Dependence of Leydig Cell’s Mitochondrial Physiology on Luteinizing Hormone Signaling
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. In Vivo Experiments
2.3. Collection of Testicular Interstitial Fluid (TIF) and Preparation of Purified Leydig Cells
2.4. Ex Vivo Experiments
2.5. Testosterone and cAMP Measurements
2.6. ATP Level, Mitochondrial Membrane Potential and O2 Consumption Measurements
2.7. Genomic DNA Purification, RNA Isolation, cDNA Synthesis and Real-Time Polymerase Chain Reaction (PCR) Relative Quantitative Analysis
2.8. Protein Extraction and Western Blot Analysis
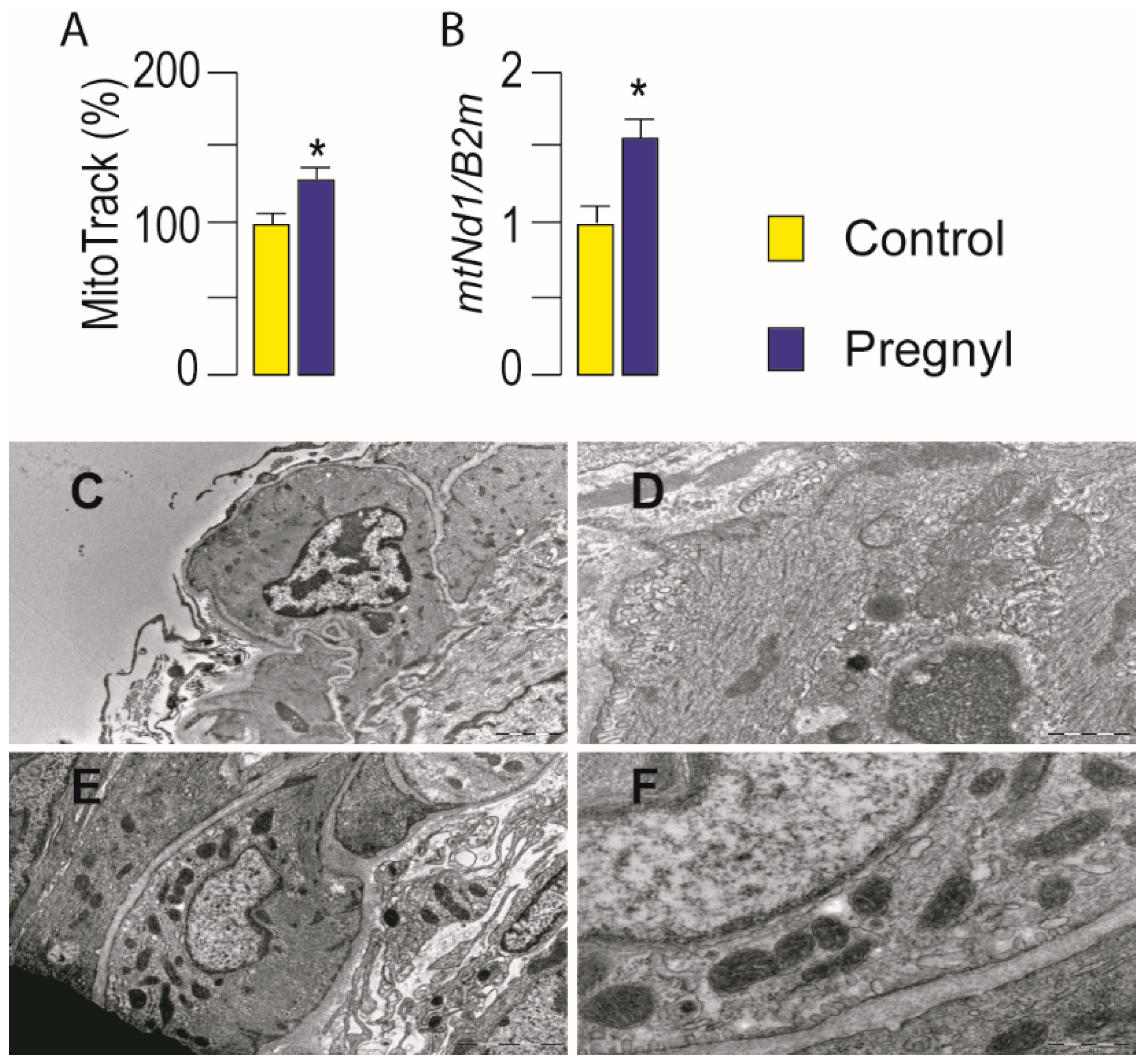
2.9. Transmission Electron Microscope (TEM) Analysis of Leydig Cell Mitochondria
2.10. Statistical Analysis
3. Results
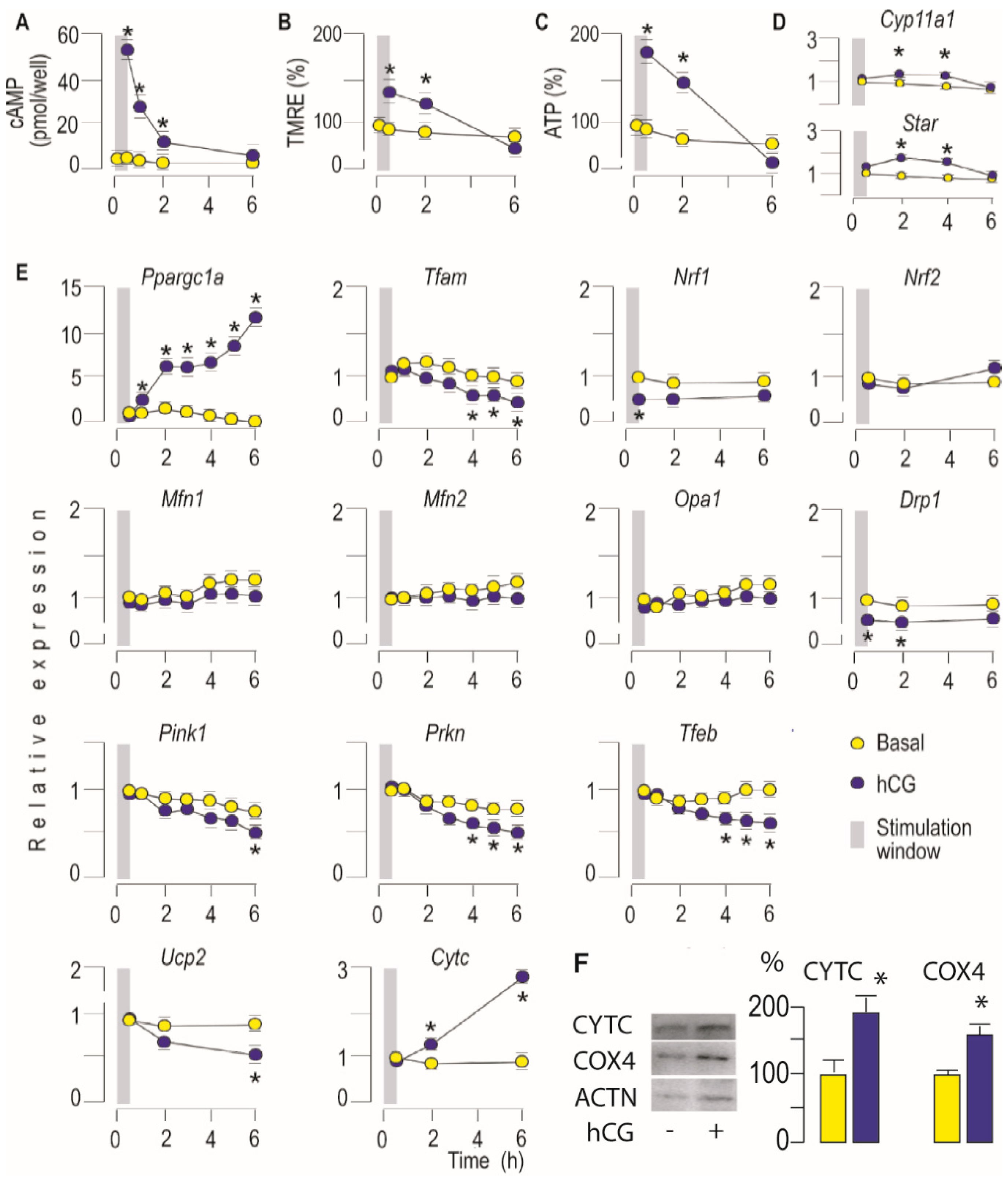
3.1. Activation of LHR Increases Mitochondrial Bioenergetics and Steroidogenic Function in Leydig Cells Ex Vivo
3.2. Activation of LHR Changed the Expression of Genes Important for Mitochondrial Function in Rat Leydig Cells Ex Vivo
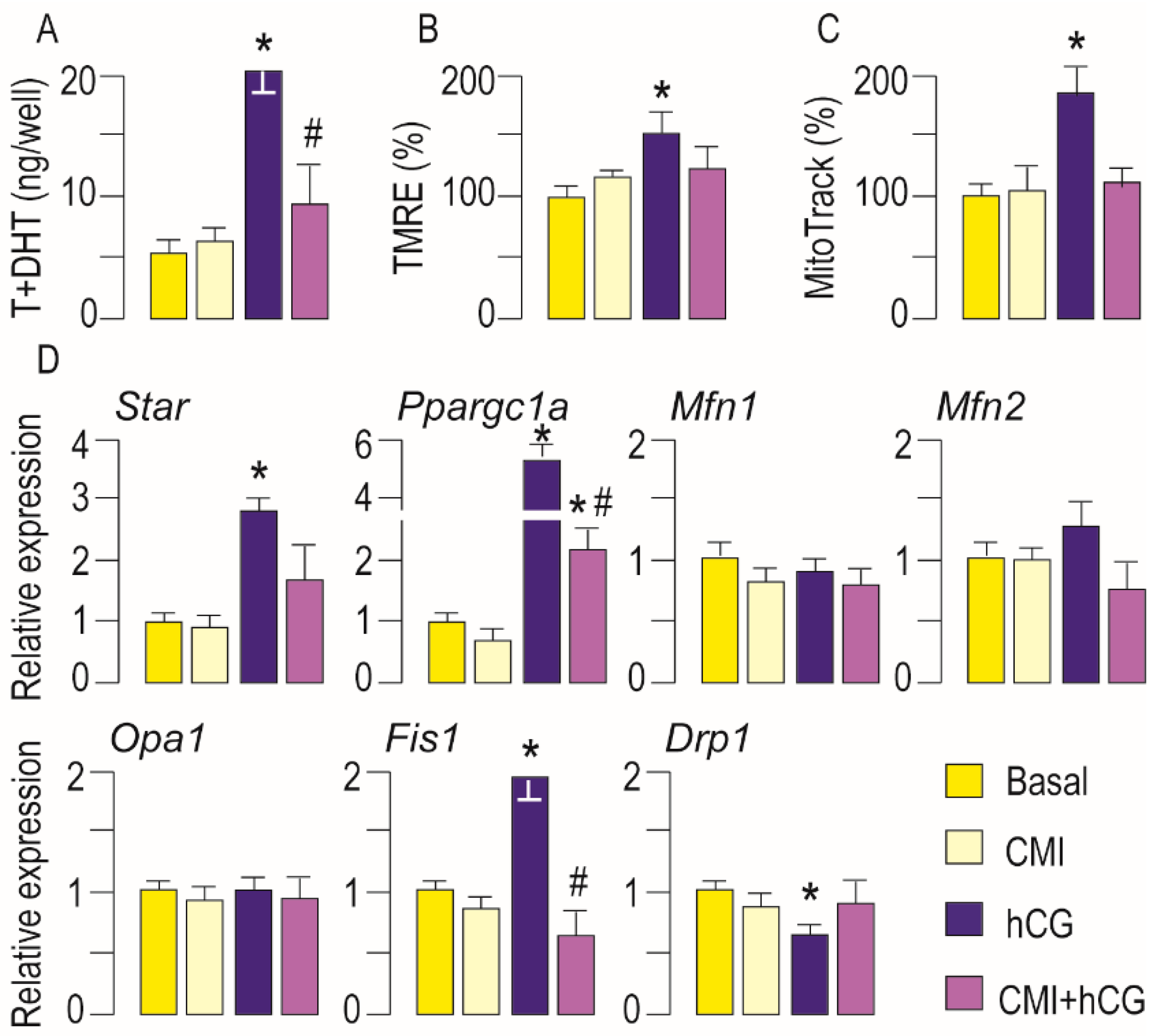
3.3. Activation of LHR Changed the Expression of Genes Important for Mitochondrial Function in Rat Leydig Cells In Vivo
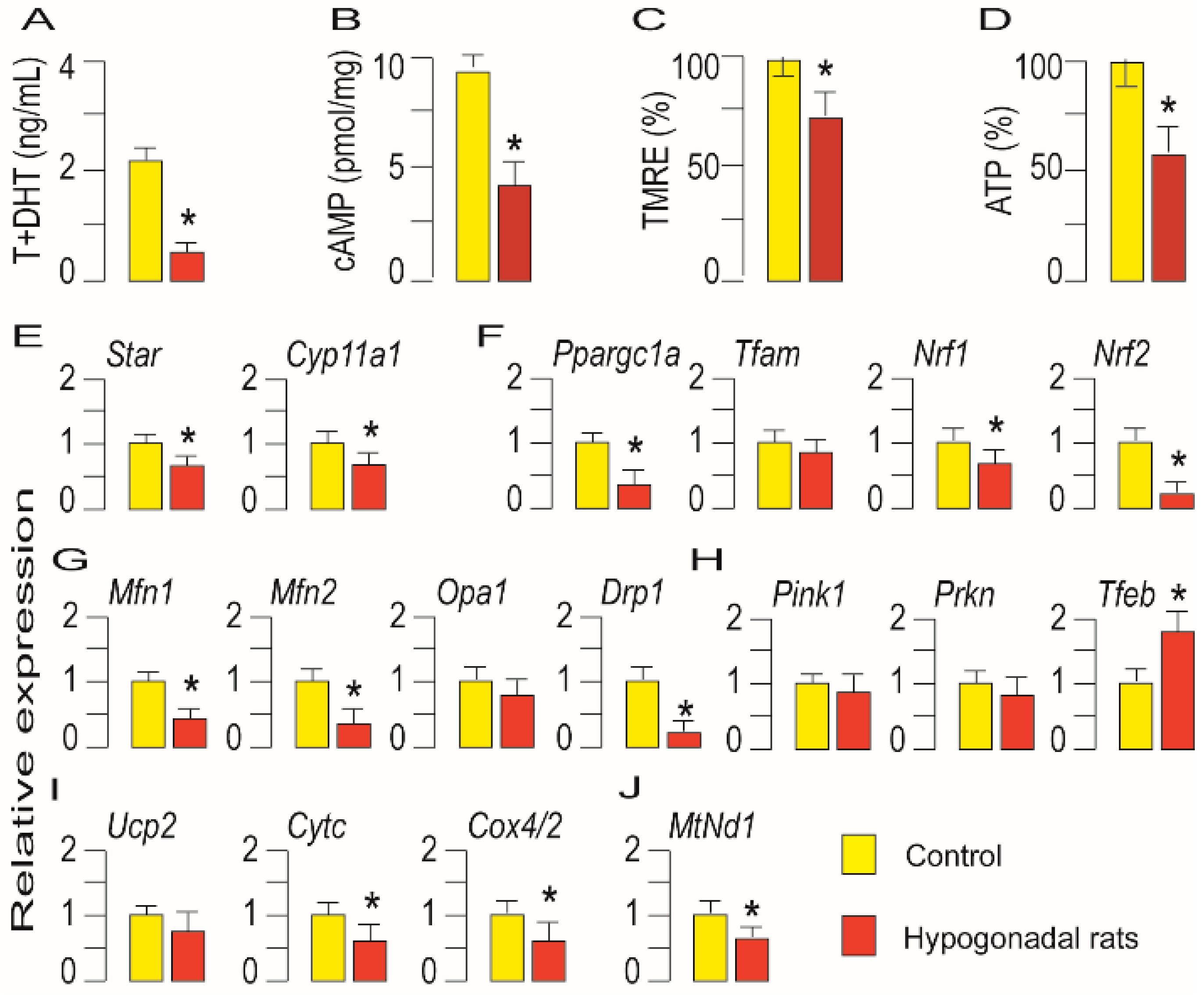
3.4. Mitochondrial Function Was Dampened in Leydig Cells from Hypogonadotropic Hypogonadal Rats
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cooper, D.M.F. Regulation and organization of adenylyl cyclases and cAMP. Biochem. J. 2003, 375, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Dufau, M.L. The luteinizing hormone receptor. Annu. Rev. Physiol. 1998, 60, 461–496. [Google Scholar] [CrossRef]
