Abstract
Aims: The aim of the work was to establish potential biomarkers or drug targets by analysing changes in miRNA concentration among patients with Gaucher disease (GD) compared to in healthy subjects. Methods: This study was an observational, cross-sectional analysis of 30 adult participants: 10 controls and 20 adults with GD type 1. Patients with GD type 1 were treated with enzyme replacement therapy (ERT) for at least two years. The control group was composed of healthy volunteers, unrelated to the patients, adjusted with age, sex and body mass index (BMI). The miRNA alteration between these groups was examined. After obtaining preliminary results on a group of six GD patients by the high-output method (TaqMan low-density array (TLDA)), potential miRNAs were selected for confirming the results by using the qRT-PCR method. With Diane Tools, we analysed miRNAs of which differential expression is most significant and their potential role in GD pathophysiology. We also determined the essential pathways these miRNAs are involved in. Results: 266 dysregulated miRNAs were found among 753 tested. Seventy-eight miRNAs were downregulated, and 188 were upregulated. Thirty miRNAs were significantly altered; all of them were upregulated. The analysis of pathways regulated by the selected miRNAs showed an effect on bone development, inflammation or regulation of axonal transmission in association with Parkinson’s disease. Conclusions: We revealed few miRNAs, like miR-26-5p, which are highly altered and fit the GD pathophysiological model, might be considered as novel biomarkers of disease progression but need further evaluation.
1. Introduction
Among lysosomal storage diseases, Gaucher disease (GD) is one of the most common. It is estimated that around 1 per 57,000 newborns is affected worldwide [1]. The disease’s phenotype is associated with a mutation in the lysosomal glucosylceramidase (GBA) gene; however, it differs remarkably even in patients with the same type of mutation [2,3,4]. Three types of GD are recognized upon central nervous system (CNS) involvement: non-neuronopathic (GD type 1: OMIM #230800), acute neuronopathic (GD type 2: OMIM #230900) and chronic neuronopathic (GD type 3: OMIM #231000). The most common form of GD, covering approximately 94% of cases, is type 1, which can manifest itself at any age [5]. It is characterised by haematological changes such as thrombocytopenia, anaemia and highly pronounced splenomegaly. Sometimes, hepatomegaly may coexist, and changes occurring in the skeletal system are also important [5]. Type 2 with a very severe course, regardless of the treatment attempts, leads to death around two years of age, while type 3, in addition to organ changes, is also associated with the involvement of the central nervous system, characterised by the presence of drug-resistant epilepsy; however, in this case, patients live to adulthood [5].
GD patients are also more susceptible to some diseases like heamatological neoplasms, Parkinson’s disease (PD) or central nervous involvement than the normal population [6].
One of the suspected reasons for that variability is an epigenetic mechanism that can modulate the course of the illness [7]. That is carried by, among others, microRNAs (miRNA), which are small noncoding RNA molecules found in almost all life forms and are responsible for RNA silencing and post-transcriptional regulation of gene expression. Therefore, to delve into it, we marked the expression of 753 miRNAs in GD patients, compared it to that in the control, healthy group, and for those miRNAs that were altered, we evaluated their potential role, compared with available data and analysed molecular pathways that they are involved in. All of the tested miRNAs are shown in the Appendix A (Table A1).
Currently, the possible role of organ-specific miRNAs in predicting the development of metabolic diseases such as diabetes and its complications or the development of complications in the course of lysosomal storage diseases (LSDs) is widely discussed [8,9]. Due to the stability of miRNAs in body fluids and distribution in specific tissues, it may turn these particles into a desirable biomarker of certain diseases [9]. Siebert, in his work on fibroblast cell lines from GD patients, showed that some miRNAs had a strong effect on the regulation of β-glucocerebrosidase (GCase) activity [10]. Two miRNAs (miR-195-5p and miR-16-5p) strongly stimulated GBA1 gene expansion, while miR-127-5p, miR-19a-5p and miR-1262 inhibited the expression of SCARB2, a membrane receptor regulating GCase activity and availability [10]. MicroRNAs were shown to downregulate GBA and GBAP1 (miR-22-3p) [11]. Some miRNAs like miR-let7b, miR-29b or miR-142 were associated with proinflammatory events [12]. Recently, Watson et al. described an unsuccessful ablation of miR-155, which was identified in knocked-down gba1 zebrafish (in vivo GD model) as a proinflammatory master regulator [13]. However, the work on the role of miRNAs in GD only focused on the modifying role of this epigenetic mechanism in GBA gene expression [10]. It is worth remembering that most of these works concerned in vivo and in vitro observation, and we identified only one human observation [10].
Nevertheless, the big picture is still missing. Therefore, our work aimed to investigate changes in miRNAs concentration in patients with GD who were treated with enzyme replacement therapy (ERT) and to try finding new diagnostic markers or potential targets for treatment.
2. Material and Methods
This study was an observational, cross-sectional analysis of 30 adult participants: 20 adults with GD type 1 and 10 controls. First, samples from 8 random persons (4 from the GD group and 4 from the control one) were used for the TaqMan low-density array (TLDA), a high-throughput method for identifying alterations in miRNAs expression levels. Based on these preliminary results, potential miRNAs were selected for a confirmation on a whole group (30 persons) using the more precise qRT-PCR method. Patients with GD type 1 were treated in the Metabolic Diseases and Diabetes Department in the Jagiellonian University Hospital in Krakow, part of EU reference centres—MetabERN. The study included adult people with GD, treated in our centre, who agreed to participate in the study and signed up an informed consent. GD diagnosis was established on the positive genetic or/and enzymatic tests and, in two cases, additional histopathological examination. All these patients underwent ERT for at least two years. The exclusion factors for the analysis were as follows: neoplastic diseases, PD, autoimmune diseases, pregnancy, infection, fever or lack of patient’s consent to participate in the study. The control group was composed of healthy volunteers, not related to the patients, adjusted with age, sex and BMI.
All the participants underwent a detailed physical examination (age, weight, height, BMI, age and any disease history). Fasting venous blood was used for molecular biology tests. Molecular tests were performed at the Clinical Biochemistry Department of the Jagiellonian University. The clinical characteristic of groups is presented in Table 1.

Table 1.
Clinical characteristics of study participants with GD (n = 20) and the control group (n = 10).
3. The miRNA Analysis
3.1. RNA Isolation
The purification of total RNA from human whole blood was performed using the PAXgene Blood RNA Kit (Qiagen, Germantown, MD, USA), following the manufacturer’s protocol. The RNA quality was analysed using the Tapestation 2200 instrument (Agilent Technologies, Santa Clara, CA, USA) and quantified by spectrophotometry on the NanoDrop (Thermo Fisher Scientific, Wilmington, DE, USA).
3.2. TLDA
The initial expression analysis of human microRNAs (753 unique assays) was performed by a high-throughput method using the TLDAs Panel v3.0 (Applied Biosystems, Foster City, CA, USA), as described by the manufacturer. Total RNA enriched with microRNAs was reverse-transcribed using stem-loop primers. In order to detect low abundant microRNAs, a preamplification step was performed. The preamplified product was loaded into a TLDA, and amplification signal detection was performed in the 7900 FAST real-time thermal cycler (ABI) (Thermo Fisher Scientific, Wilmington, DE, USA). Data were exported to the DataAssist software version 3.01 (Life Technologies, now Thermo Fisher Scientific, Waltham, MA, USA) and normalised using the small-nucleolar RNA U6snRNA. Mean relative quantity (RQ) was calculated, and microRNAs differentially expressed between groups were defined as those with a fold change of >1.5 and a p-value of <0.05. The differentially expressed miRNAs based on the TLDAs results were verified using microRNA assays (TaqMan Array Human MicroRNA Panel v3.0).
3.3. qRT-PCR
Subsequently, to validate the microarray experiment, qRT-PCR was conducted on 20 patients with GD type 1 and 10 volunteers from the control group. Total RNA was reverse-transcribed using a specific stem-loop RT primer for each microRNA and the MultiScribeTM reverse transcriptase (Thermo Fisher Scientific, Wilmington, DE, USA). Then, diluted retro-transcription reaction (1:15) was mixed with the Universal PCR Master Mix, No AmpErase R UNG (2X), in the presence of individual PCR microRNAs. The PCR reactions were done in a GeneAmp R PCR System 9700 (Applied Biosystems, Foster City, CA, USA), using the following program: 95 °C for 10 min, 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The relative expression of microRNAs was measured by qRT-PCR using the comparative Ct (∆∆CT) method. A two-tailed Student’s t-test comparing the 2(–ΔCt) values of the two groups was performed, and the Benjamini–Hochberg-adjusted p-value was calculated. The U6 snoRNA was used as an internal control for data normalisation.
3.4. Pathway Enrichment
MicroRNA targets were predicted using a microRNA target prediction program: DIANA-microT (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index). Biological pathways of predicted target-genes microRNAs were identified using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (http://www.genome.jp/kegg/pathway.html) and mirPath v.3 (http://snf-515788.vm.okeanos.grnet.gr/).
Briefly, the DIANA-mirPath utilised predicted miRNA targets provided by the miRNA-gene interactions derived from experimentally supported DIANA-TarBase v.7.0 (Department of Electrical and Computer Engineering, 382 21 Volos, Greece) or microT-CDS target prediction, if there were no hits for DIANA-TarBase. The KEGG pathways union was used to identify the main pathways targeted by predicted genes (discovered miRNA targets).
3.5. Statistical Analysis of Patients’ Baseline Characteristics
For the analysis of the characteristics of the study groups, IBM SPSS Statistic Software was used. The Shapiro–Wilk test was performed to check the normality. Differences between groups were assessed using the T-test or U-test for normally and non-normally distributed continuous variables, respectively, and by the chi-square or Fischer’s exact test for categorical variables. Continuous variables are presented as mean ± SD or median (interquartile range). Statistical significance was set at p < 0.05.
