Efficient Isolation of Bacterial RNAs Using Silica-Based Materials Modified with Ionic Liquids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Chemical Modification of Silica with Ionic Liquids
2.3. Characterization of SILs
2.3.1. Solid-State 13C NMR Experiments
2.3.2. Elemental Analysis
2.3.3. Zeta Potential
2.3.4. Scanning Electron Microscopy
2.4. Escherichia coli DH5α Culture for Nucleic Acids Production
2.5. Isolation of Nucleic Acids from Escherichia coli DH5α
2.6. Agarose Gel Electrophoresis
2.7. RNA Binding Studies
2.8. Adsorption Kinetics
2.9. Binding Experiments of Bacterial Nucleic Acids in the IL-Functionalized Supports in Batch Mode
3. Results
3.1. Characterization of IL-Functionalized Silica Supports
3.1.1. Solid-State 13C NMR Analysis
3.1.2. Elemental Analysis
3.1.3. Zeta Potential Analysis
3.1.4. Scanning Electron Microscopy
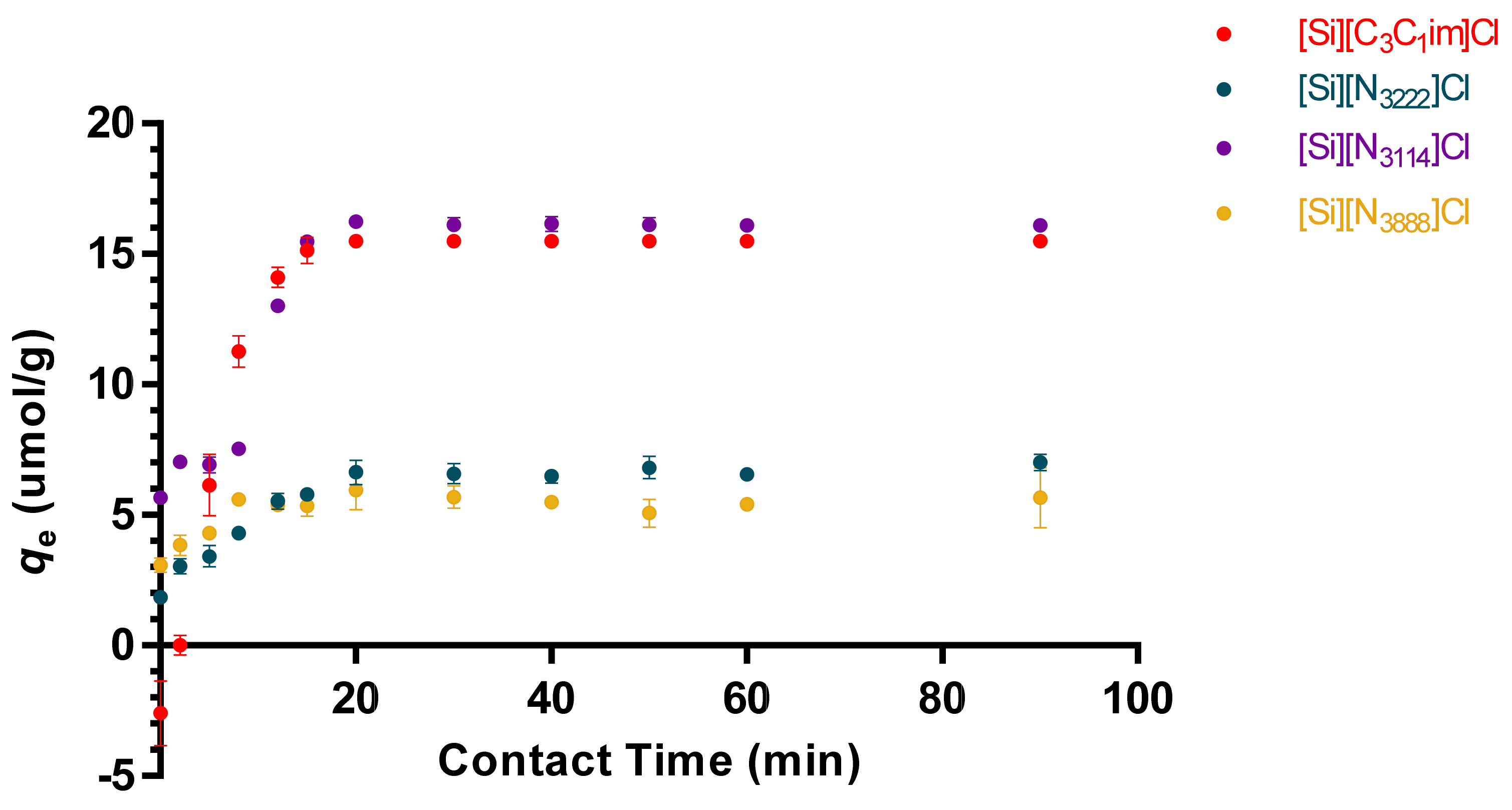