- Stocco, D.M.; Wang, X.; Jo, Y.; Manna, P.R. Multiple Signaling Pathways Regulating Steroidogenesis and Steroidogenic Acute Regulatory Protein Expression: More Complicated than We Thought. Mol. Endocrinol. 2005, 19, 2647–2659. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.H.; Hales, D.B. Overview of Steroidogenic Enzymes in the Pathway from Cholesterol to Active Steroid Hormones. Endocr. Rev. 2004, 25, 947–970. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.L. Steroid hormone synthesis in mitochondria. Mol. Cell. Endocrinol. 2013, 379, 62–73. [Google Scholar] [CrossRef]
- Midzak, A.; Papadopoulos, V. Adrenal Mitochondria and Steroidogenesis: From Individual Proteins to Functional Protein Assemblies. Front. Endocrinol. 2016, 7, 106. [Google Scholar] [CrossRef]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Zheng, J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef]
- Andric, S.A.; Janjic, M.M.; Stojkov, N.J.; Kostic, T.S. Protein kinase G-mediated stimulation of basal Leydig cell steroidogenesis. Am. J. Physiol. Metab. 2007, 293, E1399–E1408. [Google Scholar] [CrossRef]
- Midzak, A.S.; Chen, H.; Aon, M.A.; Papadopoulos, V.; Zirkin, B.R. ATP Synthesis, Mitochondrial Function, and Steroid Biosynthesis in Rodent Primary and Tumor Leydig Cells. Biol. Reprod. 2011, 84, 976–985. [Google Scholar] [CrossRef]