4. Results
The analysed groups: control and GD patients were comparable in gender distribution, age and anthropometric parameters. To unify patients’ group as much as possible, all of the patients with GD type 1 were treated with ERT.
Using the TLDA method, we were able to perform an accurate quantitation of 753 microRNAs in the study group. U6 comparison data were used for normalization. Alterations in miRNAs expression were determined in 266 cases, among which 78 were downregulated and 188 were upregulated. Eventually, among all tested miRNAs, we identified 30 miRNAs, of which the expression significantly differed between GD patients and our control data (Table 2). Interestingly, all these miRNAs were upregulated in relation to the control group.

Table 2.
30 significantly altered microRNA level in GD patient’s group vs control; U6 was used as a reference.
The most prominent alteration in the GD group included miR-26b-5p, miR-31-5p, miR-29a-3p, miR-454-3p, miR-660-5p and miR-148a-3p. The miRNAs were mapped for molecular pathways to identify those, which could be specifically modified in GD patients (Table 3). Among pathways, we chose the most noticeable ones according to the p-value, the proportion of miRNAs to controlled genes and actual knowledge about the pathophysiology of the lysosomal storage disease.

Table 3.
KEGG pathway analysis of miRNA target genes, significantly altered with p < 0.05.
The analysis using a heatmap (Figure A1) showed a strong relationship between the TGF-beta signal transmission pathway and miR-106a-5p. In addition, a significant association was found between pathways of fatty acid synthesis and metabolism, with miR-16-5p and miR-195-5p. Interestingly, these miRNAs played also a significant role in the regulation of the Akt-signalling pathway, focal adhesion or lysine degradation pathways. Figure 1 Additionally, we noted an important impact of differentially expressed miRNA on a very complex pathway of extracellular matrix (ECM)–receptor interaction, namely miR-29a-3p or miR-let-7g-5p.
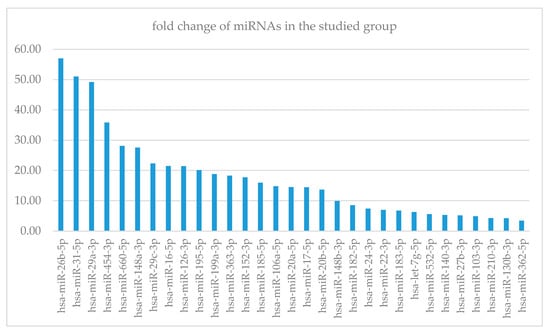
Figure 1.
Fold change of miRNA concentration between GD group and control. X-axis—miRNAs, Y-axis—fold change.
5. Discussion
5.1. Comparison to Available Data
The biological purpose of miRNAs is related to their conservative function, common for many lifeforms, to control groups of genes by muting them when needed. They influence biochemical pathways on many levels to create a complex and tangled network of dependencies and controlling apparatus.
Nowadays, research to recognise miRNA in-depth role in the ability to regulate genes expression is blooming. As for GD, only a few miRNAs analyses have been described so far. In 2014, Siebert reported data for two miRNAs: 195-5p and 16-5p concentration changes in GD and linked them to the regulation (increase) of GCase activity [10]. Our data showed the upregulation of these two. In the presence of GCase insufficiency, the results suggested a secondary hyperconcentration of regulators and may confirm in vivo the importance of miR-195-5p and miR-16-5p in GCase secretion.
5.2. GD and Malignancies
miR-195-5p was also recently found to be downregulated in a broad spectrum of solid cancers. It was described as a cancer suppressor [14], which mediates its effect by complex and still vogue pathways. Similarly, miR-16-5p was described as a protective factor in breast carcinoma [15], and again, we showed significant upregulation of this miRNA.
Highly elevated levels of miR-195-5p and 16-5p may provide a protective role in GD in terms of solid tumours [16,17,18]. Until now, GD patients were found to be more susceptible primarily for haematologic neoplasms [19]. However, the data of breast cancer in GD are inconsistent. In 2015, European Registry did not report any incidence of breast cancer; neither did Shiran in 1993 [19,20]. In 2009, Taddei reported nine cases of breast cancer, with a risk ratio of 1.84 [21]. Therefore, these observations require further studies.
For miR-148b-3p, the connection between high concentration and poor survival in breast cancer was established [22]. Our data showed a 27.5-fold increase in this miRNA in the study group, but again, there are no available studies that investigated the mentioned issue.
Few papers described an increased number of cases with hepatocellular carcinoma (HCC) [23]. Zhao in 2017 revealed that the expression of miR-31-5p was significantly upregulated in HCC tissues [24]. This information is consistent with our analysis showing highly altered miR-31-5p that was more than 50 times higher than in the comparison group. In our tested groups, there was no HCC incidence. The question remains: is miR-31-5p just a marker for HCC or does it present the properties for the highly sensitive biomarker of the liver lesion which could reveal the destruction of hepatic cells, even when classical tests such as alanine aminotransferase (ALT) and aspartate aminotransferase (AST) are within a normal range?
All the patients in the study group were on ERT for at least two years. This treatment could influence the concentrations of many miRNAs and change the course of the disease, including complication occurrence. However, the data from European Registry [20] did not reveal differences in cancer incidence between patients ever treated with ERT and those never treated with ERT, but still, the impact of ERT on miRNAs, which are associated with malignancies, is limited as the data are missing. There are also no data on the dependence of the miRNA concentration on the ERT presence.
5.3. Other Highly Upregulated miRNA
The most prominent upregulated miRNA was found to be miR-26b-5p, which was linked with TRPS1 gene, which is responsible for regulating genes involved in the growth of bone and cartilage. Therefore TRPS1 controls chondrocyte proliferation and differentiation. MiR-26b-5p controls also bone morphogenetic protein 2 (BMP-2) that affects osteoblasts and regulates proliferation, differentiation and apoptosis [25]. In patients with GD, bone involvement is one of the main symptoms and consists of distortion of the vertebra and distension of long bones, known as Erlenmeyer flask bone deformity, with infiltration of bone marrow. It has been demonstrated that miR-26b-5p with a 66-fold increase of expression in our study group is a potential biomarker of bone involvement.
5.4. miRNA Pathways Analysis
To put altered miRNAs into the bigger picture, we used the DIANA-mirPath v3 tool for analysing miRNA-predicted targets with KEGG pathways. Nineteen pathways were significantly altered. Those, most noticeable influenced and possibly important in GD pathophysiology are “TGF-beta signalling pathway”, “focal adhesion”, “FoxO signalling pathway”, “axon guidance” and “ECM–receptor interaction”.
The TGF-beta signalling pathway is found in numerous human disorders. As it controls the growth inhibitors, the alteration of its activity is observed in cancers [26], which are more frequent in GD. In addition, TGF-beta stimulates ECM deposition that may lead to fibrosis and scarring [27]. The higher prevalence of liver fibrosis in GD was also shown [28].
The focal adhesion pathway focuses on the regulation of call–matrix interaction. The central point of that pathway—focal adhesion kinase (FAK) was roughly studied in terms of cancer progression [29,30,31]. Likewise, FAK was found to promote proinflammatory gene expression via TNF-α or IL-1β [32].
The FoxO signalling pathway, on the other hand, is involved in oxidation stress and may play an important role in neurodegenerative disease like Parkinson’s [33,34].
The axon guidance pathways involve, among others, miR-20a-5p, 106a-5p, 27b-3p or 182-5p as regulators, which in our analysis showed significant overexpression. Among these, 182-5p was found to interact with cofilin-1 (CFL1) and further axon growth [34]. As axon guidance is responsible for controlling neurons morphogenesis and signalling processes [35], its malfunction may contribute to neuropathy in GD patients.
These data confirmed the previously described role of inflammatory processes and the disrupted pathways of cell migration and interaction with the ECM, that consists of a mixture of structural and macromolecules and serves an important role not only in tissue morphogenesis but also in the regulation of cell and tissue structure and function.
5.5. miRNA and PD
Since the relationship between PD and GD was revealed, many studies were trying to establish the underlying pathophysiology. It was shown that patients with PD had a decreased level of GCase in the brain tissue. Moreover, even carries of the single GBA gene mutation present tendency towards PD [36]. This finding showed that GD patients should be more susceptible to the occurrence of PD, which was confirmed in [36]. Research on the expression of genes related to neuroinflammation and synaptic plasticity are ongoing [12]. MiR-26b and miR-106a-5p acting through HSPA8 [37], as well as miR-7 and miRNA-153, are found to increase the concentration of alpha-synuclein [38], the primary accumulation material in PD. We noted a 14-fold increase of miR-106-5p in our study group, which is consistent with the known data.
6. Conclusions
GD patients are still in need of new biomarkers and therapeutic targets that can help evaluate disease progression or treatment effectiveness. We highlighted some miRNA compounds that showed remarkable alteration in those patients and might be potential biomarkers. However, due to scarce data on GD, these findings need further studies.
Author Contributions
Ł.P., M.S., E.T. and M.K. performed the research. B.S., M.S. and A.P. analysed data. B.K.-W. designed the research study and interpreted the data. Ł.P. and B.K.-W. wrote the paper. B.K.-W. and Ł.P. are the guarantors of this work. All authors have read and agreed to the published version of the manuscript.
Funding
The study was funded by the Jagiellonian University Scientific Grant to B.K.-W. (No. N41/DBS/000069).
Institutional Review Board Statement
All procedures performed in this study, involving human participants, were in accordance with the ethical standards of the Jagiellonian University Bioethical Committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards (KBET 122.6120.90.2015).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Acknowledgments
Several authors of this publication are members of the European Reference Network for Rare Hereditary Metabolic Disorders (MetabERN)—Project ID No 739543.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
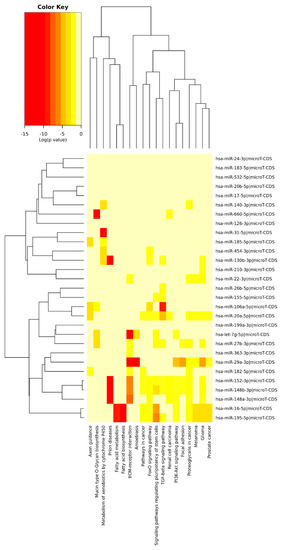
Figure A1.