3.2. Adsorption Kinetics and Diffusion Models of tRNA in IL-Modified Supports
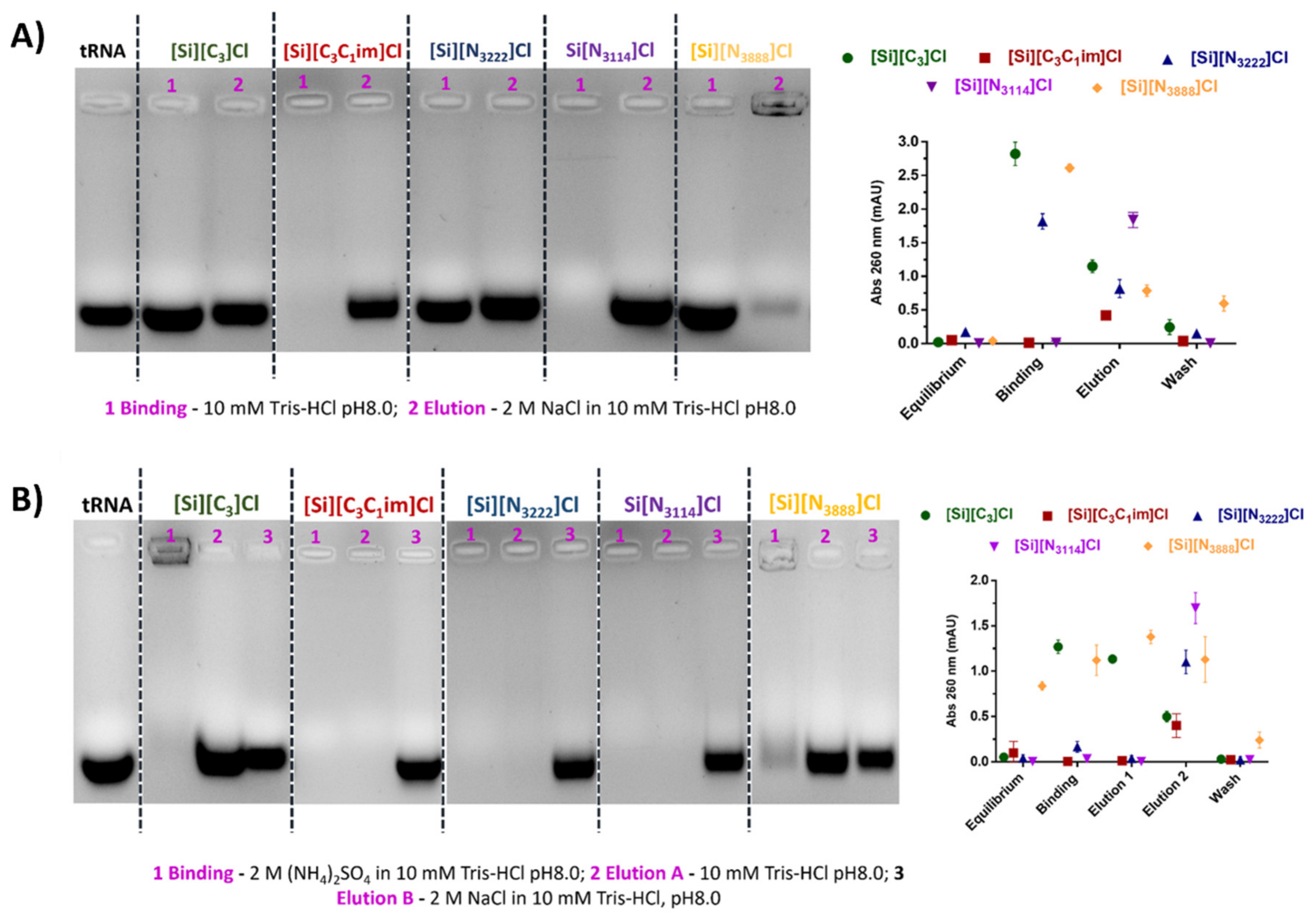
3.3. Binding/Elution Experiments of tRNA Using IL-Functionalized Silica Supports in Batch Mode
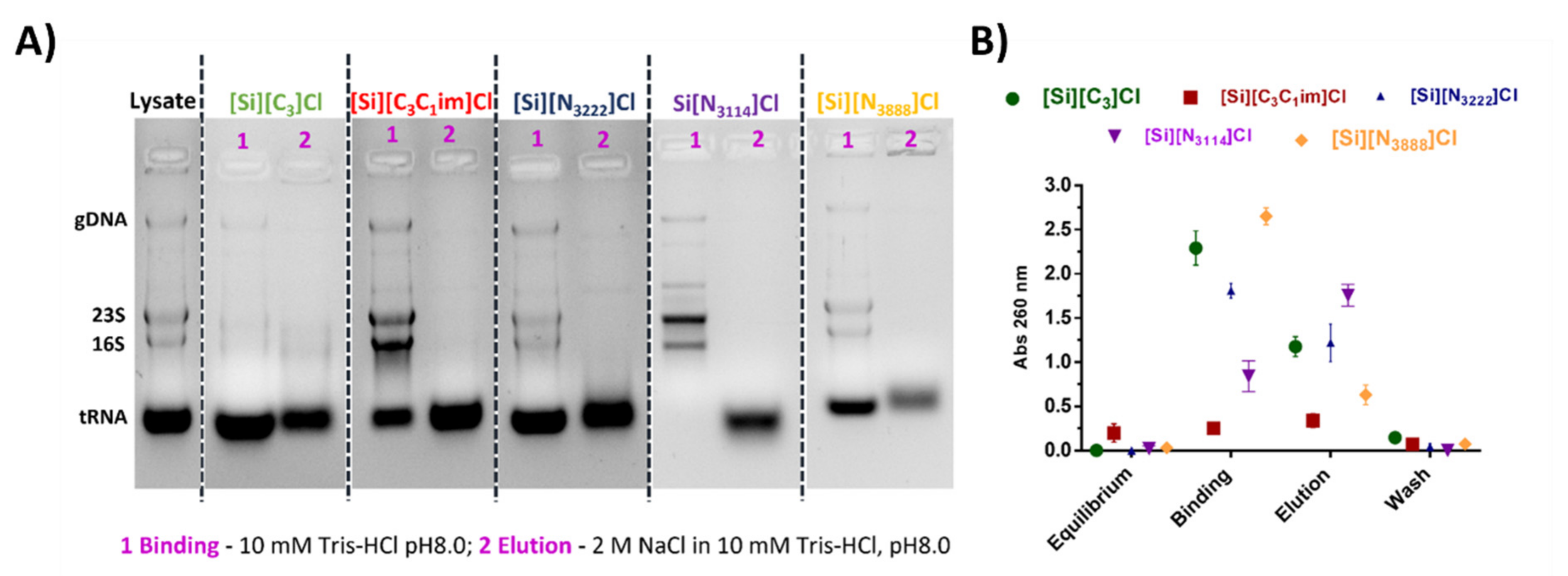
3.4. Isolation of tRNA from an Escherichia coli DH5α Extract Using IL-Functionalized Silica Supports
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crommelin, D.J.; Storm, G.; Verrijk, R.; de Leede, L.; Jiskoot, W.; Hennink, W.E. Shifting paradigms: Biopharmaceuticals versus low molecular weight drugs. Int. J. Pharm. 2003, 266, 3–16. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Platt, R.; Zhang, F. Therapeutic genome editing: Prospects and challenges. Nat. Med. 2015, 21, 121–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Moreira, E.D.; Zerbini, C.; et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef]
- Martins, R.; Queiroz, J.; Sousa, F. Ribonucleic acid purification. J. Chromatogr. A 2014, 1355, 1–14. [Google Scholar] [CrossRef]
- Rathore, A.S.; Kumar, D.; Kateja, N. Recent developments in chromatographic purification of biopharmaceuticals. Biotechnol. Lett. 2018, 40, 895–905. [Google Scholar] [CrossRef]
- Husain, S.R.; Han, J.; Au, P.; Shannon, K.; Puri, R.K. Gene therapy for cancer: Regulatory considerations for approval. Cancer Gene Ther. 2015, 22, 554–563. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. WHO/RNA/DRAFT/22 DECEMBER 2020—Evaluation of the Quality, Safety and Efficacy of RNA-Based Prophylactic Vaccines for Infectious Diseases: Regulatory Considerations. Available online: https://www.who.int/docs/default-source/biologicals/ecbs/reg-considerations-on-rna-vaccines_1st-draft_pc_tz_22122020.pdf?sfvrsn=c13e1e20_3 (accessed on 9 October 2021).