- Duarte, A.; Poderoso, C.; Cooke, M.; Soria, G.; Maciel, F.C.; Gottifredi, V.; Podesta, E.J. Mitochondrial Fusion Is Essential for Steroid Biosynthesis. PLoS ONE 2012, 7, e45829. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.N.; Leuthner, T.C.; Luz, A.L. Mitochondrial fusion, fission, and mitochondrial toxicity. Toxicology 2017, 391, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Westermann, B. Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 1833–1838. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.R.; Nunnari, J. Mitochondrial form and function. Nature 2014, 505, 335–343. [Google Scholar] [CrossRef]
- Dominy, J.E.; Puigserver, P. Mitochondrial Biogenesis through Activation of Nuclear Signaling Proteins. Cold Spring Harb. Perspect. Biol. 2013, 5, a015008. [Google Scholar] [CrossRef]
- Li, P.A.; Hou, X.; Hao, S. Mitochondrial biogenesis in neurodegeneration. J. Neurosci. Res. 2017, 95, 2025–2029. [Google Scholar] [CrossRef]
- Palikaras, K.; Lionaki, E.; Tavernarakis, N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 2018, 20, 1013–1022. [Google Scholar] [CrossRef]
- Schmidt, O.; Harbauer, A.B.; Rao, S.; Eyrich, B.; Zahedi, R.P.; Stojanovski, D.; Schönfisch, B.; Guiard, B.; Sickmann, A.; Pfanner, N.; et al. Regulation of Mitochondrial Protein Import by Cytosolic Kinases. Cell 2011, 144, 227–239. [Google Scholar] [CrossRef]
- Opalińska, M.; Meisinger, C. Metabolic control via the mitochondrial protein import machinery. Curr. Opin. Cell Biol. 2015, 33, 42–48. [Google Scholar] [CrossRef]
- Moujalled, D.; Weston, R.; Anderton, H.; Ninnis, R.; Goel, P.; Coley, A.; Huang, D.C.S.; Wu, L.; Strasser, A.; Puthalakath, H. Cyclic-AMP-dependent protein kinase A regulates apoptosis by stabilizing the BH3-only protein Bim. EMBO Rep. 2011, 12, 77–83. [Google Scholar] [CrossRef]