Heatmap presenting miRNA impact in specific pathways.

Table A1.
List of all of the tested miRNAs. For 266 altered miRNAs: fold change, p-value and SD data are presented.
Table A1.
List of all of the tested miRNAs. For 266 altered miRNAs: fold change, p-value and SD data are presented.
| miRNA | Fold Change | p-Value | SD |
|---|---|---|---|
| hsa-miR-152-000475 | 18.1382 | 0.0003 | 1.956398 |
| hsa-miR-454-002323 | 38.5767 | 0.0004 | 0.887427 |
| mmu-miR-140-001187 | 32.7431 | 0.0022 | 0.796695 |
| hsa-miR-532-001518 | 6.7361 | 0.0068 | 1.858579 |
| hsa-miR-148b-000471 | 11.4268 | 0.0084 | 0.930063 |
| hsa-miR-17-002308 | 16.9061 | 0.0088 | 1.105731 |
| hsa-miR-148a-000470 | 32.9918 | 0.0089 | 1.748115 |
| mmu-miR-93-001090 | 5.7093 | 0.0089 | 0.81462 |
| hsa-miR-106a-002169 | 17.3487 | 0.0095 | 1.104199 |
| hsa-miR-20a-000580 | 17.0211 | 0.0097 | 1.115662 |
| hsa-miR-324-5p-000539 | 26.0859 | 0.01 | 0.863699 |
| hsa-miR-16-000391 | 24.6779 | 0.0102 | 1.212765 |
| hsa-miR-660-001515 | 32.8325 | 0.0105 | 1.537621 |
| hsa-miR-126-002228 | 24.5953 | 0.0106 | 1.214868 |
| hsa-miR-26b-000407 | 66.9029 | 0.011 | 0.894591 |
| hsa-miR-29c-000587 | 22.6399 | 0.0111 | 0.729712 |
| hsa-miR-140-3p-002234 | 6.0836 | 0.0112 | 1.477748 |
| hsa-miR-30b-000602 | 4.2752 | 0.0156 | 1.025196 |
| hsa-miR-106b-000442 | 34.8823 | 0.0158 | 1.020234 |
| hsa-miR-146b-001097 | 28.6668 | 0.0171 | 0.745407 |
| mmu-miR-451-001141 | 22.3568 | 0.0173 | 1.281826 |
| hsa-miR-212-000515 | 4.014 | 0.0191 | 1.005979 |
| hsa-miR-31-002279 | 61.5757 | 0.0225 | 3.805273 |
| hsa-miR-182-002334 | 10.9175 | 0.0269 | 1.014663 |
| hsa-miR-20b-001014 | 15.102 | 0.0281 | 0.968618 |
| hsa-miR-185-002271 | 19.9198 | 0.0291 | 1.583043 |
| hsa-miR-27a-000408 | 22.6143 | 0.0302 | 1.593832 |
| hsa-miR-22-000398 | 8.934 | 0.0317 | 1.16643 |
| hsa-miR-26a-000405 | 19.104 | 0.0328 | 1.213354 |
| mmu-miR-374-5p-001319 | 10.4528 | 0.0329 | 1.260503 |
| hsa-miR-128a-002216 | 15.2816 | 0.033 | 2.046559 |
| hsa-miR-18a-002422 | 40.5059 | 0.0367 | 1.138078 |
| hsa-miR-210-000512 | 5.1833 | 0.0379 | 1.053872 |
| hsa-miR-194-000493 | 30.5951 | 0.0388 | 1.389726 |
| hsa-miR-192-000491 | 31.244 | 0.0389 | 0.902125 |
| hsa-miR-191-002299 | 5.5793 | 0.0401 | 1.600253 |
| hsa-miR-28-000411 | 19.4532 | 0.0405 | 0.901813 |
| hsa-miR-146a-000468 | 30.847 | 0.0429 | 1.382615 |
| hsa-miR-130b-000456 | 5.3197 | 0.0433 | 1.578003 |
| hsa-miR-184-000485 | 5.5476 | 0.0444 | 0.8337 |
| hsa-miR-362-001273 | 3.8332 | 0.0449 | 0.995367 |
| hsa-miR-130a-000454 | 37.3076 | 0.0469 | 1.578769 |
| hsa-miR-500-002428 | 2.8525 | 0.047 | 1.113499 |
| hsa-miR-195-000494 | 26.5732 | 0.0487 | 1.167158 |
| hsa-miR-199a-3p-002304 | 23.1246 | 0.0534 | 1.014663 |
| hsa-miR-502-3p-002083 | 3.3767 | 0.0548 | 0.812816 |
| hsa-miR-155-002623 | 9.7213 | 0.0577 | 1.37917 |
| hsa-miR-590-5p-001984 | 324.1959 | 0.0578 | 0.642024 |
| hsa-miR-24-000402 | 9.9502 | 0.0581 | 1.384917 |
| hsa-miR-374-000563 | 73.6336 | 0.0587 | 0.699017 |
| hsa-miR-339-3p-002184 | 13.3932 | 0.0609 | 1.538581 |
| hsa-miR-425-5p-001516 | 3.957 | 0.061 | 1.162153 |
| hsa-miR-27b-000409 | 6.8449 | 0.0625 | 1.167805 |
| hsa-miR-25-000403 | 7.1621 | 0.063 | 1.363864 |
| hsa-miR-29a-002112 | 62.4411 | 0.0656 | 1.976845 |
| hsa-miR-345-002186 | 7.4086 | 0.066 | 1.380509 |
| hsa-miR-183-002269 | 9.2849 | 0.0686 | 0.889644 |
| hsa-miR-331-000545 | 3.9872 | 0.0687 | 1.152766 |
| hsa-miR-103-000439 | 6.6069 | 0.0699 | 1.220862 |
| hsa-miR-101-002253 | 19.0521 | 0.0701 | 0.592054 |
| hsa-miR-30c-000419 | 3.1992 | 0.0723 | 1.025552 |
| hsa-miR-133a-002246 | 9.0033 | 0.0726 | 1.228076 |
| hsa-miR-223-002295 | 16.2644 | 0.0772 | 1.533045 |
| hsa-miR-744-002324 | 2.6774 | 0.0798 | 1.38857 |
| hsa-miR-221-000524 | 2.9226 | 0.0807 | 1.312939 |
| hsa-miR-363-001271 | 25.8516 | 0.0846 | 1.616082 |
| hsa-miR-376c-002122 | 33.9427 | 0.0911 | 0.474342 |
| hsa-miR-139-5p-002289 | 7.0336 | 0.0913 | 1.214447 |
| hsa-miR-132-000457 | 3.8428 | 0.096 | 1.014592 |
| hsa-miR-301-000528 | 64.8911 | 0.0976 | 0.509057 |
| hsa-miR-181c-000482 | 4.708 | 0.1 | 1.0808 |
| hsa-let-7g-002282 | 8.7654 | 0.1014 | 1.107341 |
| hsa-let-7f-000382 | 9.0417 | 0.1016 | 0.968148 |
| hsa-miR-361-000554 | 18.4177 | 0.1029 | 2.978323 |
| hsa-miR-21-000397 | 48.0159 | 0.103 | 0.939588 |
| hsa-miR-200c-002300 | 3.3312 | 0.1051 | 1.512568 |
| hsa-miR-186-002285 | 16.6037 | 0.1108 | 0.786326 |
| hsa-miR-27a#-002445 | 7.3705 | 0.1117 | 0.965802 |
| hsa-miR-502-001109 | 2.6428 | 0.1137 | 0.975355 |
| hsa-miR-501-001047 | 2.5867 | 0.1185 | 1.056212 |
| hsa-miR-222-002276 | 11.368 | 0.1195 | 1.742912 |
| hsa-miR-342-5p-002147 | 3.5973 | 0.1248 | 1.323263 |
| hsa-miR-126#-000451 | 40.9923 | 0.126 | 0.935753 |
| hsa-miR-19b-000396 | 91.4227 | 0.1261 | 0.75054 |
| hsa-miR-19a-000395 | 190.411 | 0.1311 | 0.747321 |
| hsa-let-7e-002406 | 4.4222 | 0.1334 | 1.317315 |
| hsa-miR-28-3p-002446 | 2.6331 | 0.1365 | 0.962861 |
| hsa-miR-539-001286 | 10.361 | 0.1371 | 0.73037 |
| hsa-miR-1233-002768 | 0.1814 | 0.1381 | 1.095964 |
| hsa-miR-376a-000565 | 6.0198 | 0.1459 | 0.517776 |
| hsa-miR-629-002436 | 4.9459 | 0.1487 | 1.014592 |
| hsa-miR-324-3p-002161 | 3.153 | 0.1501 | 1.014592 |
| hsa-miR-1255B-002801 | 9.2195 | 0.1519 | 1.384534 |
| hsa-miR-335-000546 | 57.1738 | 0.1536 | 0.403013 |
| hsa-miR-142-3p-000464 | 140.5412 | 0.1621 | 0.664435 |
| hsa-miR-181a-000480 | 3.2078 | 0.1621 | 1.395227 |
| hsa-miR-1227-002769 | 0.1024 | 0.1633 | 0.966204 |
| hsa-let-7a-000377 | 3.2189 | 0.1692 | 1.31003 |
| hsa-let-7d-002283 | 3.2535 | 0.1707 | 1.318228 |
| hsa-miR-204-000508 | 13.0355 | 0.1763 | 1.520136 |
| hsa-miR-10a-000387 | 5.052 | 0.1827 | 0.691164 |
| hsa-miR-1260-002896 | 0.1645 | 0.1841 | 1.057165 |
| hsa-miR-628-3p-002434 | 5.0633 | 0.1875 | 1.168615 |
| hsa-miR-365-001020 | 2.3993 | 0.19 | 1.761739 |
| hsa-miR-744#-002325 | 4.6279 | 0.1909 | 2.144439 |
| hsa-miR-1290-002863 | 0.1228 | 0.1913 | 0.690733 |
| hsa-miR-370-002275 | 5.138 | 0.1967 | 0.96788 |
| hsa-miR-520c-3p-002400 | 0.2261 | 0.197 | 1.062601 |
| hsa-miR-1238-002927 | 0.0698 | 0.2007 | 1.014592 |
| hsa-miR-26b#-002444 | 6.0929 | 0.2032 | 1.40211 |
| rno-miR-7#-001338 | 79.9503 | 0.2054 | 3.010285 |
| hsa-miR-1282-002803 | 0.1537 | 0.2059 | 2.198623 |
