A Maximum Flow-Based Approach to Prioritize Drugs for Drug Repurposing of Chronic Diseases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.1.1. Protein–Protein Interaction (PPI) Network
2.1.2. Drug-Target Interactions (DTIs) Network
2.1.3. Risk Genes
2.2. The Maximum Flow Algorithm for Drug Repurposing
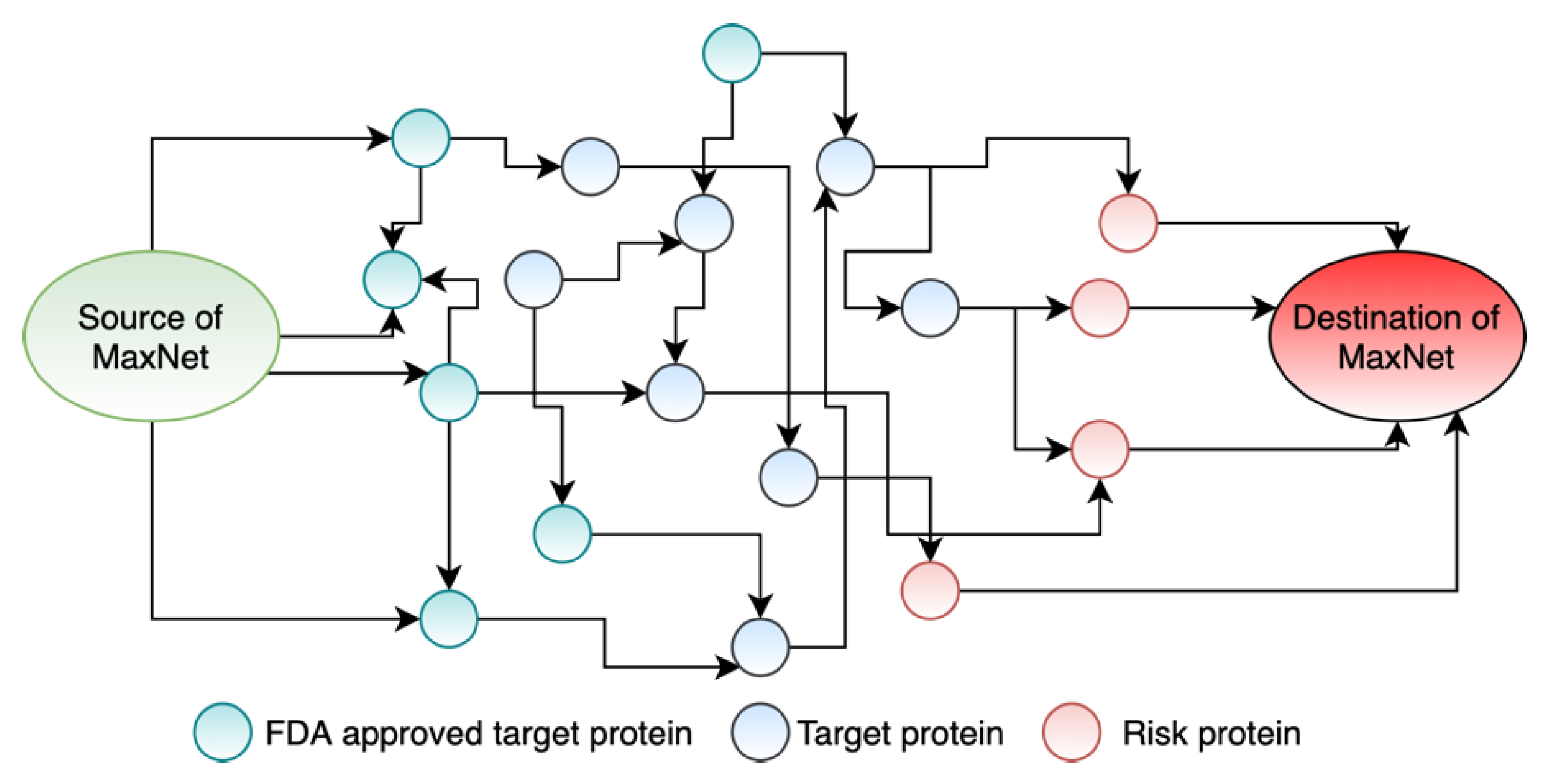
2.2.1. Constructing the Maximum Flow Network
2.2.2. Push-Relabel Maximum Flow Algorithm
| Algorithm 1 Push-Relabel_MaximumFlow_Algorithm [28]. |
| Input: PPI, Capacity = C, N = unique nodes of PPI, start_node = SDN, destination_node = DDN. Output: Maximum flow between SDN and DDN (1) FOR i = 1 to length [N]: a. HeightV [i] = 0//HeightV is height of every vertex b. FlowV [i] = 0//FlowV is the flow of every vertex (2) HeightV [start_node] = length [N] (3) FOR i = 1 to length [PPI]: a. FlowE [i] = 0//FlowE is the flow of every edge in the PPI (4) V = adjacentVetex[start_node] (5) FOR i = 1 to length [V]: a. FlowV [V[i]] = Capacity [V[i]] b. excessFlow [V[i]] = Capacity [V[i]] (6) PUSH: FOR i = 1 to length [N]: If excessFlow [N[i]] ≠ 0: (in the residual graph) tmpV = adjacentVetex[N[i]] if HeightV [N[i]] > lowest_height[tmpV] Push_flow from N[i] to lower height vertices (7) RELABEL: FOR i = 1 to length [N]: If excessFlow [N[i]] ≠ 0: (in the residual graph) tmpV = adjacentVetex[N[i]] if HeightV [N[i]] ≤ lowest_height[tmpV] HeightV [N[i]] = minimumHeight[tmp] |
2.2.3. Drug Repurposing from Maximum Flow Values
| Algorithm 2 Pipeline of the maximum flow-based drug repurposing. |
| Input: PPI = all the PPIs, FDA_DT = all the FDA approved DTs in PPIs network, DTI = DTIs for FDA_DT, RG = risk genes, W = flow capacity of edges. Output: CD = candidate drugs for repurposing for the treatment of breast cancer. 1. FOR i = 1 to length [PPI]: a. Calculate flow capacity of the edge using Equation (1): C[i] = TOMSimilarity (PPI[i]) 2. CREATE two dummy nodes: a. source dummy node = SDN and destination dummy node = DDN 3. FOR i = 1 to length [FDA_DT]: a. Index = length [PPI] + 1 b. CONNECT SDN to FDA_DT[i] and add this interaction in PPI[index] c. W[index] = sum of the capacities of the outgoing edges from PPI[index] 4. FOR i = 1 to the length of RG: a. Index = length of PPI + 1 b. CONNECT RG[i] to DDN and add this interaction in PPI[index] c. C[index] = sum