- Gomes, L.C.; Di Benedetto, G.; Scorrano, L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011, 13, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Akabane, S.; Uno, M.; Tani, N.; Shimazaki, S.; Ebara, N.; Kato, H.; Kosako, H.; Oka, T. PKA Regulates PINK1 Stability and Parkin Recruitment to Damaged Mitochondria through Phosphorylation of MIC60. Mol. Cell 2016, 62, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Lefkimmiatis, K. cAMP signalling meets mitochondrial compartments. Biochem. Soc. Trans. 2014, 42, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.; Castillo, F.; Podesta, E.J.; Poderoso, C. Mitochondrial Fusion and ERK Activity Regulate Steroidogenic Acute Regulatory Protein Localization in Mitochondria. PLoS ONE 2014, 9, e100387. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Kim, Y.-J.; Lee, S.G.; Kim, J.Y.; Chung, J.-Y.; Jeong, S.-Y.; Koh, H.; Yun, J.; Park, H.T.; Yoo, Y.H.; et al. Drp1 Phosphorylation Is Indispensable for Steroidogenesis in Leydig Cells. Endocrinology 2019, 160, 729–743. [Google Scholar] [CrossRef] [PubMed]
- De Rasmo, D.; Signorile, A.; Santeramo, A.; Larizza, M.; Lattanzio, P.; Capitanio, G.; Papa, S. Intramitochondrial adenylyl cyclase controls the turnover of nuclear-encoded subunits and activity of mammalian complex I of the respiratory chain. Biochim. Biophys. Acta Bioenerg. 2015, 1853, 183–191. [Google Scholar] [CrossRef]