| hsa-miR-1285-002822 | 6.123 | 0.206 | 1.266722 |
| hsa-miR-1249-002868 | 0.1254 | 0.2069 | 0.976979 |
| hsa-miR-190b-002263 | 6.6002 | 0.209 | 1.199389 |
| hsa-miR-193a-5p-002281 | 2.3905 | 0.2092 | 1.38934 |
| hsa-miR-151-5P-002642 | 5.9735 | 0.2108 | 1.25475 |
| hsa-miR-211-000514 | 3.3212 | 0.2137 | 2.352509 |
| hsa-miR-486-001278 | 0.1804 | 0.2163 | 1.319325 |
| hsa-miR-505-002089 | 3.0798 | 0.2207 | 1.078406 |
| U6 snRNA-001973 | 1.6133 | 0.2224 | 0.81169 |
| hsa-miR-223#-002098 | 13.2203 | 0.225 | 1.208402 |
| hsa-miR-1224-3P-002752 | 0.0546 | 0.2252 | 1.175195 |
| hsa-miR-26a-1#-002443 | 11.4173 | 0.2262 | 1.014592 |
| hsa-miR-655-001612 | 2.2436 | 0.2283 | 1.230803 |
| hsa-miR-15b-000390 | 3.0261 | 0.2297 | 1.222556 |
| hsa-miR-618-001593 | 2.3227 | 0.2301 | 0.922167 |
| hsa-miR-100-000437 | 5.4736 | 0.2319 | 0.585037 |
| hsa-miR-1236-002761 | 0.0953 | 0.2334 | 1.026049 |
| hsa-miR-196b-002215 | 9.1066 | 0.2345 | 1.099845 |
| hsa-miR-139-3p-002313 | 2.3313 | 0.2359 | 1.557573 |
| hsa-miR-197-000497 | 0.1018 | 0.2361 | 1.170318 |
| hsa-miR-425#-002302 | 3.2533 | 0.2364 | 1.391461 |
| hsa-miR-99a-000435 | 3.9774 | 0.2402 | 0.919231 |
| hsa-miR-494-002365 | 2.5905 | 0.2431 | 1.178213 |
| hsa-miR-148b#-002160 | 21.1813 | 0.2489 | 0.967343 |
| hsa-miR-149-002255 | 0.2668 | 0.25 | 1.386839 |
| hsa-miR-340#-002259 | 16.3674 | 0.2523 | 1.117907 |
| hsa-miR-15b#-002173 | 114.3628 | 0.2532 | 1.582275 |
| hsa-miR-454#-001996 | 6.9324 | 0.2536 | 0.891743 |
| hsa-miR-144#-002148 | 179.9935 | 0.255 | 1.021295 |
| hsa-miR-296-000527 | 0.23 | 0.2556 | 1.161589 |
| hsa-miR-584-001624 | 3.8238 | 0.2574 | 0.922039 |
| hsa-miR-550-002410 | 3.6444 | 0.265 | 1.583152 |
| hsa-miR-30d-000420 | 5.3738 | 0.2702 | 1.231059 |
| hsa-miR-29b-2#-002166 | 4.3903 | 0.2736 | 1.164088 |
| hsa-miR-328-000543 | 0.1902 | 0.2799 | 0.890693 |
| mmu-miR-134-001186 | 5.3946 | 0.2847 | 0.644924 |
| hsa-miR-625-002431 | 2.1806 | 0.2892 | 1.320697 |
| hsa-miR-378-002243 | 11.6798 | 0.2899 | 1.533789 |
| hsa-miR-192#-002272 | 4.5328 | 0.2919 | 0.890693 |
| hsa-miR-22#-002301 | 9.6977 | 0.2946 | 0.896205 |
| hsa-miR-1253-002894 | 0.1835 | 0.2951 | 1.154926 |
| hsa-miR-935-002178 | 0.0435 | 0.2961 | 1.020729 |
| hsa-miR-30a-5p-000417 | 8.8831 | 0.2968 | 1.22731 |
| hsa-let-7c-000379 | 2.681 | 0.2996 | 1.06157 |
| hsa-miR-629-001562 | 0.4522 | 0.302 | 1.308125 |
| hsa-miR-574-3p-002349 | 0.3825 | 0.3115 | 0.928903 |
| hsa-miR-342-3p-002260 | 1.8466 | 0.3137 | 1.487201 |
| hsa-miR-769-5p-001998 | 3.0669 | 0.3139 | 0.922231 |
| hsa-miR-1183-002841 | 0.2419 | 0.3154 | 1.205056 |
| rno-miR-29c#-001818 | 25.3746 | 0.321 | 1.22748 |
| hsa-miR-1178-002777 | 2.7188 | 0.3247 | 1.084252 |
| hsa-miR-1274A-002883 | 4.1151 | 0.3266 | 0.916877 |
| hsa-miR-483-3p-002339 | 0.0888 | 0.3271 | 1.396001 |
| hsa-miR-942-002187 | 5.4029 | 0.3287 | 1.014592 |
| hsa-miR-361-3p-002116 | 3.0612 | 0.3291 | 1.195654 |
| hsa-miR-335#-002185 | 7.5039 | 0.3302 | 1.063264 |
| hsa-miR-652-002352 | 2.3486 | 0.3309 | 2.001248 |
| hsa-miR-151-3p-002254 | 14.1994 | 0.3345 | 1.027401 |
| hsa-miR-30e-3p-000422 | 14.3748 | 0.3347 | 1.0208 |
| hsa-miR-766-001986 | 0.4831 | 0.3352 | 1.162234 |
| hsa-miR-19b-1#-002425 | 10.9526 | 0.336 | 1.023137 |
| hsa-miR-636-002088 | 0.398 | 0.3372 | 1.014592 |
| hsa-miR-616-001589 | 4.8155 | 0.3429 | 1.223403 |
| hsa-miR-30a-3p-000416 | 11.0253 | 0.3433 | 0.960262 |
| hsa-miR-375-000564 | 3.193 | 0.3442 | 0.95482 |
| dme-miR-7-000268 | 13.7859 | 0.3474 | 0.683352 |
| hsa-miR-213-000516 | 8.8128 | 0.3514 | 1.946524 |
| RNU44-001094 | 0.6987 | 0.3528 | 1.078181 |
| hsa-miR-202-002363 | 0.2041 | 0.353 | 0.975491 |
| hsa-miR-668-001992 | 0.4798 | 0.3633 | 0.511569 |
| hsa-miR-589-001543 | 2.1132 | 0.3735 | 0.811071 |
| hsa-miR-92a-000431 | 0.4571 | 0.3768 | 1.014592 |
| hsa-miR-1274B-002884 | 3.8258 | 0.3852 | 1.028969 |
| hsa-miR-382-000572 | 3.4752 | 0.3988 | 0.281596 |
| hsa-miR-206-000510 | 0.1562 | 0.407 | 1.024273 |
| hsa-miR-885-5p-002296 | 0.4147 | 0.4116 | 0.890569 |
| hsa-miR-433-001028 | 0.2961 | 0.4183 | 0.394829 |
| hsa-miR-1254-002818 | 0.5042 | 0.4233 | 0.967612 |
| hsa-let-7b-002619 | 1.6905 | 0.4289 | 1.39175 |
| hsa-miR-886-5p-002193 | 1.9418 | 0.4298 | 0.921784 |
| hsa-miR-642-001592 | 0.4673 | 0.4354 | 0.966271 |
| hsa-miR-1208-002880 | 0.1609 | 0.4369 | 0.724069 |
| hsa-miR-564-001531 | 0.1813 | 0.4433 | 1.924922 |
| hsa-miR-485-3p-001277 | 0.466 | 0.4481 | 0.722816 |
| hsa-miR-1201-002781 | 2.7108 | 0.4534 | 2.211308 |
| hsa-miR-1248-002870 | 0.4995 | 0.4543 | 1.096496 |
| hsa-miR-1271-002779 | 2.5278 | 0.4558 | 1.014592 |
| hsa-miR-518d-001159 | 0.2429 | 0.4564 | 1.010872 |
| hsa-miR-191#-002678 | 2.4958 | 0.4632 | 1.393391 |
| hsa-miR-505#-002087 | 0.5571 | 0.4665 | 1.014663 |
| hsa-miR-424#-002309 | 1.9373 | 0.473 | 2.100889 |
| hsa-miR-484-001821 | 1.4584 | 0.4773 | 1.327121 |
| hsa-miR-939-002182 | 0.3419 | 0.4916 | 0.858149 |
| hsa-miR-34b-002102 | 1.4845 | 0.4924 | 1.288328 |
| hsa-miR-645-001597 | 0.1907 | 0.4934 | 0.938026 |
| hsa-miR-1300-002902 | 0.6216 | 0.4978 | 1.225525 |
| hsa-miR-25#-002442 | 0.5651 | 0.5019 | 1.001804 |
| mmu-miR-491-001630 | 1.5858 | 0.5058 | 1.014592 |
| hsa-miR-504-002084 | 0.1667 | 0.5178 | 1.20882 |
| hsa-miR-605-001568 | 0.4642 | 0.533 | 1.336259 |
| hsa-miR-486-3p-002093 | 0.1683 | 0.5418 | 1.014874 |
| hsa-miR-625#-002432 | 0.6167 | 0.5519 | 1.020305 |
| hsa-miR-409-3p-002332 | 1.2673 | 0.5525 | 0.693323 |
| hsa-miR-320-002277 | 1.5302 | 0.5542 | 0.925112 |
| hsa-miR-720-002895 | 0.5745 | 0.5651 | 0.776361 |
| hsa-miR-1303-002792 | 0.265 | 0.5794 | 1.087262 |
| hsa-miR-99b-000436 | 0.6432 | 0.5833 | 1.373636 |
| hsa-let-7e#-002407 | 0.51 | 0.5843 | 1.084778 |
| hsa-miR-423-5p-002340 | 0.5315 | 0.5853 | 0.89199 |
| hsa-miR-320B-002844 | 0.5947 | 0.5889 | 0.967746 |
| hsa-miR-671-3p-002322 | 0.6999 | 0.5962 | 1.382711 |
| hsa-miR-338-5P-002658 | 0.6849 | 0.6055 | 1.014592 |
| hsa-miR-326-000542 | 1.6869 | 0.6102 | 1.116203 |
| hsa-miR-616-002414 | 1.6343 | 0.6257 | 1.015366 |
| hsa-miR-483-5p-002338 | 0.3713 | 0.6263 | 0.741439 |
| hsa-miR-601-001558 | 0.4504 | 0.642 | 0.742159 |
| hsa-miR-18a#-002423 | 0.7197 | 0.6483 | 1.391268 |
| hsa-miR-181a-2#-002317 | 1.4607 | 0.6512 | 1.404932 |