of the capacities of the incoming edges from PPI[index] 5. The nodes in PPIs and their associated outgoing flow value = Push-Relabel_MaximumFlow_Algorithm (PPI, C, SDN, DDN) 6. prioritized_DTs = sort the nodes in PPI in decreasing order of their outgoing flows 7. CD = sort drugs in DTI using prioritized_DTs |
3. Experimental Results
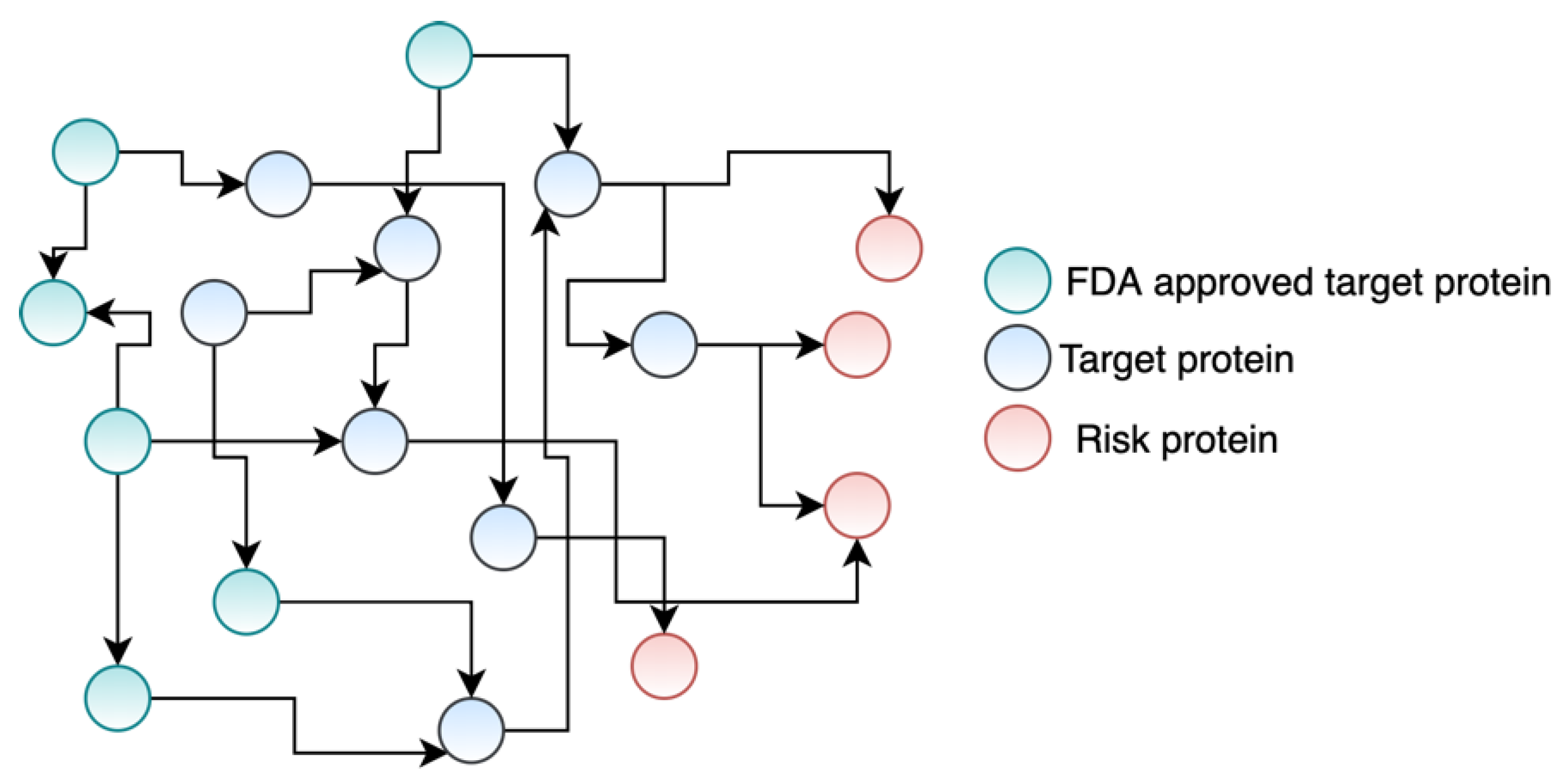
3.1. Mapping Drug Targets and Disease-Specific Risk Genes to the PPIs Network
3.2. Weights of the Interactions in PPIs Network
3.3. Formulating Drug Repurposing as a Maximum Flow Network
3.4. Drug Repurposing for Breast Cancer, IBD, and COPD
3.5. Performance Evaluation
3.6. Performance Comparison with Other Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Breast Cancer Foundation. Available online: https://www.nationalbreastcancer.org/ (accessed on 3 September 2021).
- Vogelstein, B.; Kinzler, K.W. Cancer Genes and the Pathways They Control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef]
- Cancer Statistics—National Cancer Institute. Available online: https://www.cancer.gov/about-cancer/understanding/statistics (accessed on 3 September 2021).