- De Rasmo, D.; Micelli, L.; Santeramo, A.; Signorile, A.; Lattanzio, P.; Papa, S. cAMP regulates the functional activity, coupling efficiency and structural organization of mammalian F O F 1 ATP synthase. Biochim. Biophys. Acta Gen. Subj. 2016, 1857, 350–358. [Google Scholar] [CrossRef]
- Acin-Perez, R.; Salazar, E.; Kamenetsky, M.; Buck, J.; Levin, L.R.; Manfredi, G. Cyclic AMP Produced inside Mitochondria Regulates Oxidative Phosphorylation. Cell Metab. 2009, 9, 265–276. [Google Scholar] [CrossRef]
- Di Benedetto, G.; Gerbino, A.; Lefkimmiatis, K. Shaping mitochondrial dynamics: The role of cAMP signalling. Biochem. Biophys. Res. Commun. 2018, 500, 65–74. [Google Scholar] [CrossRef]
- Di Benedetto, G.; Scalzotto, E.; Mongillo, M.; Pozzan, T. Mitochondrial Ca2+ Uptake Induces Cyclic AMP Generation in the Matrix and Modulates Organelle ATP Levels. Cell Metab. 2013, 17, 965–975. [Google Scholar] [CrossRef]
- Andric, S.A.; Kojic, Z.; Bjelic, M.M.; Mihajlović, A.I.; Baburski, A.Z.; Sokanovic, S.J.; Janjic, M.M.; Stojkov, N.J.; Stojilkovic, S.S.; Kostic, T.S. The opposite roles of glucocorticoid and α1-adrenergic receptors in stress triggered apoptosis of rat Leydig cells. Am. J. Physiol. Metab. 2013, 304, E51–E59. [Google Scholar] [CrossRef] [PubMed]
- Baburski, A.Z.; Andric, S.A.; Kostic, T.S. Luteinizing hormone signaling is involved in synchronization of Leydig cell’s clock and is crucial for rhythm robustness of testosterone production. Biol. Reprod. 2019, 100, 1406–1415. [Google Scholar] [CrossRef] [PubMed]