| hsa-miR-331-5p-002233 | 0.43 | 0.6549 | 0.884724 |
| hsa-miR-130b#-002114 | 1.4513 | 0.6589 | 1.20072 |
| hsa-miR-129#-002298 | 0.7193 | 0.6649 | 1.014522 |
| hsa-miR-589-002409 | 1.5415 | 0.6777 | 0.962128 |
| hsa-miR-543-002376 | 0.7598 | 0.6838 | 0.972116 |
| hsa-miR-127-000452 | 0.7153 | 0.6839 | 0.714547 |
| RNU48-001006 | 1.1272 | 0.6986 | 1.259106 |
| hsa-miR-1270-002807 | 0.6817 | 0.7017 | 1.219509 |
| hsa-miR-422a-002297 | 1.338 | 0.7159 | 0.966204 |
| hsa-miR-93#-002139 | 1.2568 | 0.7245 | 1.161428 |
| hsa-miR-654-001611 | 0.5941 | 0.7281 | 1.353411 |
| hsa-miR-329-001101 | 0.5892 | 0.7404 | 0.888658 |
| hsa-miR-363#-001283 | 0.7238 | 0.7439 | 1.09013 |
| hsa-miR-532-3p-002355 | 0.7953 | 0.7439 | 1.014663 |
| hsa-miR-875-5p-002203 | 1.4066 | 0.7492 | 0.504876 |
| hsa-miR-125a-5p-002198 | 0.811 | 0.7623 | 1.374112 |
| hsa-miR-664-002897 | 0.8292 | 0.7715 | 1.378787 |
| hsa-miR-145-002278 | 1.1687 | 0.7947 | 1.385878 |
| hsa-miR-150-000473 | 1.1539 | 0.8032 | 1.696075 |
| hsa-miR-1180-002847 | 0.8312 | 0.8039 | 1.014522 |
| hsa-miR-432-001026 | 0.821 | 0.8238 | 0.87043 |
| hsa-miR-106b#-002380 | 1.1768 | 0.8323 | 0.965735 |
| hsa-miR-146b-3p-002361 | 1.2251 | 0.855 | 1.25562 |
| hsa-miR-125b-000449 | 1.1266 | 0.8657 | 0.929612 |
| hsa-miR-378-000567 | 1.1219 | 0.876 | 1.168777 |
| hsa-miR-17#-002421 | 1.1894 | 0.8838 | 0.814508 |
| hsa-miR-183#-002270 | 1.1154 | 0.884 | 1.014944 |
| hsa-miR-323-3p-002227 | 0.8646 | 0.8974 | 1.334038 |
| hsa-miR-571-001613 | 0.8796 | 0.9149 | 1.038931 |
| hsa-miR-193b-002367 | 1.0645 | 0.9241 | 1.387897 |
| hsa-miR-501-3p-002435 | 0.9202 | 0.9298 | 2.597277 |
| hsa-miR-550-001544 | 1.0424 | 0.9392 | 1.389051 |
| hsa-miR-1275-002840 | 0.9498 | 0.9519 | 1.065256 |
| hsa-miR-330-000544 | 0.9717 | 0.9652 | 1.383862 |
| hsa-miR-339-5p-002257 | 0.9757 | 0.9707 | 1.168858 |
| hsa-miR-148a#-002134 | 1.04 | 0.9717 | 0.916305 |
| mmu-miR-495-001663 | 0.9631 | 0.9763 | 0.684537 |
| hsa-miR-941-002183 | 0.9924 | 0.9897 | 1.16546 |
| hsa-miR-20a#-002437 | 0.9973 | 0.9984 | 15.10234 |
| ath-miR159a-000338 | |||
| hsa-let-7a#-002307 | |||
| hsa-let-7b#-002404 | |||
| hsa-let-7c#-002405 | |||
| hsa-let-7f-1#-002417 | |||
| hsa-let-7f-2#-002418 | |||
| hsa-let-7g#-002118 | 1.4648 | ||
| hsa-let-7i#-002172 | 107.2379 | ||
| hsa-miR-100#-002142 | |||
| hsa-miR-1-002222 | |||
| hsa-miR-101#-002143 | |||
| hsa-miR-105#-002168 | |||
| hsa-miR-105-002167 | |||
| hsa-miR-106a#-002170 | |||
| hsa-miR-107-000443 | |||
| hsa-miR-10a#-002288 | |||
| hsa-miR-10b#-002315 | |||
| hsa-miR-10b-002218 | |||
| hsa-miR-1179-002776 | |||
| hsa-miR-1182-002830 | |||
| hsa-miR-1184-002842 | |||
| hsa-miR-1197-002810 | |||
| hsa-miR-1200-002829 | |||
| hsa-miR-1203-002877 | |||
| hsa-miR-1204-002872 | |||
| hsa-miR-1205-002778 | |||
| hsa-miR-1206-002878 | |||
| hsa-miR-122#-002130 | |||
| hsa-miR-122-002245 | 0.5039 | ||
| hsa-miR-1225-3P-002766 | |||
| hsa-miR-1226#-002758 | |||
| hsa-miR-1228#-002763 | |||
| hsa-miR-124#-002197 | |||
| hsa-miR-1243-002854 | |||
| hsa-miR-1244-002791 | 3.1723 | ||
| hsa-miR-1245-002823 | |||
| hsa-miR-1247-002893 | 0.0859 | ||
| hsa-miR-1250-002887 | |||
| hsa-miR-1251-002820 | |||
| hsa-miR-1252-002860 | |||
| hsa-miR-1255A-002805 | |||
| hsa-miR-1256-002850 | |||
| hsa-miR-1257-002910 | |||
| hsa-miR-1259-002796 | |||
| hsa-miR-125a-3p-002199 | 0.531 | ||
| hsa-miR-125b-1#-002378 | |||
| hsa-miR-125b-2#-002158 | |||
| hsa-miR-1262-002852 | |||
| hsa-miR-1263-002784 | |||
| hsa-miR-1264-002799 | |||
| hsa-miR-1265-002790 | |||
| hsa-miR-1267-002885 | |||
| hsa-miR-1269-002789 | |||
| hsa-miR-1272-002845 | |||
| hsa-miR-127-5p-002229 | |||
| hsa-miR-1276-002843 | |||
| hsa-miR-1278-002851 | |||
| hsa-miR-1283-002890 | |||
| hsa-miR-1284-002903 | |||
| hsa-miR-1286-002773 | |||
| hsa-miR-1288-002832 | |||
| hsa-miR-1289-002871 | |||
| hsa-miR-129-000590 | |||
| hsa-miR-1291-002838 | 6.764 | ||
| hsa-miR-1292-002824 | |||
| hsa-miR-1293-002905 | |||
| hsa-miR-1294-002785 | |||
| hsa-miR-1296-002908 | 5.0843 | ||
| hsa-miR-1298-002861 | |||
| hsa-miR-1301-002827 | |||
| hsa-miR-1302-002901 | |||
| hsa-miR-1304-002874 | |||
| hsa-miR-1305-002867 | |||
| hsa-miR-130a#-002131 | |||
| hsa-miR-132#-002132 | |||
| hsa-miR-1324-002815 | |||
| hsa-miR-133b-002247 | 88.5947 | ||
| hsa-miR-135a-000460 | |||
| hsa-miR-135b#-002159 | |||
| hsa-miR-135b-002261 | 0.1506 | ||
| hsa-miR-136#-002100 | |||
| hsa-miR-136-000592 | |||
| hsa-miR-138-002284 | |||
| hsa-miR-138-2#-002144 | |||
| hsa-miR-141#-002145 | |||
| hsa-miR-141-000463 | |||
| hsa-miR-142-5p-002248 | |||
| hsa-miR-143#-002146 | |||
| hsa-miR-143-002249 | |||
| hsa-miR-144-002676 | |||
| hsa-miR-145#-002149 | |||
| hsa-miR-146a#-002163 | |||
| hsa-miR-147-000469 | |||
| hsa-miR-147b-002262 | |||
| hsa-miR-149#-002164 | |||
| hsa-miR-154#-000478 | |||
| hsa-miR-154-000477 | |||
| hsa-miR-155#-002287 | |||
| hsa-miR-15a#-002419 | 107.6923 | ||
| hsa-miR-15a-000389 | |||
| hsa-miR-16-1#-002420 | 218.8413 | ||
| hsa-miR-16-2#-002171 | |||
| hsa-miR-181c#-002333 | |||
| hsa-miR-182#-000483 | |||
| hsa-miR-1825-002907 | |||
| hsa-miR-1826-002873 | |||
| hsa-miR-185#-002104 | |||
| hsa-miR-186#-002105 | |||
| hsa-miR-188-3p-002106 | |||
| hsa-miR-18b#-002310 | |||
| hsa-miR-18b-002217 | 211.9679 | ||
| hsa-miR-190-000489 | |||
| hsa-miR-193a-3p-002250 | |||
| hsa-miR-193b#-002366 | 0.2068 | ||
| hsa-miR-194#-002379 | 0.1097 | ||
| hsa-miR-195#-002107 | |||
| hsa-miR-196a#-002336 | |||
| hsa-miR-198-002273 | |||
| hsa-miR-199a-000498 | |||
| hsa-miR-199b-000500 | |||
| hsa-miR-19a#-002424 | |||
| hsa-miR-200a#-001011 | |||
| hsa-miR-200a-000502 | |||
| hsa-miR-200b#-002274 | |||
| hsa-miR-200b-002251 | 21.2685 | ||
| hsa-miR-200c#-002286 | |||
| hsa-miR-202#-002362 | 9.7132 | ||
| hsa-miR-203-000507 | |||
| hsa-miR-205-000509 | 10.5591 | ||
| hsa-miR-208-000511 | |||
| hsa-miR-208b-002290 | |||
| hsa-miR-20b#-002311 | |||
| hsa-miR-21#-002438 | 11.3163 | ||
| hsa-miR-214#-002293 | |||
| hsa-miR-214-002306 | 0.0964 | ||
| hsa-miR-215-000518 | |||
| hsa-miR-216a-002220 | |||
| hsa-miR-216b-002326 | |||
| hsa-miR-217-002337 | |||
| hsa-miR-218-000521 | 170.283 | ||
| hsa-miR-218-1#-002094 | |||
| hsa-miR-218-2#-002294 | |||
| hsa-miR-219-000522 | |||