- Breast Cancer: Types of Treatment|Cancer.Net. Available online: https://www.cancer.net/cancer-types/breast-cancer/types-treatment (accessed on 10 October 2021).
- Inflammatory Bowel Disease. Available online: https://kidshealth.org/en/teens/ibd.html (accessed on 3 September 2021).
- Sakornsakolpat, P.; Prokopenko, D.; Lamontagne, M.; Reeve, N.F.; Guyatt, A.L.; Jackson, V.E.; Shrine, N.; Qiao, D. Expanded Genetic Landscape of Chronic Obstructive Pulmonary Disease Reveals Heterogeneous Cell Type and Phenotype Associations. BioRxiv 2018, 355644. [Google Scholar] [CrossRef] [Green Version]
- Chornic Obstructive Pulmonary Disease. Available online: https://lung.ca/copd (accessed on 28 August 2021).
- Lipman, A.G. Drug Repurposing and Repositioning: Workshop Summary. J. Pain Palliat. Care Pharmacother. 2015, 29, 81. [Google Scholar] [CrossRef]
- Hui, D.S.; Azhar, E.I.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The Continuing 2019-NCoV Epidemic Threat of Novel Coronaviruses to Global Health—The Latest 2019 Novel Coronavirus Outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A Novel Coronavirus Outbreak of Global Health Concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Singhal, T. A Review of Coronavirus Disease-2019 (COVID-19). Indian J. Pediatr. 2020, 87, 281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabi, F.A.; Al Zoubi, M.S.; Kasasbeh, G.A.; Salameh, D.M.; Al-Nasser, A.D. SARS-CoV-2 and Coronavirus Disease 2019: What We Know So Far. Pathogens 2020, 9, 231. [Google Scholar] [CrossRef]
- Yu, L.; Huang, J.; Ma, Z.; Zhang, J.; Zou, Y.; Gao, L. Inferring Drug-Disease Associations Based on Known Protein Complexes. BMC Med. Genom. 2015, 8, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-Based Drug Repurposing for Novel Coronavirus 2019-NCoV/SARS-CoV-2. Cell Discov. 2020, 6, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Schmitt-Ulms, G.; Sato, C.; Xi, Z.; Zhang, Y.; Zhou, Y.; George-Hyslop, P.S.; Rogaeva, E. Drug Repositioning for Alzheimer’s Disease Based on Systematic “omics” Data Mining. PLoS ONE 2016, 11, e0168812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, S.; Hug, C.; Todorov, P.; Moret, N.; Boswell, S.A.; Evans, K.; Zhou, G.; Johnson, N.T.; Hyman, B.T.; Sorger, P.K.; et al. Machine Learning Identifies Candidates for Drug Repurposing in Alzheimer’s Disease. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Liu, C.; Jiang, J.; Lu, W.; Li, W.; Liu, G.; Zhou, W.; Huang, J.; Tang, Y. Prediction of Drug-Target Interactions and Drug Repositioning via Network-Based Inference. PLoS Comput. Biol. 2012, 8, e1002503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, S.H.; Yeh, H.Y.; Soo, V.W. A Network Flow Approach to Predict Drug Targets from Microarray Data, Disease Genes and Interactome Network—Case Study on Prostate Cancer. J. Clin. Bioinforma. 2012, 2, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demeter, J.; Beauheim, C.; Gollub, J.; Hernandez-Boussard, T.; Jin, H.; Maier, D.; Matese, J.C.; Nitzberg, M.; Wymore, F.; Zachariah, Z.K.; et al. The Stanford Microarray Database. Nucleic Acids Res. 2001, 29, 152–155. [Google Scholar] [CrossRef] [Green Version]
