arroyo-Peña, B. Are fundamental niches larger than the realized? Testing a 50-year-old prediction by Hutchinson. PLoS ONE 2017, 12, e0175138. [Google Scholar] [CrossRef]
- Angilletta, M.J.; Sears, M.W.; Levy, O.; Youngblood, J.; VandenBrooks, J.M. Fundamental flaws with the fundamental niche. Integr. Comp. Biol. 2019, 59, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Escobar, L.E.; Craft, M.E. Advances and limitations of disease biogeography using ecological niche modeling. Front. Microbiol. 2016, 7, 1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escobar, L.E. Ecological Niche Modeling: An Introduction for Veterinarians and Epidemiologists. Front. Vet. Sci. 2020, 7, 713. [Google Scholar] [CrossRef] [PubMed]
- Hugh-Jones, M.; Blackburn, J. The ecology of Bacillus anthracis. Mol. Asp. Med. 2009, 30, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.G.; Mor, S.M.; Hossain, S. The elephant–livestock interface modulates anthrax suitability in India. Proc. R. Soc. B 2019, 286, 20190179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mwakapeje, E.R.; Ndimuligo, S.A.; Mosomtai, G.; Ayebare, S.; Nyakarahuka, L.; Nonga, H.E.; Mdegela, R.H.; Skjerve, E. Ecological niche modeling as a tool for prediction of the potential geographic distribution of Bacillus anthracis spores in Tanzania. Int. J. Infect. Dis. 2019, 79, 142–151. [Google Scholar] [CrossRef] [Green Version]
- Sowers, K.R. Methanogenesis. In Encyclopedia of Microbiology, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 53–73. [Google Scholar]
- Liu, Y.; Beer, L.L.; Whitman, W.B. Methanogens: A window into ancient sulfur metabolism. Trends Microbiol. 2012, 20, 251–258. [Google Scholar] [CrossRef]
- Ferry, J.G. The chemical biology of methanogenesis. Planet. Space Sci. 2010, 58, 1775–1783. [Google Scholar] [CrossRef]
- Ueno, Y.; Yamada, K.; Yoshida, N.; Maruyama, S.; Isozaki, Y. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature 2006, 440, 516. [Google Scholar] [CrossRef]
- Battistuzzi, F.U.; Feijao, A.; Hedges, S.B. A genomic timescale of prokaryote evolution: Insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 2004, 4, 44. [Google Scholar] [CrossRef] [Green Version]
- Taubner, R.S.; Schleper, C.; Firneis, M.G.; Rittmann, S.K.M. Assessing the ecophysiology of methanogens in the context of recent astrobiological and planetological studies. Life 2015, 5, 1652–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, P.N.; Parks, D.H.; Chadwick, G.L.; Robbins, S.J.; Orphan, V.J.; Golding, S.D.; Tyson, G.W. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 2015, 350, 434–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Whitman, W.B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef]
- Whitman, W.B.; Bowen, T.L.; Boone, D.R. The methanogenic bacteria. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2006; pp. 165–207. [Google Scholar]
- McLain, J. Archaea. In Encyclopedia of Soils in the Environment; Elsevier: Amsterdam, The Netherlands, 2005; pp. 88–94. [Google Scholar]
- Malina, J. Design of Anaerobic Processes for Treatment of Industrial and Muncipal Waste, Volume VII; Routledge: Oxfordshire, UK, 2017. [Google Scholar]
- Lever, M.A. A new era of methanogenesis research. Trends Microbiol. 2016, 24, 84–86. [Google Scholar] [CrossRef] [PubMed]
- Boone, D.R. Methanobacterium. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–8. [Google Scholar]
- Brugère, J.F.; Hania, W.B.; Arnal, M.E.; Ribière, C.; Ballet, N.; Vandeckerkove, P.; Ollivier, B.; O’Toole, P.W. Archaea: Microbial Candidates in Next-generation Probiotics Development. J. Clin. Gastroenterol. 2018, 52, S71–S73. [Google Scholar] [CrossRef] [Green Version]
- Canfield, D.E.; Kristensen, E.; Thamdrup, B. Systematics and Phylogeny. In Advances in Marine Biology; Elsevier: Amsterdam, The Netherlands, 2005; Volume 48, pp. 1–21. [Google Scholar]
- Affholder, A.; Guyot, F.; Sauterey, B.; Ferrière, R.; Mazevet, S. Bayesian analysis of Enceladus’s plume data to assess methanogenesis. Nat. Astron. 2021, 5, 805–814. [Google Scholar] [CrossRef]
- Porco, C.C.; Dones, L.; Mitchell, C. Could it be snowing microbes on Enceladus? Assessing conditions in its plume and implications for future missions. Astrobiology 2017, 17, 876–901. [Google Scholar] [CrossRef] [Green Version]