- Lalor, F.; Fitzpatrick, J.; Sage, C.; Byrne, E. Sustainability in the biopharmaceutical industry: Seeking a holistic perspective. Biotechnol. Adv. 2019, 37, 698–707. [Google Scholar] [CrossRef]
- Kesik-Brodacka, M. Progress in biopharmaceutical development. Biotechnol. Appl. Biochem. 2018, 65, 306–322. [Google Scholar] [CrossRef] [Green Version]
- Lowe, C.R.; Lowe, A.; Gupta, G. New developments in affinity chromatography with potential application in the production of biopharmaceuticals. J. Biochem. Biophys. Methods 2001, 49, 561–574. [Google Scholar] [CrossRef]
- Ferreira, G.N.; Monteiro, G.; Prazeres, D.M.; Cabral, J.M. Downstream processing of plasmid DNA for gene therapy and DNA vaccine applications. Trends Biotechnol. 2000, 18, 380–388. [Google Scholar] [CrossRef]
- Pereira, P.; Queiroz, J.A.; Figueiras, A.; Sousa, F. Affinity approaches in RNAi-based therapeutics purification. J. Chromatogr. B 2016, 1021, 45–56. [Google Scholar] [CrossRef]
- Freire, M.G.; Cláudio, A.F.M.; Araújo, J.M.M.; Coutinho, J.A.P.; Marrucho, I.M.; Lopes, J.N.C.; Rebelo, L.P.N. Aqueous biphasic systems: A boost brought about by using ionic liquids. Chem. Soc. Rev. 2012, 41, 4966–4995. [Google Scholar] [CrossRef]
- Ventura, S.; Silva, F.; Quental, M.J.; Mondal, D.; Freire, M.; Coutinho, J.A.P. Ionic-Liquid-Mediated Extraction and Separation Processes for Bioactive Compounds: Past, Present, and Future Trends. Chem. Rev. 2017, 117, 6984–7052. [Google Scholar] [CrossRef]
- Hallett, J.P.; Welton, T. Room-Temperature Ionic Liquids: Solvents for Synthesis and Catalysis. 2. Chem. Rev. 2011, 111, 3508–3576. [Google Scholar] [CrossRef]
- Skoda-Földes, R. The Use of Supported Acidic Ionic Liquids in Organic Synthesis. Molecules 2014, 19, 8840–8884. [Google Scholar] [CrossRef]
- Du, P.; Liu, S.; Wu, P.; Cai, C. Preparation and characterization of room temperature ionic liquid/single-walled carbon nanotube nanocomposites and their application to the direct electrochemistry of heme-containing proteins/enzymes. Electrochim. Acta 2007, 52, 6534–6547. [Google Scholar] [CrossRef]