- Gak, I.A.; Radovic, S.M.; Dukic, A.R.; Janjic, M.M.; Stojkov-Mimic, N.J.; Kostic, T.S.; Andric, S.A. Stress triggers mitochondrial biogenesis to preserve steroidogenesis in Leydig cells. Biochim. Biophys. Acta Bioenerg. 2015, 1853, 2217–2227. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.H.; Downing, J.R.; Wong, K.-L. Luteinizing Hormone Receptors and Testosterone Synthesis in Two Distinct Populations of Ley dig Cells. Endocrinology 1980, 106, 1424–1429. [Google Scholar] [CrossRef]
- Kostic, T.S.; Andric, S.A.; Stojilkovic, S.S. Receptor-Controlled Phosphorylation of α1 Soluble Guanylyl Cyclase Enhances Nitric Oxide-Dependent Cyclic Guanosine 5′-Monophosphate Production in Pituitary Cells. Mol. Endocrinol. 2004, 18, 458–470. [Google Scholar] [CrossRef][Green Version]
- Sokanovic, S.J.; Baburski, A.Z.; Kojic, Z.; Medar, M.L.J.; Andric, S.A.; Kostic, T.S. Aging-Related Increase of cGMP Disrupts Mitochondrial Homeostasis in Leydig Cells. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2020. [Google Scholar] [CrossRef]
- Rato, L.; Duarte, A.I.; Tomás, G.D.; Santos, M.S.; Moreira, P.I.; Socorro, S.; Cavaco, J.E.; Alves, M.G.; Oliveira, P.F. Pre-diabetes alters testicular PGC1-α/SIRT3 axis modulating mitochondrial bioenergetics and oxidative stress. Biochim. Biophys. Acta Bioenerg. 2014, 1837, 335–344. [Google Scholar] [CrossRef]
- Allen, J.A.; Shankara, T.; Janus, P.; Buck, S.; Diemer, T.; Hales, K.H.; Hales, D.B. Energized, Polarized, and Actively Respiring Mitochondria Are Required for Acute Leydig Cell Steroidogenesis. Endocrinology 2006, 147, 3924–3935. [Google Scholar] [CrossRef]