- Steel, E.L.; Davila, A.; McKay, C.P. Abiotic and biotic formation of amino acids in the Enceladus ocean. Astrobiology 2017, 17, 862–875. [Google Scholar] [CrossRef]
- Taubner, R.S.; Pappenreiter, P.; Zwicker, J.; Smrzka, D.; Pruckner, C.; Kolar, P.; Bernacchi, S.; Seifert, A.H.; Krajete, A.; Bach, W.; et al. Biological methane production under putative Enceladus-like conditions. Nat. Commun. 2018, 9, 748. [Google Scholar] [CrossRef] [Green Version]
- McKay, C.; Khare, B.N.; Amin, R.; Klasson, M.; Kral, T.A. Possible sources for methane and C2–C5 organics in the plume of Enceladus. Planet. Space Sci. 2012, 71, 73–79. [Google Scholar] [CrossRef] [Green Version]
- McKay, C.P.; Porco, C.C.; Altheide, T.; Davis, W.L.; Kral, T.A. The possible origin and persistence of life on Enceladus and detection of biomarkers in the plume. Astrobiology 2008, 8, 909–919. [Google Scholar] [CrossRef] [Green Version]
- Monod, J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef] [Green Version]
- Van Proosdij, A.S.; Sosef, M.S.; Wieringa, J.J.; Raes, N. Minimum required number of specimen records to develop accurate species distribution models. Ecography 2016, 39, 542–552. [Google Scholar] [CrossRef]
- Soberon, J.; Peterson, A.T. Interpretation of Models of Fundamental Ecological Niches and Species’ Distributional Areas; University of Kansas: Lawrence, KS, USA, 2005. [Google Scholar]
- Jones, P.; Gladkov, A. FloraMap: A Computer Tool for Predicting the Distribution of Plants and Other Organisms in the Wild; Annie, L., Ed.; Version 1; Jones. CIAT CD-ROM Series; Centro Internacional de Agricultura Tropical: Cali, Colombia, 1999. [Google Scholar]
- Carpenter, G.; Gillison, A.; Winter, J. DOMAIN: A flexible modelling procedure for mapping potential distributions of plants and animals. Biodivers. Conserv. 1993, 2, 667–680. [Google Scholar] [CrossRef]
- GBIF.org. GBIF Occurrence Download. 2019. Available online: https://doi.org/10.15468/dl.wfrylx (accessed on 24 October 2019).
- GBIF.org. GBIF Occurrence Download. 2019. Available online: https://doi.org/10.15468/dl.r6btuw (accessed on 24 October 2019).
- GBIF.org. GBIF Occurrence Download. 2019. Available online: https://doi.org/10.15468/dl.endhc9 (accessed on 24 October 2019).
- Allioux, M.; Yvenou, S.; Slobodkina, G.; Slobodkin, A.; Shao, Z.; Jebbar, M.; Alain, K. Genomic Characterization and Environmental Distribution of a Thermophilic Anaerobe Dissulfurirhabdus thermomarina SH388T Involved in Disproportionation of Sulfur Compounds in Shallow Sea Hydrothermal Vents. Microorganisms 2020, 8, 1132. [Google Scholar] [CrossRef]
- Williams, R.A.; Owens, H.L.; Clamp, J.; Peterson, A.T.; Warren, A.; Martín-Cereceda, M. Endemicity and climatic niche differentiation in three marine ciliated protists. Limnol. Oceanogr. 2018, 63, 2727–2736. [Google Scholar] [CrossRef] [Green Version]
- Selama, O.; James, P.; Nateche, F.; Wellington, E.M.; Hacène, H. The world bacterial biogeography and biodiversity through databases: A case study of NCBI Nucleotide Database and GBIF Database. BioMed Res. Int. 2013, 2013, 240175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Troudet, J.; Grandcolas, P.; Blin, A.; Vignes-Lebbe, R.; Legendre, F. Taxonomic bias in biodiversity data and societal preferences. Sci. Rep. 2017, 7, 9132. [Google Scholar] [CrossRef] [PubMed]
- Tyberghein, L.; Verbruggen, H.; Pauly, K.; Troupin, C.; Mineur, F.; De Clerck, O. Bio-ORACLE: A global environmental dataset for marine species distribution modelling. Glob. Ecol. Biogeogr. 2012, 21, 272–281. [Google Scholar] [CrossRef]
- QGISDevelopmentTeam. QGIS Geographic Information System. Open Source Geospatial Foundation Project. 2019. Available online: http://qgis.osgeo.org (accessed on 24 October 2019).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Soberón, J.; Peterson, A.T. What is the shape of the fundamental Grinnellian niche? Theor. Ecol. 2019, 13, 105–115. [Google Scholar] [CrossRef]
- Osorio, L. NicheToolBox. 2019. Available online: https://github.com/luismurao/nichetoolbox (accessed on 24 October 2019).
- Nix, H.; Busby, J. BIOCLIM, a Bioclimatic Analysis and Prediction System; Division of Water and Land Resources: Canberra, Australia, 1986.