- Singh, N.; Sharma, M.; Mondal, D.; Pereira, M.M.; Prasad, K. Very High Concentration Solubility and Long-Term Stability of DNA in an Ammonium-Based Ionic Liquid: A Suitable Medium for Nucleic Acid Packaging and Preservation. ACS Sustain. Chem. Eng. 2016, 5, 1998–2005. [Google Scholar] [CrossRef]
- Tateishi-Karimata, H.; Sugimoto, N. Structure, stability and behaviour of nucleic acids in ionic liquids. Nucleic Acids Res. 2014, 42, 8831–8844. [Google Scholar] [CrossRef]
- Kumar, A.; Venkatesu, P. Innovative aspects of protein stability in ionic liquid mixtures. Biophys. Rev. 2018, 10, 841–846. [Google Scholar] [CrossRef]
- Schindl, A.; Hagen, M.L.; Muzammal, S.; Gunasekera, H.A.D.; Croft, A.K. Proteins in Ionic Liquids: Reactions, Applications, and Futures. Front. Chem. 2019, 7, 347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazid, R.R.; Divisekera, U.; Yang, W.; Ranganathan, V.; MacFarlane, D.R.; Cortez-Jugo, C.; Cheng, W. Biological stability and activity of siRNA in ionic liquids. Chem. Commun. 2014, 50, 13457–13460. [Google Scholar] [CrossRef]
- Quental, M.V.; Pedro, A.Q.; Pereira, P.; Sharma, M.; Queiroz, J.A.; Coutinho, J.A.P.; Sousa, F.; Freire, M.G. Integrated Extraction-Preservation Strategies for RNA Using Biobased Ionic Liquids. ACS Sustain. Chem. Eng. 2019, 7, 9439–9448. [Google Scholar] [CrossRef]
- Pedro, A.; Pereira, P.; Quental, M.J.; Carvalho, M.; Santos, S.M.; Queiroz, J.A.; Sousa, F.; Freire, M.G. Cholinium-Based Good’s Buffers Ionic Liquids as Remarkable Stabilizers and Recyclable Preservation Media for Recombinant Small RNAs. ACS Sustain. Chem. Eng. 2018, 6, 16645–16656. [Google Scholar] [CrossRef] [PubMed]
- Fehrmann, R.; Haumann, M.; Riisager, A. Introduction. In Supported Ionic Liquids; John Wiley & Sons: New York, NY, USA, 2014; pp. 1–10. [Google Scholar]
- Campisciano, V.; Giacalone, F.; Gruttadauria, M. Supported Ionic Liquids: A Versatile and Useful Class of Materials. Chem. Rec. 2017, 17, 918–938. [Google Scholar] [CrossRef]
- Almeida, H.F.; Neves, M.C.; Trindade, T.; Marrucho, I.; Freire, M.G. Supported ionic liquids as efficient materials to remove non-steroidal anti-inflammatory drugs from aqueous media. Chem. Eng. J. 2020, 381, 122616. [Google Scholar] [CrossRef]