| hsa-miR-219-1-3p-002095 | 30.3606 | ||
| hsa-miR-219-2-3p-002390 | |||
| hsa-miR-220-000523 | |||
| hsa-miR-220b-002206 | |||
| hsa-miR-220c-002211 | |||
| hsa-miR-221#-002096 | |||
| hsa-miR-222#-002097 | 43.7736 | ||
| hsa-miR-224-002099 | |||
| hsa-miR-23a#-002439 | |||
| hsa-miR-23a-000399 | 38.5634 | ||
| hsa-miR-23b#-002126 | |||
| hsa-miR-23b-000400 | 0.082 | ||
| hsa-miR-24-1#-002440 | |||
| hsa-miR-24-2#-002441 | |||
| hsa-miR-26a-2#-002115 | |||
| hsa-miR-27b#-002174 | |||
| hsa-miR-296-3p-002101 | |||
| hsa-miR-298-002190 | |||
| hsa-miR-299-3p-001015 | |||
| hsa-miR-299-5p-000600 | |||
| hsa-miR-29a#-002447 | |||
| hsa-miR-29b-000413 | |||
| hsa-miR-29b-1#-002165 | |||
| hsa-miR-301b-002392 | |||
| hsa-miR-302a#-002381 | |||
| hsa-miR-302a-000529 | |||
| hsa-miR-302b#-002119 | |||
| hsa-miR-302b-000531 | |||
| hsa-miR-302c#-000534 | |||
| hsa-miR-302c-000533 | |||
| hsa-miR-302d#-002120 | |||
| hsa-miR-302d-000535 | |||
| hsa-miR-30b#-002129 | |||
| hsa-miR-30c-1#-002108 | |||
| hsa-miR-30c-2#-002110 | |||
| hsa-miR-30d#-002305 | |||
| hsa-miR-31#-002113 | 11.1358 | ||
| hsa-miR-32#-002111 | |||
| hsa-miR-32-002109 | |||
| hsa-miR-325-000540 | |||
| hsa-miR-330-5p-002230 | |||
| hsa-miR-337-3p-002157 | 0.881 | ||
| hsa-miR-337-5p-002156 | |||
| hsa-miR-338-3p-002252 | 0.8708 | ||
| hsa-miR-33a#-002136 | |||
| hsa-miR-33a-002135 | |||
| hsa-miR-33b-002085 | |||
| hsa-miR-340-002258 | 26.6065 | ||
| hsa-miR-346-000553 | |||
| hsa-miR-34a#-002316 | |||
| hsa-miR-34a-000426 | |||
| hsa-miR-34b-000427 | |||
| hsa-miR-34c-000428 | |||
| hsa-miR-362-3p-002117 | 107.0275 | ||
| hsa-miR-367#-002121 | |||
| hsa-miR-367-000555 | |||
| hsa-miR-369-3p-000557 | |||
| hsa-miR-369-5p-001021 | |||
| hsa-miR-371-3p-002124 | |||
| hsa-miR-372-000560 | |||
| hsa-miR-373-000561 | |||
| hsa-miR-374a#-002125 | |||
| hsa-miR-374b#-002391 | |||
| hsa-miR-376a#-002127 | |||
| hsa-miR-376b-001102 | |||
| hsa-miR-377#-002128 | 0.8371 | ||
| hsa-miR-377-000566 | |||
| hsa-miR-380-3p-000569 | |||
| hsa-miR-380-5p-000570 | |||
| hsa-miR-381-000571 | |||
| hsa-miR-383-000573 | |||
| hsa-miR-384-000574 | |||
| hsa-miR-409-5p-002331 | |||
| hsa-miR-410-001274 | |||
| hsa-miR-411#-002238 | |||
| hsa-miR-411-001610 | |||
| hsa-miR-412-001023 | |||
| hsa-miR-424-000604 | |||
| hsa-miR-429-001024 | |||
| hsa-miR-431#-002312 | |||
| hsa-miR-431-001979 | |||
| hsa-miR-432#-001027 | 1.8383 | ||
| hsa-miR-448-001029 | |||
| hsa-miR-449-001030 | 125.8155 | ||
| hsa-miR-449b-001608 | |||
| hsa-miR-450a-002303 | |||
| hsa-miR-450b-3p-002208 | |||
| hsa-miR-450b-5p-002207 | |||
| hsa-miR-452#-002330 | |||
| hsa-miR-452-002329 | |||
| hsa-miR-453-002318 | |||
| hsa-miR-455-001280 | |||
| hsa-miR-455-3p-002244 | |||
| hsa-miR-485-5p-001036 | |||
| hsa-miR-487a-001279 | 0.0523 | ||
| hsa-miR-487b-001285 | 94.345 | ||
| hsa-miR-488-001106 | |||
| hsa-miR-488-002357 | |||
| hsa-miR-489-002358 | |||
| hsa-miR-490-001037 | |||
| hsa-miR-491-3p-002360 | |||
| hsa-miR-492-001039 | |||
| hsa-miR-493-002364 | |||
| hsa-miR-497#-002368 | |||
| hsa-miR-497-001043 | |||
| hsa-miR-499-3p-002427 | |||
| hsa-miR-500-001046 | 3.8468 | ||
| hsa-miR-503-001048 | |||
| hsa-miR-506-001050 | 23.0067 | ||
| hsa-miR-507-001051 | |||
| hsa-miR-508-001052 | 0.631 | ||
| hsa-miR-508-5p-002092 | |||
| hsa-miR-509-3-5p-002155 | |||
| hsa-miR-509-5p-002235 | |||
| hsa-miR-510-002241 | |||
| hsa-miR-511-001111 | |||
| hsa-miR-512-3p-001823 | |||
| hsa-miR-512-5p-001145 | |||
| hsa-miR-513-5p-002090 | |||
| hsa-miR-513B-002757 | |||
| hsa-miR-513C-002756 | |||
| hsa-miR-515-3p-002369 | |||
| hsa-miR-515-5p-001112 | |||
| hsa-miR-516-3p-001149 | 0.2851 | ||
| hsa-miR-516a-5p-002416 | |||
| hsa-miR-516b-001150 | |||
| hsa-miR-517#-001113 | |||
| hsa-miR-517a-002402 | |||
| hsa-miR-517b-001152 | |||
| hsa-miR-517c-001153 | |||
| hsa-miR-518a-3p-002397 | |||
| hsa-miR-518a-5p-002396 | |||
| hsa-miR-518b-001156 | |||
| hsa-miR-518c#-001158 | |||
| hsa-miR-518c-002401 | |||
| hsa-miR-518d-5p-002389 | |||
| hsa-miR-518e#-002371 | |||
| hsa-miR-518e-002395 | |||
| hsa-miR-518f#-002387 | |||
| hsa-miR-518f-002388 | |||
| hsa-miR-519a-002415 | |||
| hsa-miR-519b-3p-002384 | |||
| hsa-miR-519c-001163 | |||
| hsa-miR-519d-002403 | |||
| hsa-miR-519e#-001166 | |||
| hsa-miR-519e-002370 | |||
| hsa-miR-520a#-001168 | |||
| hsa-miR-520a-001167 | |||
| hsa-miR-520b-001116 | |||
| hsa-miR-520D-3P-002743 | |||
| hsa-miR-520d-5p-002393 | |||
| hsa-miR-520e-001119 | |||
| hsa-miR-520f-001120 | |||
| hsa-miR-520g-001121 | |||
| hsa-miR-520h-001170 | |||
| hsa-miR-521-001122 | |||
| hsa-miR-522-002413 | |||
| hsa-miR-523-002386 | |||
| hsa-miR-524-001173 | |||
| hsa-miR-524-5p-001982 | |||
| hsa-miR-525-001174 | |||
| hsa-miR-525-3p-002385 | |||
| hsa-miR-526b-002382 | |||
| hsa-miR-541#-002200 | |||
| hsa-miR-541-002201 | |||
| hsa-miR-542-3p-001284 | |||
| hsa-miR-542-5p-002240 | |||
| hsa-miR-544-002265 | |||
| hsa-miR-545#-002266 | 0.4262 | ||
| hsa-miR-545-002267 | |||
| hsa-miR-548a-001538 | 0.0416 | ||
| hsa-miR-548a-5p-002412 | |||
| hsa-miR-548b-001541 | |||
| hsa-miR-548b-5p-002408 | |||
| hsa-miR-548c-001590 | 5.3989 | ||
| hsa-miR-548c-5p-002429 | 38.7768 | ||
| hsa-miR-548d-001605 | 29.9772 | ||
| hsa-miR-548d-5p-002237 | |||
| hsa-miR-548E-002881 | |||
| hsa-miR-548G-002879 | |||
| hsa-miR-548H-002816 | |||
| hsa-miR-548I-002909 | |||
| hsa-miR-548J-002783 | 1.8774 | ||
| hsa-miR-548K-002819 | |||
| hsa-miR-548L-002904 | |||
| hsa-miR-548M-002775 | |||
| hsa-miR-548N-002888 | |||
| hsa-miR-548P-002798 | |||
| hsa-miR-549-001511 | |||
| hsa-miR-551a-001519 | 1.4235 | ||
| hsa-miR-551b#-002346 | |||
| hsa-miR-551b-001535 | |||
| hsa-miR-552-001520 | |||
| hsa-miR-553-001521 | |||
| hsa-miR-554-001522 | |||
| hsa-miR-555-001523 | |||
| hsa-miR-556-3p-002345 | |||
| hsa-miR-556-5p-002344 | |||
| hsa-miR-557-001525 | |||
| hsa-miR-558-001526 | |||
| hsa-miR-559-001527 | |||
| hsa-miR-561-001528 | |||
| hsa-miR-562-001529 | |||
| hsa-miR-563-001530 | |||
| hsa-miR-566-001533 | |||
| hsa-miR-567-001534 | |||
| hsa-miR-569-001536 | |||
| hsa-miR-570-002347 | |||
| hsa-miR-572-001614 | 0.437 | ||
| hsa-miR-573-001615 | |||
| hsa-miR-575-001617 | |||
| hsa-miR-576-3p-002351 | 3.0672 | ||
| hsa-miR-576-5p-002350 | |||