- Midzak, A.S.; Chen, H.; Papadopoulos, V.; Zirkin, B.R. Leydig cell aging and the mechanisms of reduced testosterone synthesis. Mol. Cell. Endocrinol. 2009, 299, 23–31. [Google Scholar] [CrossRef]
- Salin, K.; Villasevil, E.M.; Anderson, G.J.; Selman, C.; Chinopoulos, C.; Metcalfe, N.B. The RCR and ATP/O Indices Can Give Contradictory Messages about Mitochondrial Efficiency. Integr. Comp. Biol. 2018, 58, 486–494. [Google Scholar] [CrossRef]
- Palorini, R.; De Rasmo, D.; Gaviraghi, M.; Danna, L.S.; Signorile, A.; Cirulli, C.; Chiaradonna, F.; Alberghina, L.; Papa, S. Oncogenic K-ras expression is associated with derangement of the cAMP/PKA pathway and forskolin-reversible alterations of mitochondrial dynamics and respiration. Oncogene 2012, 32, 352–362. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Yang, G.; Hou, Y.; Tang, X.; Wu, C.; Wu, X.-A.; Guo, L.; Zhu, Q.; Luo, H.; Du, Y.-E.; et al. Cytoplasmic GPER translocation in cancer-associated fibroblasts mediates cAMP/PKA/CREB/glycolytic axis to confer tumor cells with multidrug resistance. Oncogene 2017, 36, 2131–2145. [Google Scholar] [CrossRef] [PubMed]
- Glancy, B.; Kane, D.A.; Kavazis, A.N.; Goodwin, M.L.; Willis, W.T.; Gladden, L.B. Mitochondrial lactate metabolism: History and implications for exercise and disease. J. Physiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chan, D.C. Emerging functions of mammalian mitochondrial fusion and fission. Hum. Mol. Genet. 2005, 14, R283–R289. [Google Scholar] [CrossRef]