- Neves, M.; Pereira, P.; Pedro, A.; Martins, J.; Trindade, T.; Queiroz, J.; Freire, M.; Sousa, F. Improved ionic-liquid-functionalized macroporous supports able to purify nucleic acids in one step. Mater. Today Bio 2020, 8, 100086. [Google Scholar] [CrossRef]
- Bernardo, S.C.; Araújo, B.R.; Sousa, A.C.A.; Barros, R.A.; Cristovão, A.C.; Neves, M.C.; Freire, M.G. Supported Ionic Liquids for the Efficient Removal of Acetylsalicylic Acid from Aqueous Solutions. Eur. J. Inorg. Chem. 2020, 2020, 2380–2389. [Google Scholar] [CrossRef]
- Smoluchowski, M.V. Über Brownsche Molekularbewegung unter Einwirkung äußerer Kräfte und deren Zusammenhang mit der verallgemeinerten Diffusionsgleichung. Ann. Phys. 1916, 353, 1103–1112. [Google Scholar] [CrossRef] [Green Version]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- Cheung, C.; Porter, J.; Mckay, G. Sorption kinetic analysis for the removal of cadmium ions from effluents using bone char. Water Res. 2001, 35, 605–612. [Google Scholar] [CrossRef]
- Alothman, Z.A. A Review: Fundamental Aspects of Silicate Mesoporous Materials. Materials 2012, 5, 2874–2902. [Google Scholar] [CrossRef] [Green Version]
- Santos, T.; Proença, Z.; Queiroz, J.; Tomaz, C.; Cruz, C. Plasmid purification by using a new naphthalene tripodal support. Sep. Purif. Technol. 2017, 188, 81–89. [Google Scholar] [CrossRef]
- Pereira, P.; Sousa, Â.; Queiroz, J.; Correia, I.; Figueiras, A.; Sousa, F. Purification of pre-miR-29 by arginine-affinity chromatography. J. Chromatogr. B 2014, 951–952, 16–23. [Google Scholar] [CrossRef]
- Freitas, S.; Sousa, F.; Azzoni, A.R.; Prazeres, D.M.F.; Queiroz, J. Selective purification of supercoiled plasmid DNA from clarified cell lysates with a single histidine–agarose chromatography step. Biotechnol. Appl. Biochem. 2006, 45, 131–140. [Google Scholar] [CrossRef]

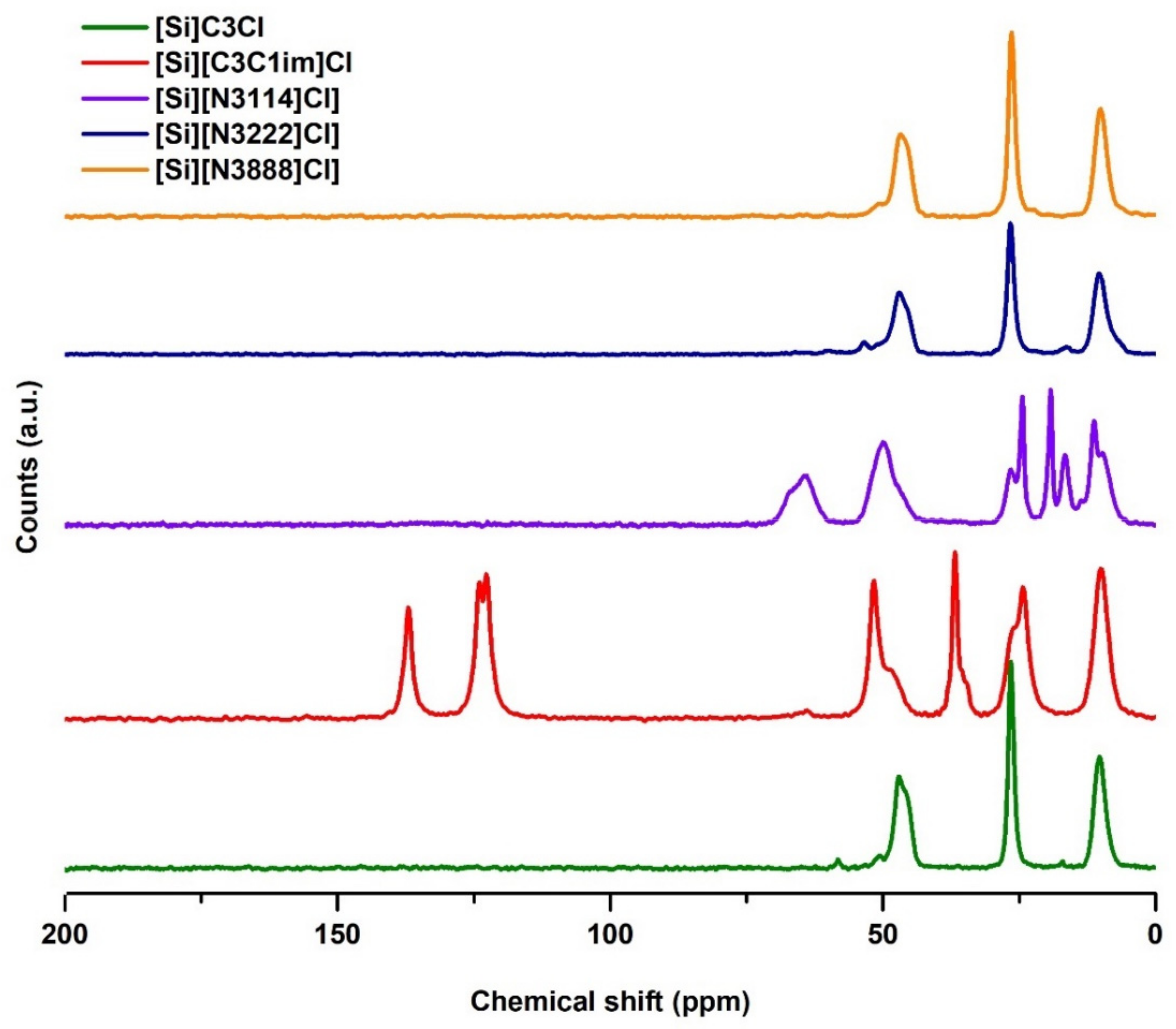
| Sample | wt% C | wt% H | wt% N | Ligand Density (nmol IL per g of Silica) 1 | PZC |
|---|---|---|---|---|---|
| [SiO2] | - | - | - | - | 3.4 |