| hsa-miR-577-002675 | |||
| hsa-miR-578-001619 | |||
| hsa-miR-579-002398 | 130.8115 | ||
| hsa-miR-580-001621 | |||
| hsa-miR-581-001622 | |||
| hsa-miR-582-3p-002399 | |||
| hsa-miR-582-5p-001983 | |||
| hsa-miR-583-001623 | |||
| hsa-miR-585-001625 | |||
| hsa-miR-586-001539 | 0.0471 | ||
| hsa-miR-587-001540 | |||
| hsa-miR-588-001542 | |||
| hsa-miR-590-3P-002677 | |||
| hsa-miR-591-001545 | |||
| hsa-miR-592-001546 | |||
| hsa-miR-593-001547 | |||
| hsa-miR-593-002411 | |||
| hsa-miR-595-001987 | |||
| hsa-miR-596-001550 | |||
| hsa-miR-597-001551 | |||
| hsa-miR-598-001988 | 155.6581 | ||
| hsa-miR-599-001554 | |||
| hsa-miR-600-001556 | |||
| hsa-miR-603-001566 | |||
| hsa-miR-604-001567 | |||
| hsa-miR-606-001569 | |||
| hsa-miR-607-001570 | |||
| hsa-miR-608-001571 | |||
| hsa-miR-609-001573 | |||
| hsa-miR-613-001586 | |||
| hsa-miR-614-001587 | |||
| hsa-miR-615-5p-002353 | |||
| hsa-miR-617-001591 | |||
| hsa-miR-620-002672 | |||
| hsa-miR-621-001598 | |||
| hsa-miR-622-001553 | |||
| hsa-miR-623-001555 | |||
| hsa-miR-624-001557 | 0.9465 | ||
| hsa-miR-624-002430 | |||
| hsa-miR-626-001559 | |||
| hsa-miR-627-001560 | 228.4357 | ||
| hsa-miR-628-5p-002433 | 271.071 | ||
| hsa-miR-630-001563 | |||
| hsa-miR-631-001564 | |||
| hsa-miR-633-001574 | |||
| hsa-miR-634-001576 | |||
| hsa-miR-635-001578 | |||
| hsa-miR-637-001581 | |||
| hsa-miR-638-001582 | 0.746 | ||
| hsa-miR-639-001583 | 0.4233 | ||
| hsa-miR-640-001584 | |||
| hsa-miR-641-001585 | |||
| hsa-miR-643-001594 | |||
| hsa-miR-644-001596 | |||
| hsa-miR-646-001599 | |||
| hsa-miR-647-001600 | |||
| hsa-miR-648-001601 | |||
| hsa-miR-649-001602 | |||
| hsa-miR-650-001603 | |||
| hsa-miR-651-001604 | |||
| hsa-miR-653-002292 | |||
| hsa-miR-654-3p-002239 | |||
| hsa-miR-656-001510 | |||
| hsa-miR-657-001512 | |||
| hsa-miR-658-001513 | |||
| hsa-miR-659-001514 | |||
| hsa-miR-661-001606 | 2.7873 | ||
| hsa-miR-662-001607 | |||
| hsa-miR-663B-002857 | 0.1863 | ||
| hsa-miR-665-002681 | |||
| hsa-miR-672-002327 | |||
| hsa-miR-674-002021 | |||
| hsa-miR-675-002005 | |||
| hsa-miR-708#-002342 | |||
| hsa-miR-708-002341 | 0.4321 | ||
| hsa-miR-7-2#-002314 | 26.3744 | ||
| hsa-miR-758-001990 | 0.2387 | ||
| hsa-miR-765-002643 | |||
| hsa-miR-767-3p-001995 | |||
| hsa-miR-767-5p-001993 | |||
| hsa-miR-769-3p-002003 | 2.3061 | ||
| hsa-miR-770-5p-002002 | |||
| hsa-miR-802-002004 | |||
| hsa-miR-871-002354 | |||
| hsa-miR-872-002264 | |||
| hsa-miR-873-002356 | |||
| hsa-miR-874-002268 | 7.9572 | ||
| hsa-miR-875-3p-002204 | |||
| hsa-miR-876-3p-002225 | |||
| hsa-miR-876-5p-002205 | |||
| hsa-miR-885-3p-002372 | |||
| hsa-miR-886-3p-002194 | 1.2122 | ||
| hsa-miR-887-002374 | |||
| hsa-miR-888#-002213 | |||
| hsa-miR-888-002212 | |||
| hsa-miR-889-002202 | |||
| hsa-miR-890-002209 | |||
| hsa-miR-891a-002191 | 4.6329 | ||
| hsa-miR-891b-002210 | |||
| hsa-miR-892a-002195 | |||
| hsa-miR-892b-002214 | |||
| hsa-miR-9#-002231 | |||
| hsa-miR-9-000583 | |||
| hsa-miR-920-002150 | |||
| hsa-miR-921-002151 | |||
| hsa-miR-922-002152 | |||
| hsa-miR-924-002154 | |||
| hsa-miR-92a-1#-002137 | |||
| hsa-miR-92a-2#-002138 | |||
| hsa-miR-92b#-002343 | |||
| hsa-miR-933-002176 | |||
| hsa-miR-934-002177 | |||
| hsa-miR-936-002179 | |||
| hsa-miR-937-002180 | |||
| hsa-miR-938-002181 | |||
| hsa-miR-943-002188 | |||
| hsa-miR-944-002189 | |||
| hsa-miR-95-000433 | |||
| hsa-miR-96#-002140 | |||
| hsa-miR-98-000577 | |||
| hsa-miR-99a#-002141 | 0.0374 | ||
| hsa-miR-99b#-002196 | 0.0378 | ||
| mmu-let-7d#-001178 | |||
| mmu-miR-124a-001182 | |||
| mmu-miR-129-3p-001184 | 0.2208 | ||
| mmu-miR-137-001129 | |||
| mmu-miR-153-001191 | |||
| mmu-miR-187-001193 | |||
| mmu-miR-379-001138 | |||
| mmu-miR-496-001953 | |||
| mmu-miR-499-001352 |
References
- Meikle, P.J.; Hopwood, J.J.; Clague, A.E.; Carey, W.F. Prevalence of lysosomal storage disorders. J. Am. Med. Assoc. 1999, 281, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Amaral, O.; Fortuna, A.M.; Lacerda, L.; Pinto, R.; Sa Miranda, M.C. Molecular characterisation of type 1 Gaucher disease families and patients: Intrafamilial heterogeneity at the clinical level. J. Med. Genet. 1994, 31, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Goker-Alpan, O.; Hruska, K.S.; Orvisky, E.; Kishnani, P.S.; Stubblefield, B.K.; Schiffmann, R.; Sidransky, E. Divergent phenotypes in Gaucher disease implicate the role of modifiers. J. Med. Genet. 2005, 42, e37. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, R.H.; Grant, I.R.; Halsall, D.; Cox, T.M. Twin pairs showing discordance of phenotype in adult Gaucher’s disease. QJM Mon. J. Assoc. Phys. 2004, 97, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Baris, H.N.; Cohen, I.J.; Mistry, P.K. Gaucher disease: The metabolic defect, pathophysiology, phenotypes and natural history. Pediatr. Endocrinol. Rev. 2014, 12, 72–81. [Google Scholar]
- Balestrino, R.; Schapira, A.H.V. Glucocerebrosidase and Parkinson Disease: Molecular, Clinical, and Therapeutic Implications. Neuroscientist 2018, 24, 540–559. [Google Scholar] [CrossRef]
- Hassan, S.; Sidransky, E.; Tayebi, N. The role of epigenetics in lysosomal storage disorders: Uncharted territory. Mol. Genet. Metab. 2017, 122, 10–18. [Google Scholar] [CrossRef]
- Wegner, M.; Neddermann, D.; Piorunska-Stolzmann, M.; Jagodzinski, P.P. Role of epigenetic mechanisms in the development of chronic complications of diabetes. Diabetes Res. Clin. Pract. 2014, 105, 164–175. [Google Scholar] [CrossRef]
- Gilad, S.; Meiri, E.; Yogev, Y.; Benjamin, S.; Lebanony, D.; Yerushalmi, N.; Benjamin, H.; Kushnir, M.; Cholakh, H.; Melamed, N.; et al. Serum microRNAs are promising novel biomarkers. PLoS ONE 2008, 3, e3148. [Google Scholar] [CrossRef]
- Siebert, M.; Westbroek, W.; Chen, Y.-C.; Moaven, N.; Li, Y.; Velayati, A.; Saraiva-Pereira, M.L.; Martin, S.E.; Sidransky, E. Identification of miRNAs that modulate glucocerebrosidase activity in Gaucher disease cells. RNA Biol. 2014, 11, 1291–1300. [Google Scholar] [CrossRef]