- Twig, G.; Elorza, A.; Molina, A.A.J.; Mohamed, H.; Wikstrom, J.D.; Walzer, G.; Stiles, L.; Haigh, S.E.; Katz, S.; Las, G.; et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008, 27, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shachar, D. The interplay between mitochondrial complex I, dopamine and Sp1 in schizophrenia. J. Neural Transm. 2009, 116, 1383–1396. [Google Scholar] [CrossRef]
- Wallace, D.C. The epigenome and the mitochondrion: Bioenergetics and the environment. Genes Dev. 2010, 24, 1571–1573. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medar, M.L.J.; Marinkovic, D.Z.; Kojic, Z.; Becin, A.P.; Starovlah, I.M.; Kravic-Stevovic, T.; Andric, S.A.; Kostic, T.S. Dependence of Leydig Cell’s Mitochondrial Physiology on Luteinizing Hormone Signaling. Life 2021, 11, 19. https://doi.org/10.3390/life11010019
Medar MLJ, Marinkovic DZ, Kojic Z, Becin AP, Starovlah IM, Kravic-Stevovic T, Andric SA, Kostic TS. Dependence of Leydig Cell’s Mitochondrial Physiology on Luteinizing Hormone Signaling. Life. 2021; 11(1):19. https://doi.org/10.3390/life11010019
Chicago/Turabian StyleMedar, Marija L. J., Dijana Z. Marinkovic, Zvezdana Kojic, Alisa P. Becin, Isidora M. Starovlah, Tamara Kravic-Stevovic, Silvana A. Andric, and Tatjana S. Kostic. 2021. "Dependence of Leydig Cell’s Mitochondrial Physiology on Luteinizing Hormone Signaling" Life 11, no. 1: 19. https://doi.org/10.3390/life11010019
APA StyleMedar, M. L. J., Marinkovic, D. Z., Kojic, Z., Becin, A. P., Starovlah, I. M., Kravic-Stevovic, T., Andric, S. A., & Kostic, T. S. (2021). Dependence of Leydig Cell’s Mitochondrial Physiology on Luteinizing Hormone Signaling. Life, 11(1), 19. https://doi.org/10.3390/life11010019