- Lapointe, J.; Li, C.; Higgins, J.P.; van de Rijn, M.; Bair, E.; Montgomery, K.; Ferrari, M.; Egevad, L.; Rayford, W.; Bergerheim, U.; et al. Gene Expression Profiling Identifies Clinically Relevant Subtypes of Prostate Cancer. Proc. Natl. Acad. Sci. USA 2004, 101, 811–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floyd, R.W. Algorithm 97: Shortest Path. Commun. ACM 1962, 5, 345. [Google Scholar] [CrossRef]
- Melak, T.; Gakkhar, S. Maximum Flow Approach to Prioritize Potential Drug Targets of Mycobacterium Tuberculosis H37Rv from Protein-Protein Interaction Network. Clin. Transl. Med. 2015, 4, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Menche, J.; Sharma, A.; Kitsak, M.; Ghiassian, S.D.; Vidal, M.; Loscalzo, J.; Barabási, A.L. Uncovering Disease-Disease Relationships through the Incomplete Interactome. Science 2015, 347, 1257601. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Baxter, J.S.; Leavy, O.C.; Dryden, N.H.; Maguire, S.; Johnson, N.; Fedele, V.; Simigdala, N.; Martin, L.A.; Andrews, S.; Wingett, S.W.; et al. Capture Hi-C Identifies Putative Target Genes at 33 Breast Cancer Risk Loci. Nat. Commun. 2018, 9, 1–3. [Google Scholar] [CrossRef]
- Wu, L.; Shi, W.; Long, J.; Guo, X.; Michailidou, K.; Beesley, J.; Bolla, M.K.; Shu, X.O.; Lu, Y.; Cai, Q.; et al. A Transcriptome-Wide Association Study of 229,000 Women Identifies New Candidate Susceptibility Genes for Breast Cancer. Nat. Genet. 2018, 50, 968–978. [Google Scholar] [CrossRef]
- De Lange, K.M.; Moutsianas, L.; Lee, J.C.; Lamb, C.A.; Luo, Y.; Kennedy, N.A.; Jostins, L.; Rice, D.L.; Gutierrez-Achury, J.; Ji, S.G.; et al. Genome-Wide Association Study Implicates Immune Activation of Multiple Integrin Genes in Inflammatory Bowel Disease. Nat. Genet. 2017, 49, 256–261. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, A.V.; Tarjan, R.E. A New Approach to the Maximum-Flow Problem. J. ACM 1988, 35, 921–940. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. DrugBank: A Knowledgebase for Drugs, Drug Actions and Drug Targets. Nucleic Acids Res. 2008, 36 (Suppl. S1), 901–906. [Google Scholar] [CrossRef]
- Meurling, L.; Marquez, M.; Nilsson, S.; Holmberg, A.R. Polymer-Conjugated Guanidine Is a Potentially Useful Anti-Tumor Agent. Int. J. Oncol. 2009, 35, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Srivastava, S.K. Antitumor Activity of Phenethyl Isothiocyanate in HER2-Positive Breast Cancer Models. BMC Med. 2012, 10, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pantziarka, P.; Sukhtame, V.; Meheus, L.; Sukhatme, V.P.V.V.; Bouche, G.; Meheus, L.; Sukhatme, V.P.V.V.; Bouche, G. Repurposing Non-Cancer Drugs in Oncology—How Many Drugs Are out There? bioRxiv 2017, 197434. [Google Scholar] [CrossRef] [Green Version]
- Tamoxifen. Available online: https://www.webmd.com/drugs/2/drug-4497/tamoxifen-oral/details (accessed on 29 August 2021).