- Booth, T.H.; Nix, H.A.; Busby, J.R.; Hutchinson, M.F. BIOCLIM: The first species distribution modelling package, its early applications and relevance to most current MAXENT studies. Divers. Distrib. 2014, 20, 1–9. [Google Scholar] [CrossRef]
- Loyola, T.F.R.R.D. Labeling ecological niche models. Nat. Conserv. 2012, 10, 119–126. [Google Scholar]
- Mbogga, M.S.; Wang, X.; Hamann, A. Bioclimate envelope model predictions for natural resource management: Dealing with uncertainty. J. Appl. Ecol. 2010, 47, 731–740. [Google Scholar] [CrossRef]
- Peterson, A.T.; Soberón, J.; Pearson, R.G.; Anderson, R.P.; Martínez-Meyer, E.; Nakamura, M.; Araújo, M.B. Ecological Niches and Geographic Distributions (MPB-49); Princeton University Press: Princeton, NJ, USA, 2011; Volume 56. [Google Scholar]
- Liu, S. Bioprocess Engineering: Kinetics, Sustainability, and Reactor Design; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Senn, H.; Lendenmann, U.; Snozzi, M.; Hamer, G.; Egli, T. The growth of Escherichia coli in glucose-limited chemostat cultures: A re-examination of the kinetics. Biochim. Biophys. Acta Gen. Subj. 1994, 1201, 424–436. [Google Scholar] [CrossRef]
- Chezeau, B.; Vial, C. Modeling and Simulation of the Biohydrogen Production Processes. In Biohydrogen; Elsevier: Amsterdam, The Netherlands, 2019; pp. 445–483. [Google Scholar]
- Lendenmann, U.; Egli, T. Kinetic models for the growth of Escherichia coli with mixtures of sugars under carbon-limited conditions. Biotechnol. Bioeng. 1998, 59, 99–107. [Google Scholar] [CrossRef]
- Thomas, P.; Tajeddine, R.; Tiscareno, M.; Burns, J.; Joseph, J.; Loredo, T.; Helfenstein, P.; Porco, C. Enceladus’s measured physical libration requires a global subsurface ocean. Icarus 2016, 264, 37–47. [Google Scholar] [CrossRef] [Green Version]
- Schönheit, P.; Moll, J.; Thauer, R.K. Growth parameters (K s, μ max, Y s) of Methanobacterium thermoautotrophicum. Arch. Microbiol. 1980, 127, 59–65. [Google Scholar] [CrossRef]
- Porco, C.; Helfenstein, P.; Thomas, P.; Ingersoll, A.; Wisdom, J.; West, R.; Neukum, G.; Denk, T.; Wagner, R.; Roatsch, T.; et al. Cassini observes the active south pole of Enceladus. Science 2006, 311, 1393–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Postberg, F.; Clark, R.N.; Hansen, C.J.; Coates, A.J.; Dalle Ore, C.; Scipioni, F.; Hedman, M.; Waite, J. Plume and surface composition of Enceladus. Enceladus Icy Moons Saturn 2018, 129–162. [Google Scholar] [CrossRef] [Green Version]
- Postberg, F.; Kempf, S.; Schmidt, J.; Brilliantov, N.; Beinsen, A.; Abel, B.; Buck, U.; Srama, R. Sodium salts in E-ring ice grains from an ocean below the surface of Enceladus. Nature 2009, 459, 1098–1101. [Google Scholar] [CrossRef]
- Sekine, Y.; Shibuya, T.; Postberg, F.; Hsu, H.W.; Suzuki, K.; Masaki, Y.; Kuwatani, T.; Mori, M.; Hong, P.K.; Yoshizaki, M.; et al. High-temperature water–rock interactions and hydrothermal environments in the chondrite-like core of Enceladus. Nat. Commun. 2015, 6, 8604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treguer, P.; Nelson, D.M.; Van Bennekom, A.J.; DeMaster, D.J.; Leynaert, A.; Queguiner, B. The silica balance in the world ocean: A reestimate. Science 1995, 268, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Lobry, J.; Flandrois, J.P.; Carret, G.; Pave, A. Monod’s bacterial growth model revisited. Bull. Math. Biol. 1992, 54, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Cabrol, N.A.; Grin, E.A. From Habitability to Life on Mars; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tenelanda-Osorio, L.I.; Parra, J.L.; Cuartas-Restrepo, P.; Zuluaga, J.I. Enceladus as a Potential Niche for Methanogens and Estimation of Its Biomass. Life 2021, 11, 1182. https://doi.org/10.3390/life11111182
Tenelanda-Osorio LI, Parra JL, Cuartas-Restrepo P, Zuluaga JI. Enceladus as a Potential Niche for Methanogens and Estimation of Its Biomass. Life. 2021; 11(11):1182. https://doi.org/10.3390/life11111182
Chicago/Turabian StyleTenelanda-Osorio, Laura I., Juan L. Parra, Pablo Cuartas-Restrepo, and Jorge I. Zuluaga. 2021. "Enceladus as a Potential Niche for Methanogens and Estimation of Its Biomass" Life 11, no. 11: 1182. https://doi.org/10.3390/life11111182
APA StyleTenelanda-Osorio, L. I., Parra, J. L., Cuartas-Restrepo, P., & Zuluaga, J. I. (2021). Enceladus as a Potential Niche for Methanogens and Estimation of Its Biomass. Life, 11(11), 1182. https://doi.org/10.3390/life11111182