- Straniero, L.; Rimoldi, V.; Samarani, M.; Goldwurm, S.; Di Fonzo, A.; Krüger, R.; Deleidi, M.; Aureli, M.; Soldà, G.; Duga, S.; et al. The GBAP1 pseudogene acts as a ceRNA for the glucocerebrosidase gene GBA by sponging miR-22-3p. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ginns, E.I.; Mak, S.K.-K.; Ko, N.; Karlgren, J.; Akbarian, S.; Chou, V.P.; Guo, Y.; Lim, A.; Samuelsson, S.; Lamarca, M.L.; et al. Neuroinflammation and α-synuclein accumulation in response to glucocerebrosidase deficiency are accompanied by synaptic dysfunction. Mol. Genet. Metab. 2014, 111, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Watson, L.; Keatinge, M.; Gegg, M.; Bai, Q.; Sandulescu, M.C.; Vardi, A.; Futerman, A.H.; Schapira, A.H.; Burton, E.A.; Bandmann, O. Ablation of the pro-inflammatory master regulator miR-155 does not mitigate neuroinflammation or neurodegeneration in a vertebrate model of Gaucher’s disease. Neurobiol. Dis. 2019, 127, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Liang, X.; Li, X.; Zhang, Y.; Sun, Z.; Liu, Y.; Wang, J. MicroRNA-195: A review of its role in cancers. Onco Targets Ther. 2018, 11, 7109. [Google Scholar] [CrossRef]
- Qu, Y.; Liu, H.; Lv, X.; Liu, Y.; Wang, X.; Zhang, M.; Zhang, X.; Li, Y.; Lou, Q.; Li, S.; et al. MicroRNA-16-5p overexpression suppresses proliferation and invasion as well as triggers apoptosis by targeting VEGFA expression in breast carcinoma. Oncotarget 2017, 8, 72400–72410. [Google Scholar] [CrossRef]
- Wang, N.; Wei, H.; Yin, D.; Lu, Y.; Zhang, Y.; Zhang, Q.; Ma, X.; Zhang, S. MicroRNA-195 inhibits proliferation of cervical cancer cells by targeting cyclin D1a. Tumor Biol. 2016, 37, 4711–4720. [Google Scholar] [CrossRef]
- Liu, B.; Qu, J.; Xu, F.; Guo, Y.; Wang, Y.; Yu, H.; Qian, B.-Y. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget 2015, 6, 9445–9456. [Google Scholar] [CrossRef]
- Liu, C.; Guan, H.; Wang, Y.; Chen, M.; Xu, B.; Zhang, L.; Lu, K.; Tao, T.; Zhang, X.; Huang, Y. MIR-195 inhibits emt by targeting FGF2 in prostate cancer cells. PLoS ONE 2015, 10, e0144073. [Google Scholar] [CrossRef]
- Shiran, A.; Brenner, B.; Laor, A.; Tatarsky, I. Increased risk of cancer in patients with gaucher disease. Cancer 1993, 72, 219–224. [Google Scholar] [CrossRef]
- Rosenbloom, B.E.; Weinreb, N.J.; Zimran, A.; Kacena, K.A.; Charrow, J.; Ward, E. Gaucher disease and cancer incidence: A study from the Gaucher Registry. Blood 2005, 105, 4569–4572. [Google Scholar] [CrossRef]
- Taddei, T.H.; Kacena, K.A.; Yang, M.; Yang, R.; Malhotra, A.; Boxer, M.; Aleck, K.A.; Rennert, G.; Pastores, G.M.; Mistry, P.K. The underrecognized progressive nature of N370S Gaucher disease and assessment of cancer risk in 403 patients. Am. J. Hematol. 2009, 84, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; He, J.; Zheng, L.; Bi, M.; Hu, F.; Chen, M.; Niu, H.; Yang, J.; Luo, Y.; Tang, W.; et al. miR-148b-3p, miR-190b, and miR-429 regulate cell progression and act as potential biomarkers for breast cancer. J. Breast Cancer 2019, 22, 219–236. [Google Scholar] [CrossRef] [PubMed]
- Regenboog, M.; Van Dussen, L.; Verheij, J.; Weinreb, N.J.; Santosa, D.; Dahl, S.V.; Häussinger, D.; Müller, M.N.; Canbay, A.; Rigoldi, M.; et al. Hepatocellular carcinoma in Gaucher disease: An international case series. J. Inherit. Metab. Dis. 2018, 41, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Han, C.; Zhang, Z.; Wang, L.; Xu, J. Increased expression of microRNA-31-5p inhibits cell proliferation, migration, and invasion via regulating Sp1 transcription factor in HepG2 hepatocellular carcinoma cell line. Biochem. Biophys. Res. Commun. 2017, 490, 371–377. [Google Scholar] [CrossRef]
- Marie, P.J.; Debiais, F.; Haÿ, E. Regulation of human cranial osteoblast phenotype by FGF-2, FGFR-2 and BMP-2 signaling. Histol. Histopathol. 2002, 17, 877–885. [Google Scholar] [CrossRef]
- Fynan, T.M.; Reiss, M. Resistance to inhibition of cell growth by transforming growth factor-beta and its role in oncogenesis. Crit. Rev. Oncog. 1993, 4, 493–540. [Google Scholar]
- Border, W.A.; Ruoslahti, E. Transforming growth factor-β in disease: The dark side of tissue repair. J. Clin. Investig. 1992, 90, 1–7. [Google Scholar] [CrossRef]
- Nascimbeni, F.; Cassinerio, E.; Salda, A.D.; Motta, I.; Bursi, S.; Donatiello, S.; Spina, V.; Cappellini, M.D.; Carubbi, F. Prevalence and predictors of liver fibrosis evaluated by vibration controlled transient elastography in type 1 Gaucher disease. Mol. Genet. Metab. 2018, 125, 64–72. [Google Scholar] [CrossRef]
- Mon, N.N.; Ito, S.; Senga, T.; Hamaguchi, M. FAK signaling in neoplastic disorders: A linkage between inflammation and cancer. Ann. N. Y. Acad. Sci. 2006, 1086, 199–212. [Google Scholar] [CrossRef]
- Kornberg, L.J. Focal adhesion kinase and its potential involvement in tumor invasion and metastasis. Head Neck 1998, 20, 745–752. [Google Scholar] [CrossRef]
- Zhao, J.; Guan, J.L. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009, 28, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.M.; Jeong, K.; Rodriguez, Y.A.R.; Kim, J.H.; Ahn, E.Y.E.; Lim, S.T.S. FAK and Pyk2 activity promote TNF-α and IL-1β-mediated pro-inflammatory gene expression and vascular inflammation. Sci. Rep. 2019, 9, 7617. [Google Scholar] [CrossRef] [PubMed]
- Santo, E.E.; Paik, J. FOXO in Neural Cells and Diseases of the Nervous System. Curr. Top. Develop. Biol. 2018, 127, 105–118. [Google Scholar] [CrossRef]
- Xia, S.; Wen, X.; Fan, X.; Chen, X.; Wei, Z.; Li, Q.; Sun, L. Wnt2 overexpression protects against PINK1 mutant-induced mitochondrial dysfunction and oxidative stress. Mol. Med. Rep. 2020, 21, 2633–2641. [Google Scholar] [CrossRef]
- Russell, S.A.; Bashaw, G.J. Axon guidance pathways and the control of gene expression. Dev. Dyn. 2018, 247, 571–580. [Google Scholar] [CrossRef]
- Riboldi, G.M.; Di Fonzo, A. GBA, Gaucher Disease, and Parkinson’s Disease: From Genetic to Clinic to New Therapeutic Approaches. Cells 2019, 8, 364. [Google Scholar] [CrossRef]
- Arshad, A.R.; Sulaiman, S.A.; Saperi, A.A.; Jamal, R.; Mohamed Ibrahim, N.; Abdul Murad, N.A. MicroRNAs and target genes as biomarkers for the diagnosis of early onset of parkinson disease. Front. Mol. Neurosci. 2017, 10, 352. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, Z.; Mata, I. MicroRNAs: Game changers in the regulation of α-Synuclein in Parkinson’s disease. Parkinsons Dis. 2019, 2019, 1743183. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).