| [Si][C3]Cl | 4.73 | 1.44 | 0.00 | - | 4.1 |
| [Si][C3C1im]Cl | 8.21 | 1.55 | 2.50 | 0.89 | 9.5 |
| [Si][N3114]Cl | 7.75 | 1.77 | 0.77 | 0.55 | 9.2 |
| [Si][N3222]Cl | 7.29 | 1.51 | 0.26 | 0.19 | 9.2 |
| [Si][N3888]Cl | 6.34 | 1.66 | 0.06 | 0.05 | 6.2 |
| Model/SIL | PFO | PSO | Elovich |
|---|---|---|---|
| [Si][C3C1im]Cl | qe = 6.060 mg g−1 K1 = 0.142 min−1 R2 = 0.970 | qe = 6.837 mg g−1 K2 = 0.267 g mg−1 min−1 R2 = 0.928 | β = 0.773 g mg−1 α = 3.068 mg g−1 min−1 R2 = 0.851 |
| [Si][N3114]Cl | qe = 6.607 mg g−1 K1 = 0.131 min−1 R2 = 0.981 | qe = 7.326 mg g−1 K2 = 0.265 g mg−1 min−1 R2 = 0.949 | β = 0.885 g mg−1 α = 7.787 mg g−1 min−1 R2 = 0.835 |
| [Si][N3222]Cl | qe = 6.060 mg g−1 K1 = 0.142 min−1 R2 = 0.970 | qe = 6.837 mg g−1 K2 = 0.267 g mg−1 min−1 R2 = 0.928 | β = 0.773 g mg−1 α = 3.068 mg g−1 min−1 R2 = 0.851 |
| [Si][N3888]Cl | qe = 5.424 mg g−1 K1 = 0.615 min−1 R2 = 0.988 | qe = 5.643 mg g−1 K2 = 0.212 g mg−1 min−1 R2 = 0.919 | β = 2.555 g mg−1 α = 1.428 mg g−1 min−1 R2 = 0.610 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, P.; Pedro, A.Q.; Neves, M.C.; Martins, J.C.; Rodrigues, I.; Freire, M.G.; Sousa, F. Efficient Isolation of Bacterial RNAs Using Silica-Based Materials Modified with Ionic Liquids. Life 2021, 11, 1090. https://doi.org/10.3390/life11101090
Pereira P, Pedro AQ, Neves MC, Martins JC, Rodrigues I, Freire MG, Sousa F. Efficient Isolation of Bacterial RNAs Using Silica-Based Materials Modified with Ionic Liquids. Life. 2021; 11(10):1090. https://doi.org/10.3390/life11101090
Chicago/Turabian StylePereira, Patrícia, Augusto Q. Pedro, Márcia C. Neves, João C. Martins, Inês Rodrigues, Mara G. Freire, and Fani Sousa. 2021. "Efficient Isolation of Bacterial RNAs Using Silica-Based Materials Modified with Ionic Liquids" Life 11, no. 10: 1090. https://doi.org/10.3390/life11101090
APA StylePereira, P., Pedro, A. Q., Neves, M. C., Martins, J. C., Rodrigues, I., Freire, M. G., & Sousa, F. (2021). Efficient Isolation of Bacterial RNAs Using Silica-Based Materials Modified with Ionic Liquids. Life, 11(10), 1090. https://doi.org/10.3390/life11101090