- Can, G.; Ayvaz, S.; Can, H.; Karaboğa, İ.; Demirtaş, S.; Akşit, H.; Yılmaz, B.; Korkmaz, U.; Kurt, M.; Karaca, T. The Efficacy of Tyrosine Kinase Inhibitor Dasatinib on Colonic Mucosal Damage in Murine Model of Colitis. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Dey, M.; Kuhn, P.; Ribnicky, D.; Premkumar, V.; Reuhl, K.; Raskin, I. Dietary Phenethylisothiocyanate Attenuates Bowel Inflammation in Mice. BMC Chem. Biol. 2010, 10, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.H.; Rajendran, V.M. Adenosine: An Immune Modulator of Inflammatory Bowel Diseases. World J. Gastroenterol. 2009, 15, 4491. [Google Scholar] [CrossRef] [PubMed]
- Li, T.T.; Zhang, J.F.; Fei, S.J.; Zhu, S.P.; Zhu, J.Z.; Qiao, X.; Liu, Z.B. Glutamate Microinjection into the Hypothalamic Paraventricular Nucleus Attenuates Ulcerative Colitis in Rats. Acta Pharmacol. Sin. 2014, 35, 185–194. [Google Scholar] [CrossRef] [Green Version]
- Drugbank. Available online: https://www.drugbank.ca/drugs/DB12695 (accessed on 2 September 2021).
- Sotomayor, E.A.; Teicher, B.A.; Schwartz, G.N.; Holden, S.A.; Menon, K.; Herman, T.S.; Frei, E. Minocycline in Combination with Chemotherapy or Radiation Therapy in Vitro and in Vivo. Cancer Chemother. Pharmacol. 1992, 30, 377–384. [Google Scholar] [CrossRef]
- Li, X.; Yang, H.; Sun, H.; Lu, R.; Zhang, C.; Gao, N.; Meng, Q.; Wu, S.; Wang, S.; Aschner, M.; et al. Taurine Ameliorates Particulate Matter-Induced Emphysema by Switching on Mitochondrial NADH Dehydrogenase Genes. Proc. Natl. Acad. Sci. USA 2017, 114, E9655–E9664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, M.E.J. A Measure of Betweenness Centrality Based on Random Walks. Soc. Netw. 2005, 27, 39–54. [Google Scholar] [CrossRef] [Green Version]
- Li, Y. Toward a Qualitative Search Engine. IEEE Internet Comput. 1998, 2, 24–29. [Google Scholar] [CrossRef]
- Zhao, K.; So, H.-C. A Machine Learning Approach to Drug Repositioning Based on Drug Expression Profiles: Applications to Schizophrenia and Depression/Anxiety Disorders. arXiv 2017, arXiv:1706.03014. [Google Scholar]
- Saberian, N.; Peyvandipour, A.; Donato, M.; Ansari, S.; Draghici, S. A New Computational Drug Repurposing Method Using Established Disease–Drug Pair Knowledge. Bioinformatics 2019, 35, 3672–3678. [Google Scholar] [CrossRef]
- Napolitano, F.; Zhao, Y.; Moreira, V.M.; Tagliaferri, R.; Kere, J.; D’Amato, M.; Greco, D. Drug Repositioning: A Machine-Learning Approach through Data Integration. J. Cheminform. 2013, 5, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Estrada, E. Protein Bipartivity and Essentiality in the Yeast Protein-Protein Interaction Network. J. Proteome Res. 2006, 5, 2177–2184. [Google Scholar] [CrossRef]
- Ruepp, A.; Waegele, B.; Lechner, M.; Brauner, B.; Dunger-Kaltenbach, I.; Fobo, G.; Frishman, G.; Montrone, C.; Mewes, H.W. CORUM: The Comprehensive Resource of Mammalian Protein Complexes-2009. Nucleic Acids Res. 2009, 38 (Suppl. S1), 497–501. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Zhang, B.; Horvath, S. Defining Clusters from a Hierarchical Cluster Tree: The Dynamic Tree Cut Package for R. Bioinformatics 2008, 124, 719–720. [Google Scholar] [CrossRef]
- Pseudoephedrine. Available online: https://go.drugbank.com/drugs/DB00852 (accessed on 28 August 2021).
- Sudafed Oral: Uses, Side Effects, Interactions, Pictures, Warnings & Dosing—WebMD. Available online: https://www.webmd.com/drugs/2/drug-6573/sudafed-oral/details (accessed on 28 August 2021).


| Properties | Values |
|---|---|
| Number of nodes | 13,368 |
| Number of edges | 140,899 |
| Transitivity | 0.292 |
| Average clustering coefficient | 0.173 |
| Edge density | 0.002 |
| Average degree | 21.08 |
| Total triangles | 4,105,272 |
| Drug Name | Target Protein | Target Gene | Flow Value | Status | Reference |
|---|---|---|---|---|---|
| Guanidine | P78352 | DLG4 | 0.0489 | Confirmed | [30] |
| Phenethyl Isothiocyanate | P31946 | YWHAB | 0.0389 | Confirmed | [31] |
| Caffeine | P78527 | PRKDC | 0.0363 | Confirmed | [32] |
| Tamoxifen | Q05655 | PRKCD | 0.0363 | Confirmed | [33] |
| (2S)-2-({6-[(3-Amino-5-chlorophenyl)amino]-9-isopropyl-9H-purin-2-yl}amino)-3-methyl-1-butanol | Q00534 | CDK6 | 0.03319202 |
| Drug Name | Target Protein | Target Gene | Flow Value | Status | Reference |
|---|---|---|---|---|---|
| Dasatinib | P12931 | SRC | 0.08292133 | Confirmed | [34] |
| Phenethyl Isothiocyanate | P31946 | YWHAB | 0.06112281 | Confirmed | [35] |
| Adenosine-5′ | P00558 | PGK1 | 0.04545455 | Confirmed | [36] |
| Acetylsalicylic acid | P54646 | PRKAA2 | 0.03627599 | ||
| Glutamic Acid | P07814 | EPRS | 0.03527291 | Confirmed | [37] |
| Drug Name | Target Protein | Target Gene | Flow Value | Status | Reference |
|---|---|---|---|---|---|
| Phenethyl Isothiocyanate | P31946 | YWHAB | 0.05054656 | Confirmed | [38] |
| Minocycline | P42574 | CASP3 | 0.03767546 | Confirmed | [39] |
| Pseudoephedrine | P15336 | ATF2 | 0.03201844 | Confirmed | [38] |
| Methyl 4,6-O-[(1R)-1-carboxyethylidene]-beta-D-galactopyranoside | P02743 | APCS | 0.03150388 | ||
| NADH | O43920 | NDUFS5 | 0.02409639 | Confirmed | [40] |
| Method | Number of Confirmed Candidates in Top 5 Candidate Drug List | ||
|---|---|---|---|
| Breast Cancer | IBD | COPD | |
| Degree centrality | 0 | 0 | 1 |
| Closeness centrality | 2 | 1 | 0 |
| Betweenness centrality | 0 | 0 | 2 |
| Random walk [41] | 0 | 2 | 2 |
| Page rank [42] | 2 | 2 | 2 |
| Our proposed framework | 4 | 4 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, M.M.; Wang, Y.; Hu, P. A Maximum Flow-Based Approach to Prioritize Drugs for Drug Repurposing of Chronic Diseases. Life 2021, 11, 1115. https://doi.org/10.3390/life11111115
Islam MM, Wang Y, Hu P. A Maximum Flow-Based Approach to Prioritize Drugs for Drug Repurposing of Chronic Diseases. Life. 2021; 11(11):1115. https://doi.org/10.3390/life11111115
Chicago/Turabian StyleIslam, Md. Mohaiminul, Yang Wang, and Pingzhao Hu. 2021. "A Maximum Flow-Based Approach to Prioritize Drugs for Drug Repurposing of Chronic Diseases" Life 11, no. 11: 1115. https://doi.org/10.3390/life11111115
APA StyleIslam, M. M., Wang, Y., & Hu, P. (2021). A Maximum Flow-Based Approach to Prioritize Drugs for Drug Repurposing of Chronic Diseases. Life, 11(11), 1115. https://doi.org/10.3390/life11111115






